Abstract
Intensification of food production has the potential to drive increased disease prevalence in food plants and animals. Microsporidia are diversely distributed, opportunistic, and density-dependent parasites infecting hosts from almost all known animal taxa. They are frequent in highly managed aquatic and terrestrial hosts, many of which are vulnerable to epizootics, and all of which are crucial for the stability of the animal–human food chain. Mass rearing and changes in global climate may exacerbate disease and more efficient transmission of parasites in stressed or immune-deficient hosts. Further, human microsporidiosis appears to be adventitious and primarily associated with an increasing community of immune-deficient individuals. Taken together, strong evidence exists for an increasing prevalence of microsporidiosis in animals and humans, and for sharing of pathogens across hosts and biomes.
Parasites in Food Chains
In high-income countries, approximately 70% of deaths in people over the age of 70 result from non-communicable or chronic conditions. In low-income countries almost 40% of deaths occur in children under the age of 15 and are generally associated with infectious diseases (e.g., HIV/ AIDS, malaria, diarrhea, and tuberculosis). Many of these deaths are caused by pathogens transmitted via food and water supplies [1]. Human food originating from both plants and animals is produced, processed, and marketed in intricately linked systems of primary producers (e.g., corn, cattle, fish), input and service providers (i.e., pesticides, water, veterinary drugs), transporters, processors, wholesalers, retailers, consumers, and end-users of byproducts (e.g., manure). Foodborne diseases comprise a broad range of illnesses caused by ingestion of pathogens, parasites, chemical contaminants, and biotoxins that are either naturally present in food or can contaminate food at different points in the production and preparation process [2]. Many of the 300 species of helminths and over 70 species of protists known to infect humans are transmitted via food and water [3]. Infectious life stages are acquired by ingesting tissues of infected mammals, fish, or invertebrates, as well as from contaminated food and water supplies or via contaminated fomites or fingers. Although traditionally associated with tropical outbreaks, perceptions of risk in temperate regions are changing following large outbreaks of parasitic infections due to agents such as Toxoplasma gondii [4] and Cryptosporidium spp. [5]. Globalized food trade and travel clearly have the potential to increase the risk of imported parasitoses from tropical countries [6]. Microsporidia, although not currently considered to be priority foodborne parasites, have the potential to enter the human food chain through waterborne and foodborne routes, and via exposure to the environment. As such, natural hosts of human infective microsporidia can be part of the human food chain (e.g., [7,8]). In this review we consider members of the phylum Microsporidia as agents of emergent disease in hosts from major global biomes and food production sectors (terrestrial, aquatic) and in human consumers. Further, we combine phylogenetic, ecological and immunological perspectives to propose unifying themes, under a ‘One Health’ banner, which may explain the emergence of these opportunists.
Microsporidia – What Are They and Where Did They Come From?
Microsporidia are a hyper-diverse phylum of spore-forming parasites infecting hosts from all major animal taxa in all global biomes (Box 1). The array of hosts is equally diverse, ranging from protists (in some of which Microsporidia are hyperparasites) to vertebrates including humans. Species in almost half the known microsporidian genera infect aquatic hosts, and thousands of these pathogens remain undescribed [9]. Morphological approaches to within-phylum taxonomy have generally been superseded (or at least augmented) by sequence comparisons of the ribosomal rRNA genes (e.g., [10,11]). Debate over placement of the Microsporidia within the tree of life has progressed from historical grouping with spore-forming parasites to the current molecular phylogenetics-based view that they are affiliated with the fungi [12,13]. Analysis of the first complete microsporidian genome (Encephalitozoon cuniculi) [14] confirmed that the previous phylogenies showing a deeper position, and which suggested that the microsporidia were an ancient primitive lineage, was an artifact of long branch attraction [15], a finding supported by the discovery of highly reduced mitochondria (mitosomes) within the microsporidian cytoplasm [16]. While more recent confirmation of a fungal relationship is now accepted by most, their specific relationships and their branching either within the Fungi (e.g., [17]) or outside the group [18,19] are a topic of further debate. Although phylogenetic comparison of known taxa from within the Microsporidia or the Fungi has failed to resolve this issue, the recent discovery (and phylogenetic placement) of three novel lineages, the Cryptomycota [20,21], the aphelids [22,23], and the genus Mitosporidium [24] as intermediate between Fungi and the rest of the eukaryotes has re-ignited interest. The Cryptomycota appear to branch at the base of the Fungi and contain the Microsporidia as well as the aforementioned aphelids and Mitosporidium. Discovery of the group is clarifying relationships between the Microsporidia, parasites with intermediate characteristics (such as Mitosporidium), and all other eukaryotes, at the same time revealing how their peculiar infection machinery likely evolved [25].
Box 1. Microsporidia Form and Function.
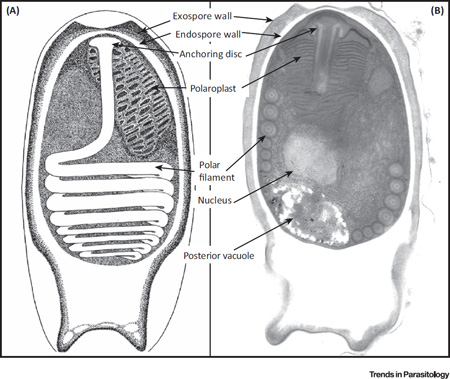
Microsporidia are single-celled, eukaryotic, spore-forming parasites, and both generalist and specialist species are found in invertebrate and vertebrate hosts. There are two main clades of microsporidia: the typical (or advanced) and atypical (or primitive) microsporidia [94]. The atypical microsporidia are a small group composed of approximately 13 genera and 42 species [95]. The majority of known microsporidia are of the typical variety, with ~190 genera and an estimated 1300–1500 species [28]. This group contains the opportunistic taxa that have simple to complex developmental sequences and life cycles. Spores of the typical microsporidia contain one or two nuclei (the diplokaryon), are most commonly oval or pyriform in shape, and average 2–8 microns in size, but can be as small as 1 micron or as large as 30 microns in length. The spore has a very complex structure that contains the extrusion apparatus for infecting the host cell. The spore wall is composed of two layers: an electron-lucent endospore layer that contains chitin, and an electron-dense exospore that is often layered. The unique infection apparatus is composed of three main parts: a long, thread-like polar filament, a multilayered polaroplast, which is a highly membranous structure that occupies the anterior half of the spore, and a posterior vacuole (Figure I). When the spore is in the appropriate host and environment, the spore germinates and the polar filament is everted to become a hollow tube. The sporoplasm travels through this tube and is inoculated into the cytoplasm of the host cell to begin replication [94,96]. Generalist species of microsporidia have a broad host-range and the ability to infect both invertebrate and vertebrate hosts [28]. Generalists are often responsible for opportunistic infections in vertebrates. Some notable genera containing species capable of infecting and developing in both arthropod and vertebrate hosts are Anncaliia, Tubulinosema, Trachipleistophora, and Encephalitozoon, although other genera have been implicated by molecular data with species in arthropod and vertebrate hosts (e.g., Enterocytozoon, Endoreticulatus) [29]. Specialists are restricted to infecting and developing within a narrow range of closely related hosts, or some species require an obligate two-host system with a definitive host and intermediate host (e.g., Amblyospora).
Figure I. The Features of a Typical Microsporidian Spore. (A) Diagram. (B) Transmission electron micrograph. The rigid spore wall is composed of the exospores, the endospore, and a sophisticated extrusion apparatus containing the polar filament, polaroplast, and the posterior vacuole.
Opportunistic Pathogens in all Major Biomes
Based on the descriptive criteria defined by the International Committee of Zoological Nomenclature (ICZN), the phylum Microsporidia currently comprises over 200 genera [26]. Phylogenetic analysis (based upon small subunit rRNA partial gene sequence) of 70 of these genera reveals five apparent clades broadly classified into three major groups according to predominant host and environment type, termed the Aquasporidia (clades 1 and 3), the Terresporidia (clades 2 and 4), and the Marinosporidia (clade 5) [27]. It is noteworthy that most of these clades contain exceptions, likely associated with either the pathogen or even the host switching to a new habitat. Host-switching may be more likely if the microsporidian parasite is a generalist or where hosts move between habitats (e.g., freshwater to marine, or freshwater to terrestrial). In the case of confirmed human-infecting taxa, representatives are observed across the phylum and include the genera Enterocytozoon, Encephalitozoon, and Vittaforma (clade 4, Terresporidia), Anncalia and Tubulinosema (clade 3, Aquasporidia), and Pleistophora (clade 5, Marinosporidia) [27]. Although not inconceivable that Homo sapiens serves as a type host for particular microsporidian taxa, given the spread of human-infecting genera across known clades and the preponderance for infection to occur in immune-compromised patients (see below), it is perhaps more likely that these infections represents zoonotic transfer from hosts inhabiting terrestrial, freshwater, andmarineenvironments.Transferinthiscasemayrelatetodirectexposuretotype host taxa (e.g., via the food chain) but also by contact with extra-hostparasite life-stages in the environment in which they reside. In this respect, the potential for susceptibility of humans to infection by other Microsporidia across the phylum appears to be significant.
Microsporidia and Immune Competence
In nature, microsporidia typically develop a balanced interaction with their host, leading to long-term subclinical infections [28]. When the immune condition of the host is compromised, infection can lead to overt signs of clinical disease, highlighting the key role of immune competence in mitigating individual- and population-level health effects of microsporidiosis [29]. Human immune deficiencies can be categorized into primary and secondary types. Primary immune deficiencies (PID) are derived from intrinsic and inherited defects in the immune system. Although PID cases are rare (an estimated 250 000 cases are currently diagnosed in the USA) (Immune Deficiency Foundation; http://primaryimmune.org/about/), microsporidian infections have been occasionally reported in PID patients [30]. More common are secondary immune deficiencies (SID) which are acquired from an array of causes including chemotherapy and/or radiation treatments for malignancies, immune-suppressive therapies (to prevent transplant rejection), malnutrition, poor sanitation, aging, and infectious diseases such as HIV/AIDS (www.uptodate.com/contents/secondary-immunodeficiency-due-to-underlying-disease-states-environmental-exposures-and-miscellaneous-causes). Prior to the HIV/AIDS pandemic in the mid-1980s, microsporidiosis was rarely reported in human patients [31]. The pandemic brought to light the opportunistic capability of microsporidia to infect humans and produce disease in virtually all organs [32,33] (Figure 1). Before common use of anti-retroviral therapies, microsporidiosis was reported in at least 15% (and up to 85%) of HIV/AIDS patients [34]. However, although prevalence declined with improved therapy, an increase in newly diagnosed cases of HIV in people over 50 years of age, coupled with an aging population of patients living with HIV, is leading to so-called HIV-associated non-AIDS (HANA) conditions that accelerate the onset of diseases normally observed in the elderly. These patients show accelerated immune senescence, leaving them susceptible to opportunistic infections, including microsporidia. Reactivation of latent microsporidian infections with age, or with subsequent use of chemotherapy or immune-suppressive treatments, has also been reported [35]. Although at least ten microsporidian genera have been associated with human patients (Table 1), the most frequently detected species is the gut-infecting Enterocytozoon bieneusi in patients with HIV/AIDS, in whom it produces chronic diarrhea [32] (Box 2).
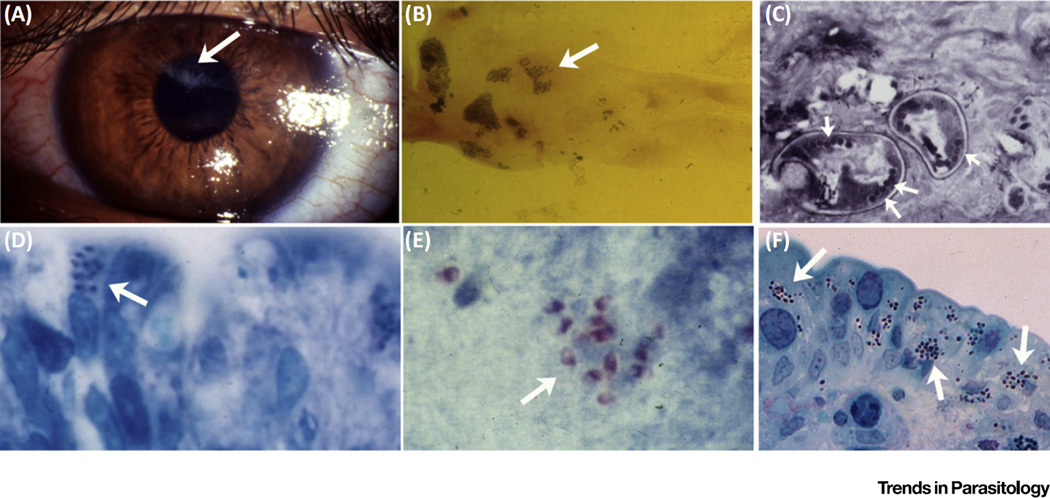
Figure 1. Microsporidiosis in Humans.
(A) Encephalitozoon hellem keratoconjunctivitis. Areas of corneal damage due to microsporidiosis (arrow). (B) Corneal scraping from a case of microsporidian keratoconjunctivitis demonstrating spores (arrow) of E. hellem. (C) Conjunctival biopsy in a case of microsporidian keratoconjunctivitis demonstrating microsporidian spores in cross-section (arrows point to polar tubes, the infective structures). The arrangement of the polar tubes is consistent with Encephalitozoon. (D) Intestinal biopsy from a patient with gastrointestinal microsporidiosis and diarrhea due to Enterocytozoon bieneusi (arrows point to spores in the apical region of an intestinal epithelial cell). (E) Stool stained with modified trichrome stain (arrows point to spores). PCR confirmed that this infection was due to E. bieneusi. (F) Intestinal biopsy from a patient with gastrointestinal microsporidiosis and diarrhea due to Encephalitozoon intestinalis.
Table 1.
Confirmed Infections of Humans by Members of the Phylum Microsporidia
| Conditions of Immune-Deficiency or Immune-Suppression | Refs | ||||
|---|---|---|---|---|---|
| Taxon | HIV | Transplant | Cancer | Other Conditions and Risk Factors |
|
|
Anncaliia (syn. Nosema, Brachiola) algerae |
N/ra | Kidney | Yes | Rheumatoid arthritis, ocular infection, steroids, Crohn’s disease, diabetes |
[97,98] |
|
Anncaliia (syn. Nosema) connori |
N/r | N/r | N/r | Athymic child | [99,100] |
|
Anncaliia (syn. Brachiola) vesicularum |
Yes | N/r | N/r | N/r | [99,101] |
| Encephalitozoon cuniculi | Yes | Kidney, bone marrow |
Yes | Children, Primary Immune Deficiency, diabetes, heart disease, ocular infection, steroids |
[29,35,102–105] |
| Encephalitozoon hellem | Yes | N/r | Yes | Ocular infection, steroids |
[106–108] |
| Encephalitozoon intestinalis | Yes | Bone marrow |
Yes | Children, ocular infection, steroids |
[29,106,108–114] |
|
Encephalitozoon sp. (undetermined) |
N/r | Pancreas, kidney |
Yes | Diabetes | [115] |
| Endoreticulatus spp. | N/r | N/r | N/r | Ocular infection, steroids |
[108] |
| Enterocytozoon bieneusi | Yes | Kidney, liver, heart, lung |
Yes | Children | [33,36,110,111,116,117] |
|
Microsporidium ceylonensis, M. africanum |
N/r | N/r | N/r | Ocular infection, trauma | [108] |
| Nosema ocularum | N/r | N/r | N/r | Ocular infection, trauma | [118] |
| Pleistophora ronneafiei | Yes | N/r | N/r | N/r | [52] |
|
Trachipleistophora anthropopthera |
Yes | N/r | N/r | Ocular infection, steroids |
[51,119] |
| Trachipleistophora hominis | Yes | N/r | N/r | Ocular infection, steroids |
[51,108] |
| Tubulinosema acridophagus | Yes | Bone marrow |
N/r | N/r | [120] |
| Vittaforma corneae | Yes | N/r | N/r | Children, ocular infection, steroids, trauma |
[35,108,121] |
| Unidentified species | N/r | N/r | Yes | Sjögren’s disease, ocular infection, immune-suppressive treatment |
[122] |
N/r – not recorded.
Box 2. Enterocytozoon bieneusi - A Zoonotic Pathogen of Humans.
Enterocytozoon bieneusi was originally described in 1985 as a cause of gastrointestinal infection presenting as chronic diarrhea in humans with advanced HIV-1 infection (i.e., AIDS) [32]. Spores are smaller (1.0 x 1.5 µm) than those of Encephalitozoon spp. (1.2 x 2.2 µm) and more difficult to find in tissue sections. E. bieneusi shows interesting intracellular developmental involving multinucleated plasmodia with characteristic electron-lucent lamellar inclusions. Inclusions associate with the nuclear envelope, the endoplasmic reticulum, or both. E. bieneusi appears to be widely distributed in both mammals and birds; it has been reported in pigs, dogs, cows, chickens, pigeons, falcons, and various primates. Family-level relatives exist in fish and other aquatic animals. Zoonotic transmission of E. bieneusi has been confirmed [57]. Infection of the epithelium of the gastrointestinal tract is the most frequent presentation of microsporidiosis, and over 90% of these infections are caused by E. bieneusi, with the remainder mostly being caused by En. intestinalis. Infection does not produce active enteritis or ulceration, although infection results in variable degrees of villous blunting and crypt hyperplasia. Infection is associated with malabsorption, perhaps as a consequence of increased villous epithelial cell turnover leading to functional immaturity of the villous epithelial cells. In humans with AIDS, E. bieneusi infection has also been associated with infection of the biliary tract and sclerosing cholangitis [33]. Hepatitis with infection of the biliary system (including the gallbladder) caused by E. bieneusi has also been described in monkeys and pigs; although systemic dissemination is rare, spores have been associated with proliferative serositis (peritonitis) in macaques (Macaca mulatta) and in the nasal mucosa of humans [123]. There are also reports of pulmonary involvement associated with chronic diarrhea, persistent cough, dyspnea, wheezing, and chest radiographs showing interstitial infiltrates, with spores being found in stool, bronchoalveolar lavage fluid, and transbronchial biopsy specimens [124], as well as a report of this organism being found in the urine of renal transplant patients.
Age, both young and old, has been associated with elevated burden of microsporidiosis. In very young children (below age six) immune immaturity coupled with inadequate hygiene practices and malnutrition have revealed surprising levels of infection (e.g., 18.2% of children in one study from Spain) [36]. Epidemiological studies of E. bieneusi specifically have revealed background prevalence ranging from 4.4% to 22.5% in HIV-negative children [37]. In the elderly, immune senescence and declining numbers of naïve T cells lead to weakened response to new infections. In one study of HIV-negative individuals with a mean age of 73.5 years, 17% of patients presenting with symptoms of diarrhea were infected with E. bieneusi [38]. Given a growing human global population aged 65 years and over (16% by 2050), immune senescence-associated microsporidiosis is likely to increase [39].
Microsporidia in Food and Water
Ingestion of contaminated food and water, either directly or indirectly (via exposure to the environment) offers the most likely route of transmission of microsporidian spores to humans [40]. To this end, the majority of human-pathogenic microsporidia detailed in Table 1 have been detected in water. Comprehensively reviewed by Fayer and Santin [34], it is considered that water, either consumed directly by drinking or indirectly via irrigating or washing foods, bathing, washing, or for recreation, provides a crucial medium for spore survival and transmission. Excretion of spores from infected humans and animals via urine and feces is the primary route of water contamination. Recalcitrance within freshwater and marine environments at a range of temperatures contributes to retention of infectivity and the potential for wide dispersal from point-sources [41]. Surveys of surface, drinking, waste, and recreational waters have consistently demonstrated the presence of microsporidian parasites. In some cases, filter-feeding molluscs have been deployed as sentinels for detection of microsporidia in surface waters, specifically demonstrating the presence of the human pathogens E. bieneusi, Encephalitozoon intestinalis, and Encephalitozoon hellem [42,43]. However, given that over 200 genotypes of E. bieneusi have so far been identified (some exclusively in human or animal hosts, and others infecting both), accurate typing of isolates detected in the water sources used by humans is an important step in understanding the true risk of exposure [44]. Furthermore, because E. bieneusi resides within a family of microsporidia otherwise exclusively infecting fish and crustacean hosts [45], future studies to investigate the potential for genotypes of E. bienuesi (or closely related taxa) to exist in a replicative form within aquatic environments are required [9].
Microsporidia have also been detected directly in food destined for human consumption. Soft fruits, vegetables, and herbs collected from markets in Poland were contaminated with E. bieneusi and En. intestinalis [46]. Milk contaminated with human pathogenic genotypes of E. bieneusi has been reported originating from herds in Korea [47]. A foodborne outbreak of gastrointestinal illness in over 100 people was associated with consumption of E. bieneusi-contaminated cucumbers in Sweden [48].
Although not directly related to food consumption, the propensity for insect-infecting microsporidia to be vectored to humans either by bite, sting, or contamination of the skin by feces of the insect host has been demonstrated. Examples include Anncalia algerae infections in the eye and musculature [49], Tubulinosema sp. infection of the tongue [50], and Trachipleistophora sp. infections of the skeletal muscle and organs [51]. Increasing contact with infected insects mass-reared for human consumption may pose a future occupational and consumption risk. Similar contact-related risks have been identified for aquatic animals associated with infections by Pleistophora sp. in the musculature of immune-suppressed patients with or without HIV/AIDS [52,53]. Furthermore, microsporidiosis has been widely reported in livestock, including infection of chickens. Genotypes of E. bieneusi [54] and Encephalitozoon spp. [55] occur in pigs and cows in China, some infected with the same genotypes of E. bieneusi that infect humans living in close proximity [56]. Contact between humans and companion animals (pets) has also revealed potential for zoonotic transfer of E. bieneusi between guinea pigs and children [57] and potentially from a human AIDS patient (infected with En. intestinalis) to a cat [58]. Clearly, the environment offers ample potential for food-, water- and contact-driven transmission of microsporidian parasites from animals to susceptible human hosts (Box 1).
Microsporidia in Major Food Production Chains
In addition to the direct risk to humans associated with consuming contaminated water or food, it is appropriate to consider how microsporidia may directly interact with hosts mass-produced for food and for use in food production chains (e.g., biological control agents), or with pollinators that provide ecosystem services to enable food production.
Microsporidia infecting terrestrial invertebrates directly impact their natural host populations, and can devastate mass-reared colonies of insects used as human or animal food, as biological control agents of agricultural pests, or agricultural pest species used to produce biological control agents. Microsporidia are known to infect more than 30 species of field-collected and mass-reared beneficial invertebrates including parasitoids, predatory insects and mites, phytophagous insects used for weed control, and beneficial nematodes. They decrease food consumption in their hosts, prolong development, impart physical deformations, reduce fecundity and longevity, and increase mortality [59]. For example, Muscidifurax raptor, a parasitoid found naturally occurring on dairy farms where they provide effective house- and stable-fly control, is mass-reared for inundative release. However, overcrowding and stress in commercial insectaries leads to high prevalence (86–100%) of Nosema muscidifuracis, a microsporidium that reduces both the lifespan and fecundity of the parasitoid and heavily impacts on fly control on the farm. Pathogen prevalence is also high (up to 84%) on farms where infected parasitoids are released [60,61]. Because microsporidian infections are typically cryptic, they may be overlooked initially in mass-reared colonies [59]. However, with increasing recognition of the potential of insects as a source of protein for the burgeoning global population [62], more controlled mass-rearing conditions, including the development of pathogen-free brood lines and appropriate legal frameworks for their trade, are now required.
Insects play pivotal roles in global food production, with wild and managed bees providing critical pollination services [63]. Apparent gaps between global crop pollination needs and the availability of large-scale pollinator populations (e.g., domesticated honeybee colonies) are due (at least in part) to the highly publicized syndromic condition termed colony collapse disorder (CCD), which has prompted focus on research about bee health and disease in recent years [64]. Despite the fact that infections by Nosema apis and Nosema ceranae have specifically been correlated to losses of honeybee colonies [65,66], definitively linking microsporidian infections per se to colony declines, in either the USA or in Europe, has not been possible [67,68]. In addition to potential shortfalls in pollination by managed pollinator populations, a global decline in wild pollinator populations has also been reported [69]. ‘Spillover’ of infectious diseases from domesticated pollinator populations to wild pollinators has been highlighted as a significant potential source of emerging infectious disease (EID) in wildlife [70]. Specifically, the propensity for honeybees to host a wide range of infectious agents (including microsporidia) [71], and the detection of parasites such as N. ceranae in bumblebees occurring in close proximity to managed honeybee colonies (e.g., [72]), provides at least some evidence for such spillover. However, lack of historical information, inconsistent application of accurate diagnostics to honeybee and bumblebee infections, and a paucity of well-designed studies to examine possible spillover make confirmation of this effect difficult [73]. Recent application of managed bumblebee colonies for greenhouse pollination has also raised questions about the potential for similar spillover effects to surrounding wildlife [74] (Box 3).
Box 3. Nosema disease in bumblebees and honeybees.
Nosema species infecting honeybees and bumblebees (Apidae) belong to the phylogenetic clade Nosema/Vairimorpha, the microsporidian taxon most frequently isolated from the Lepidoptera. Nosema bombi, reported from more than 50 species of bumblebees, is a systemic pathogen that appears to be specific to the genus Bombus. Effects of chronic infections on these essential native pollinators include reduced colony size, males with reduced sperm, decreased hibernation survival and colony establishment, fewer reproductive females, and reduced female mating capability (e.g., [125]). Some Bombus species appear to be more susceptible to N. bombi infection than others, and, although cause and effect has not been established, higher prevalences of this microsporidian infection have been reported in several North American bee species with apparently declining populations [69]. Concerns that exotic strains of N. bombi have been released into North American Bombus populations via managed pollination services have not been substantiated, but Nosema pressure on susceptible species could potentially lower resistance to other pathogens. The annual value of pollination services and hive products of the western honeybee, Apis mellifera, is estimated to exceed $200 billion globally, but anthropogenic global distribution of this species has resulted in a significant increase of parasites and pathogens that may have host-switched from other hymenopteran species. Among the most invasive is Nosema ceranae, thought to have originated from the Asian honeybee, Apis cerana [66]. Similar to Nosema apis, which is naturally occurring in A. mellifera, N. ceranae infects adult bees and has chronic effects, but this pathogen appears to be dominant and has nearly completely displaced N. apis, particularly in honey bee populations below the 50th parallel north in Europe and North America. Nosemosis now figures in many reports of colony loss. Unlike N. bombi and most other Nosema spp., both N. apis and N.ceranae are pathogens ofthe honeybee midgut tissues; however, N. apis appears to bespecific to A. mellifera while N. ceranae has been reported from three other Apis spp. and at least 14 Bombus spp. N. ceranae causes energy stress, longer and less-frequent foraging flights, and shortens the lifespan of bees (e.g., [126]). It has been reported to synergize with the deleterious effects of viruses and a variety of agricultural and apicultural pesticides, while low levels of fumagillin, used to treat nosemosis, may synergize with N. ceranae [127].
In terms of aquatic hosts, microsporidia may directly impact on the production of animals destined for human consumption, or may alter prey populations on which animals destined for human consumption (e.g., fish) rely. As mentioned above, aquatic hosts support almost half the known microsporidian genera [9]. In terms of wild (fished) populations, microsporidian epizootics have been historically associated with collapse of commercial fisheries (e.g., the North American ocean pout fishery in the 1940s) [75], while in aquaculture, species from numerous micro-sporidian taxa have impacted on production during the hatchery, grow-out (netpen), processing, and marketing phases (see [75] for context) Recently, an emergent disease condition termed ‘emaciative syndrome’ was shown to be caused by infection with Enterospora nucleophila in farmed seabream (Sparus aurata) from the Mediterranean. Disease associated with infection by this parasite is apparently associated with immune suppression in its host [76], a feature shared with several other members of the Enterocytozoon clade in which this parasite resides. Previously, immune suppression has been associated with increased severity of microsporidiosis in model fish hosts (e.g., zebrafish infected with Pseudoloma neurophilia [77]) while, in other scenarios, infections by microsporidian parasites have directly impaired immunity, presumably making their hosts more susceptible to infection by other pathogens (e.g., Nucleospora salmonis infection of salmonids [78]). It appears likely that an association between suboptimal environmental conditions, relative immune suppression, and host proximity in aquaculture settings can encourage microsporidiosis and will lead to further emergence of yield-limiting diseases in farmed fish.
Other high-profile examples exist in wild and farmed aquatic invertebrates destined for human consumption. Although parasitism is known to occur across most aquatic invertebrate phyla, the aquatic arthropods in particular, hosting over 50 known genera, appear to be the most affected by microsporidiosis [9]. In the context of the human food chain, the group containing the decapod crustaceans (shrimp, crabs, lobsters etc.) support a major economy, amounting to almost 40 billion dollars per annum from wild fisheries and aquaculture [79]. Those pathogens that target the edible musculature of crustacean hosts have the potential to render marketable meats inedible [80], while those infecting connective tissues can blight the visual esthetics and marketing of high-value captured hosts such as king crabs [81]. In aquaculture settings, farmed penaeid shrimp represent one of the highest-value traded seafood commodities (see [79]). Historically low-prevalence microsporidian infections such as Enterocytozoon hepatopenaeii have been associated with ‘slow growth’ syndromes in Penaeus monodon [82]. However, increasingly intensive farming of the congeneric penaeid Penaeus vannamei in Asia, which now dominates the global market with first sale values exceeding 10 billion dollars per annum, has led to host-switching of E. hepatopenaeii to P. vannamei, with accompanying high prevalence and high-intensity infections being observed in both hosts in association with the recently emergent and devastating syndromic condition ‘early mortality syndrome’ (EMS) [83] (Figure 2). Phylogenetic analysis placed the parasite within the Enterocytozoon clade, closest to the human gut pathogen E. bieneusi and to another intranuclear pathogen, Enterospora canceri, that infects the hepatopancreas of the European edible crab(Cancer pagurus) [84]. The rapid emergence of this microsporidian has prompted high-profile warnings to industry from regional bodies such as the Network of Aquaculture Centers in the Asia Pacific (NACA; www.enaca.org) advising that E. hepatopenaei should be added to list of pathogens screened for during production of post-larvae for eventual stocking of commercial farms. Once again, the link between microsporidiosis and either suboptimal environmental conditions within the farm, or population immune suppression associated with inbreeding, may have played a role in recent and rapid emergence across major shrimp-farming regions [85].
Figure 2. Microsporidiosis in Shrimp Farming.
Routine health-checking of shrimp stock throughout the production cycle (A,B) and the application of sensitive and specific diagnostics for known and emergent shrimp pathogens has revealed a host-switching event and emergence of clinical disease caused by the microsporidian parasite Enterocytozoon hepatopenaeii in Penaeus vannamei from in Asia. The parasite, congeneric with the human pathogen Enterocytozoon bieneusi, undergoes similar development within the gut of infected shrimp and is implicated in the multi-billion dollar yield-limiting condition known as early mortality syndrome (EMS).
Terrestrial farm animals can also be infected with microsporidia. Although no clinical cases of microsporidiosis have been reported in cows or pigs, E. bieneusi, including human-pathogenic genotypic strains, are commonly detected in the feces of dairy and beef herds [86] and swine with diarrhea [87]. En. cuniculi and En. intestinalis have also been detected in pigs, again without apparent clinical outcome for the infected host [88]. Similar associations apparently exist between En. cuniculi, goats [89], and horses [90]. Human pathogenic strains of E. bieneusi have also been detected in feces of goats [91]. The first case of non-mammalian E. bieneusi infection was detected in chickens destined for human consumption [54], and subsequently other avian hosts were shown to be susceptible [88]. Although published epidemiological studies determining zoonotic transfer of microsporidia from farm animals to humans are rare, evidence for shared genotypes in humans, cows, and pigs have been reported from rural communities in China [56]. Zoonotic transfer between region-specific food animals and humans have been reported, including guinea pig to human transfer in Peru [57] and rabbit to human transfer in New Zealand [92].
Concluding Remarks
Microsporidia are ubiquitous inhabitants of all major biomes. As hyper-diverse opportunists, they exhibit differing degrees of host specificity, life cycle complexity, and ability to infect and cause disease in almost all known invertebrate and numerous vertebrate phyla, including humans. The diseases that they impart impact upon managed pollinators, on mass-reared fish and invertebrates for food, and on hosts used in biological control of pests. The presence of free- and host-associated parasite life-stages in water, soil, and food appear to offer ample opportunity for exposure of humans to animal-infecting forms. Even though the phylogenetic range of human-infecting forms extends to only 10 of the known 200 genera at present, increasingly consistent application of molecular diagnostics to animal and human infections will undoubtedly reveal an increased potential zoonotic range, particularly as new taxa are described from terrestrial and aquatic systems. Conversely, the application of environmental DNA approaches [93] not only has the potential to uncover hitherto unknown parasite diversity but will enable research on the identification of reservoirs for human-pathogenic taxa in terrestrial and aquatic wildlife hosts.
Definitive confirmation of emergence, or even increased prevalence, of microsporidiosis has been difficult to establish for wild populations in absence of long-term monitoring programs. However, well-publicized cases of emergence, increased pathogenesis, and morbidity associated with microsporidian infections exist for widely-divergent host groups, ranging from farmed shrimp to human patients with underlying infections such as with HIV. In all such cases, emergence (including potential for host switching) appears to center on a common node of altered immune competence in these diverse host groups. In essence, the prevalence of microsporidian infection and the intensity of the diseases they cause provide a living sentinel of host immune competence that traverses both host taxonomy and the biome in which these hosts exist. Climate change and other biome-level stressors (e.g., ocean acidification, intensification of farming) may associate to impart greater disease burden on hosts from all biomes, and thus increase the contact rate between infected animals and humans. Coupled with an increasing global population of immune-compromised individuals (associated with age, those undergoing treatment for malignancies and other infectious diseases such as produced by HIV), microsporidiosis may be expected to rise in both prevalence and severity. The major transmission route between host groups is via the food chain. Broader consideration of plant/animal/ human diseases associated with environmental pressures under the One Health agenda will be increasingly required as a means to address the grand challenges associated with global sustainability (http://www.cdc.gov/onehealth) and to manage microsporidian infections in wildlife, food animals, and humans (see Outstanding Questions).
Trends.
Microsporidiosis is an emerging disease in hosts from aquatic and terrestrial biomes.
Human infections are often derived from contact with animals and the environment.
Common nodes of immune suppression allow opportunistic infection and disease.
The animal–human food chain provides a portal for transmission and emergence.
Outstanding Questions.
Are appropriate phylogenetic tools available to allow detailed molecular epidemiology of known and novel members of the phylum Microsporidia?
Can humans be infected with a broader range of microsporidian taxa than is currently recognized?
What are the conditions that allow microsporidia to cross between species, and what allows emergent infections in this phylum?
Is Enterocytozoon bieneusi able to replicate in aquatic vertebrate or invertebrate host taxa?
Is there a common node of interaction (immunological, biochemical) among microsporidia from across the phylum and their broad range of hosts?
Can common nodes of interaction be exploited to evade infection or to mitigate pathogenic outcomes in infected hosts?
Can techniques be developed to allow genetic manipulation of the microsporidia that would facilitate experiments aimed at understanding the biology of these organisms?
Acknowledgments
This review is an output from a symposium sponsored by the Organisation for Economic Cooperation and Development (OECD) Cooperative Research Programme (CRP) on Biological Resource Management for Sustainable Agricultural Systems and the Society for Invertebrate Pathology (SIP), held on 9th August 2015 at the University of British Columbia, Vancouver, BC, Canada. The symposium was entitled ‘Microsporidia in the Animal to Human Food Chain: An International Symposium To Address Chronic Epizootic Disease’. We acknowledge the generous funding provided by the OECD CRP and the SIP to speakers at this event. The lead author (G.D.S.) would like to acknowledge funding by DG SANCO of the European Commission (under contract C5473) and the UK Department for Environment, Food, and Rural Affairs (DEFRA) (under contract FB002).
References
- 1.Gajadhar AA, et al. Overview of food- and water-borne zoonotic parasites at the farm level. Rev. Sci. Tech. 2006;25:595–606. [PubMed] [Google Scholar]
- 2.WHO. First formal meeting of the Foodborne Disease Burden Epidemiology Reference Group (FERG): Implementing Strategy, Setting Priorities and Assigning the Tasks. World Health Organization; 2007. [Google Scholar]
- 3.Doyle ME. Foodborne Parasites. A Review of the Scientific Literature. University of Wisconsin-Madison: Food Research Institute; 2003. [Google Scholar]
- 4.Centers For Disease Control and Prevention. CDC Estimates of Foodborne Illness in the United States. CDC: CDC 2011 Estimates: Findings; 2011. [Google Scholar]
- 5.MacKenzie WR, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 6.Simarro PP, et al. Human African trypanosomiasis in non-endemic countries (2000–2010) J. Travel Med. 2012;19:44–53. doi: 10.1111/j.1708-8305.2011.00576.x. [DOI] [PubMed] [Google Scholar]
- 7.Slifko TR, et al. Emerging parasite zoonoses associated with water and food. Int. J. Parasitol. 2000;30:1379–1393. doi: 10.1016/s0020-7519(00)00128-4. [DOI] [PubMed] [Google Scholar]
- 8.Sak B, et al. First report of Enterocytozoon bieneusi infection on a pig farm in the Czech Republic. Vet. Parasitol. 2008;153:220–224. doi: 10.1016/j.vetpar.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Stentiford GD, et al. Microsporidia: diverse, dynamic and emergent pathogens in aquatic systems. Trends Parasitol. 2013;29:567–578. doi: 10.1016/j.pt.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Vossbrinck CR, Debrunner-Vossbrinck BA. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitol. 2005;52:131–142. doi: 10.14411/fp.2005.017. [DOI] [PubMed] [Google Scholar]
- 11.Stentiford GD, et al. Plastic parasites: extreme dimorphism creates a taxonomic conundrum in the phylum Microsporidia. Int. J. Parasitol. 2013;43:339–352. doi: 10.1016/j.ijpara.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Cavalier-Smith T. A 6-kingdom classification and a unified phylogeny. In: Schenk HEA, Schwemmler WS, editors. Endocytobiology II: Intracellular Space as Oligo-genetic. Walter de Gruyter; 1983. pp. 1027–1034. [Google Scholar]
- 13.Keeling PJ. Five things to know about Microsporidia. PLoS Pathog. 2009;5:e1000489. doi: 10.1371/journal.ppat.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 15.Thomarat F, et al. Phylogenetic analysis of the complete genome sequence of Encephalitozoon cuniculi supports the fungal origin of microsporidia and reveals a high frequency of fast-evolving genes. J. Mol. Evol. 2004;59:780–791. doi: 10.1007/s00239-004-2673-0. [DOI] [PubMed] [Google Scholar]
- 16.Williams BA, et al. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 17.Gill EE, Fast NM. Assessing the microsporidia-fung relationship: Combined phylogenetic analysis of eight genes. Gene. 2006;375:103–109. doi: 10.1016/j.gene.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe Y, et al. Are Microsporidia really related to Fungi? A reappraisal based on additional gene sequences from basa fungi. Mycol. Res. 2002;106:1380–1391. [Google Scholar]
- 19.Capella-Gutierrez S, et al. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 2012;10:47. doi: 10.1186/1741-7007-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones MD, et al. Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474:200–203. doi: 10.1038/nature09984. [DOI] [PubMed] [Google Scholar]
- 21.Jones MD, et al. Validation and justification of the phylum name Cryptomycota phyl. nov. IMA Fungus. 2011;2:173–175. doi: 10.5598/imafungus.2011.02.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpov SA, et al. Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front. Microbiol. 2014;5:112. doi: 10.3389/fmicb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karpov SA, et al. Obligately phagotrophic aphelids turned out to branch with the earliest-diverging Fungi. Protist. 2013;164:195–205. doi: 10.1016/j.protis.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Haag KL, et al. Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15480–15485. doi: 10.1073/pnas.1410442111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeling PJ. Phylogenetic place of Microsporidia in the tree of eukaryotes. In: Weiss L, Becnel JJ, editors. Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 195–202. [Google Scholar]
- 26.Becnel JJ, et al. Checklist of available generic names for microsporidia with type species and type hosts. In: Weiss L, Becnel JJ, editors. Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 671–686. [Google Scholar]
- 27.Vossbrinck OR, et al. Phylogeny of the Microsporidia. In: Weiss L, Becnel JJ, editors. Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 203–220. [Google Scholar]
- 28.Vavra J, Lukes J. Microsporidia and ‘the art of living together’. Adv. Parasitol. 2013;82:254–319. doi: 10.1016/B978-0-12-407706-5.00004-6. [DOI] [PubMed] [Google Scholar]
- 29.Didier ES, Khan IA. The immunology of microsporidiosis in mammals. In: Weiss L, Becnel JJ, editors. Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 307–325. [Google Scholar]
- 30.Bednarska M, et al. Occurrence of intestinal microspordia in immunodeficient patients in Poland. Ann. Agric. Environ. Med. 2014;21:244–248. doi: 10.5604/1232-1966.1108584. [DOI] [PubMed] [Google Scholar]
- 31.Sprague V. Nosema connori n. sp., a microsporidian parasite of man. Trans. Am. Microscop. Soc. 1974;93:400–403. [PubMed] [Google Scholar]
- 32.Desportes I, et al. Occurrence of a new microsporidan: Enterocytozoon bieneusi n.g., n. sp., in the enterocytes of a human patient with AIDS. J. Protozool. 1985;32:250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x. [DOI] [PubMed] [Google Scholar]
- 33.Orenstein JM, et al. Disseminated microsporidiosis in AIDS: are any organs spared? AIDS. 1997;11:385–386. [PubMed] [Google Scholar]
- 34.Fayer R, Santin-Duran M. Epidemiology of micro-sporidia in human infections. In: Weiss L, Becnel JJ, editors. In Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 135–164. [Google Scholar]
- 35.Sak B, et al. Latent microsporidial infection in immuno-competent individuals - a longitudinal study. PLoS Negl. Trop. Dis. 2011;5:e1162. doi: 10.1371/journal.pntd.0001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobo ML, et al. Microsporidia as emerging pathogens and the implication for public health: a 10-year study on HIV-positive and -negative patients. Int. J. Parasitol. 2012;42:197–205. doi: 10.1016/j.ijpara.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Matos O, et al. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012;2012:981424. doi: 10.1155/2012/981424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lores B, et al. Intestinal microsporidiosis due to Enter-ocytozoon bieneusi in elderly human immunodeficiency virus-negative patients from Vigo, Spain. Clin. Infect. Dis. 2002;34:918–921. doi: 10.1086/339205. [DOI] [PubMed] [Google Scholar]
- 39.National Institute of Aging WHO. Global Health and Aging (NIH Publication 11-7737) National Institutes of Health and World Health Organization; 2011. [Google Scholar]
- 40.Didier ES, Weiss LM. Microsporidiosis: not just in AIDS patients. Curr. Opin. Infect. Dis. 2011;24:490–495. doi: 10.1097/QCO.0b013e32834aa152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, et al. Infectivity of microsporidia spores stored in water at environmental temperatures. J. Parasitol. 2003;89:185–188. doi: 10.1645/0022-3395(2003)089[0185:IOMSSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 42.Graczyk TK, et al. Human waterborne parasites in zebra mussels (Dreissena polymorpha) from the Shannon River drainage area, Ireland. Parasitol. Res. 2004;93:385–391. doi: 10.1007/s00436-004-1142-4. [DOI] [PubMed] [Google Scholar]
- 43.Lucy FE, et al. Biomonitoring of surface and coastal water for Cryptosporidium, Giardia and human virulent microsporidia using molluscan shellfish. Parasitol. Res. 2008;103:1369–1375. doi: 10.1007/s00436-008-1143-9. [DOI] [PubMed] [Google Scholar]
- 44.Santin M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 2009;56:34–38. doi: 10.1111/j.1550-7408.2008.00380.x. [DOI] [PubMed] [Google Scholar]
- 45.Stentiford GD, et al. Hepatospora eriocheir (Wang & Chen, 2007) gen. et comb. nov. from European Chinese mitten crabs (Eriocheir sinensis) J. Invertebr. Pathol. 2011;108:156–166. doi: 10.1016/j.jip.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Jedrzejewski S, et al. Quantitative assessment of contamination of fresh food produce of various retail types by human-virulent microsporidian spores. Appl. Environ. Microbiol. 2007;73:4071–4073. doi: 10.1128/AEM.00477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JH. Molecular detection of Enterocytozoon bieneusi and identification of a potentially human-pathogenic genotype in milk. Appl. Environ. Microbiol. 2008;74:1664–1666. doi: 10.1128/AEM.02110-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Decreane V, et al. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol. Infect. 2012;140:519–527. doi: 10.1017/S095026881100077X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coyle CM, et al. Fatal myositis due to the microsporidian Brachiola algerae, a mosquito pathogen. N. Engl. J. Med. 2004;351:42–47. doi: 10.1056/NEJMoa032655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choudary MM, et al. Tubulinosema sp. microsporidian myositis in immunosuppressed patient. Emerg. Infect. Dis. 2011;17:1727–1730. doi: 10.3201/eid1709.101926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vávra J, et al. Microsporidia of the genus Trachipleistophora - causative agents of human microsporidiosis: description of Trachipleistophora anthropophthera n. sp. (Protozoa: Microsporidia) J. Eukaryot. Microbiol. 1998;45:273–283. doi: 10.1111/j.1550-7408.1998.tb04536.x. [DOI] [PubMed] [Google Scholar]
- 52.Cali A, Takavorian PM. Ultrastructure and development of Pleistophora ronneafiei n.sp., amicrosporidium (Protista) in the skeletal muscle of an immuno-compromised individual. J. Eukaryot. Microbiol. 2003;50:77–85. doi: 10.1111/j.1550-7408.2003.tb00237.x. [DOI] [PubMed] [Google Scholar]
- 53.Chupp GL, et al. Myositis due to Pleistophora (Microsporidia) in a patient with AIDS. Clin. Infect. Dis. 1993;16:15–21. doi: 10.1093/clinids/16.1.15. [DOI] [PubMed] [Google Scholar]
- 54.Reetz J, et al. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens) Int. J. Parasitol. 2002;32:785–787. doi: 10.1016/s0020-7519(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 55.Fayer R, et al. Detection of Encephalitozoon hellem in feces of experimentally infected chickens. J. Eukaryot. Microbiol. 2003;50:574–575. doi: 10.1111/j.1550-7408.2003.tb00634.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 2011;49:2006–2008. doi: 10.1128/JCM.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cama VA, et al. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. J. Clin. Microbiol. 2007;45:2708–2710. doi: 10.1128/JCM.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velásquez JN, et al. First case report of infection caused by Encephalitozoon intestinalis in a domestic cat and a patient with AIDS. Vet. Parasitol. 2012;190:583–586. doi: 10.1016/j.vetpar.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 59.Bjørnson S, Oi D. Microsporidia biological control agents and pathogens of beneficial insects. In: Weiss L, Becnel JJ, editors. In Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 635–670. [Google Scholar]
- 60.Geden CJ, et al. Rapid deterioration of searching behavior, host destruction, and fecundity of the parasitoid Muscidifurax raptor (Hymenoptera: Pteromalidae) in culture. Ann. Entomol. Soc. Am. 1992;85:179–187. [Google Scholar]
- 61.Geden CJ, et al. Nosema disease of the parasitoid Muscidifurax raptor (Hymenoptera: Pteromalidae): prevalence, patterns of transmission, management, and impact. Biol. Control. 1995;5:607–614. [Google Scholar]
- 62.van Huis A, et al. Edible Insects: Future Prospects for Food and Feed Security. Food and Agriculture Organization of the United Nations; 2013. [Google Scholar]
- 63.Potts SG, et al. Global pollinator declines: impacts, trends and drivers. Trends Ecol. Evol. 2010;25:345–353. doi: 10.1016/j.tree.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Cornman SR, et al. Pathogen webs in collapsing honeybee colonies. PLoS ONE. 2012;7:e43562. doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fries I. Nosema apis - a parasite in the honeybee colony. Bee World. 1993;74:5–19. [Google Scholar]
- 66.Fries I, et al. Nosema ceranae n. sp. (Microspora, Nose-matidae), morphological and molecular characterization of a microsporidian parasite of the Asian honeybee Apis cerana (Hymenoptera, Apidae) Eur. J. Protistol. 1996;32:356–365. [Google Scholar]
- 67.Van Engelsdorp D, et al. Colony collapse disorder: a descriptive study. PLoS ONE. 2009;4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dainat B, et al. Colony collapse disorder in Europe. Environ. Microbiol. Rep. 2012;4:123–125. doi: 10.1111/j.1758-2229.2011.00312.x. [DOI] [PubMed] [Google Scholar]
- 69.Cameron SA, et al. Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. U.S.A. 2011;108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daszak P. Emerging infectious diseases of wildlife - threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 71.Ratnieks FLW, Carreck NL. Clarity on honey bee collapse? Science. 2010;327:152–153. doi: 10.1126/science.1185563. [DOI] [PubMed] [Google Scholar]
- 72.Fürst MA, et al. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. 2014;506:364–366. doi: 10.1038/nature12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown MJF. The trouble with bumblebees. Nature. 2011;469:169. doi: 10.1038/469169a. [DOI] [PubMed] [Google Scholar]
- 74.Murray TE, et al. Pathogen prevalence in commercially reared bumble bees and evidence of spillover in conspecific populations. Biol. Conserv. 2013;159:269–276. doi: 10.1016/j.biocon.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kent ML, et al. Microsporidia in fish. In: Weiss L, Becnel JJ, editors. In Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 493–520. [Google Scholar]
- 76.Palenzuela O, et al. A new intranuclear microsporidium, Enterospora nucleophila n. sp., causing an emaciative syndrome in a piscine host (Sparus aurata), prompts the redescription of the family Enterocytozoonidae. Int. J. Parasitol. 2014;44:189–203. doi: 10.1016/j.ijpara.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 77.Ramsay JM, et al. Pseudoloma neurophilia infections in zebrafish Danio rerio: effects of stress on survival, growth, and reproduction. Dis. Aquat. Org. 2009;88:69–84. doi: 10.3354/dao02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wongtavatchai J, et al. Effects of the microsporidian Enterocytozoon salmonis on the immune response of chinook salmon. Vet. Immunol. Immunopathol. 1995;48:367–374. doi: 10.1016/0165-2427(95)05435-9. [DOI] [PubMed] [Google Scholar]
- 79.Stentiford GD, et al. Disease will limit future food supply from global crustacean fishery and aquaculture sectors. J. Invertebr. Pathol. 2012;110:141–147. doi: 10.1016/j.jip.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 80.Stentiford GD, et al. Myospora metanephrops (n. g.,n.sp.) from marine lobsters and aproposal for erection of a new order and family (Crustaceacida; Myosporidae) in the class Marinosporidia (Phylum Microsporidia) Int. J. Parasitol. 2010;40:1433–1446. doi: 10.1016/j.ijpara.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 81.Stentiford GD, et al. Areospora rohanae n.gn. n.sp. (Microsporidia; Areosporiidae n. fam.) elicits multi-nucleate giant-cell formation in crabs. J. Invert. Pathol. 2014;118:1–11. doi: 10.1016/j.jip.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 82.Tourtip S, et al. Enterocytozoon hepatopenaei sp. nov. (Microspora: Enterocytozoonidae), a parasite of the black tiger shrimp Penaeus monodon (Decapoda: Penaeidae):fine structure and phylogenetic relationships. J. Invert. Pathol. 2009;102:21–29. doi: 10.1016/j.jip.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 83.Tangprasittipap A, et al. The microsporidian Enterocytozoon hepatopenaei is not the cause of white feces syndrome in whiteleg shrimp Penaeus (Litopenaeus) vannamei. BMC Vet. Res. 2013;9:139. doi: 10.1186/1746-6148-9-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stentiford GD, et al. Enterospora canceri n.gen., n.sp., an intranuclear microsporidian infecting European edible crab (Cancer pagurus) Dis. Aquat. Org. 2007;75:61–72. doi: 10.3354/dao075061. [DOI] [PubMed] [Google Scholar]
- 85.Doyle RW. Inbreeding and disease in tropical shrimp aquaculture:areappraisal and caution. Aquacult. Res. 2014;2014:1–15. [Google Scholar]
- 86.Santin M, Fayer R. A longitudinal study of Enterocytozoon bieneusi in dairy cattle. Parasitol. Res. 2009;105:141–144. doi: 10.1007/s00436-009-1374-4. [DOI] [PubMed] [Google Scholar]
- 87.Jeong DK, et al. Occurrence and genotypic characteristics of Enterocytozoon bieneusi in pigs with diarrhea. Parasitol. Res. 2007;102:123–128. doi: 10.1007/s00436-007-0740-3. [DOI] [PubMed] [Google Scholar]
- 88.Snowden KF. Microsporidia in higher vertebrates. In: Weiss L, Becnel JJ, editors. In Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 469–491. [Google Scholar]
- 89.Cislakova L, et al. Prevalence of antibodies to Encephalitozoon cuniculi (Microsporidia) in angora goats - a potential risk of infection for breeders. Ann. Agric. Environ. Med. 2001;8:289–291. [PubMed] [Google Scholar]
- 90.Goodwin D, et al. Prevalence of antibodies to Encephalitozoon cuniculi in horses from Brazil. Vet. Parasitol. 2006;142:380–382. doi: 10.1016/j.vetpar.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 91.Santin M, et al. A zoonotic genotype of Enterocytozoon bieneusi in horses. J. Parasitol. 2010;96:157–161. doi: 10.1645/GE-2184.1. [DOI] [PubMed] [Google Scholar]
- 92.Ozcan O, et al. Encephalitozoonosis in New Zealand rabbits and potential transmission risk. Vet. Parasitol. 2011;179:234–237. doi: 10.1016/j.vetpar.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 93.Bass D, et al. Diverse applications of environmental DNA methods in parasitology. Trends Parasitol. 2015;31:499–513. doi: 10.1016/j.pt.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 94.Vávra J, Larsson JIR. Structure of microsporidia. In: Weiss L, Becnel JJ, editors. In Microsporidia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 1–70. [Google Scholar]
- 95.Larsson JIR. The primitive microsporidia. In: Weiss L, Becnel JJ, editors. In Microspor-idia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 605–634. [Google Scholar]
- 96.Cali A, Takvorian P. Developmental morphology and lifecycles of the microsporidia. In: Weiss LM, Becnel JJ, editors. In Pathogens of Opportunity. Wiley-Blackwell; 2014. pp. 71–133. [Google Scholar]
- 97.Cali A, et al. Human vocal cord infection with the microsporidium Anncaliia algerae. J. Eukaryot. Microbiol. 2010;57:562–567. doi: 10.1111/j.1550-7408.2010.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watts MR, et al. Anncaliia algerae microsporidial myositis. Emerg. Infect. Dis. 2014;20:185–191. doi: 10.3201/eid2002.131126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franzen C, et al. Transfer of the members of the genus Brachiola (microsporidia) to the genus Anncaliia based on ultra-structural and molecular data. J. Eukaryot. Microbiol. 2006;53:26–35. doi: 10.1111/j.1550-7408.2005.00066.x. [DOI] [PubMed] [Google Scholar]
- 100.Margileth AM, et al. Disseminated nosematosis in an immunologically compromised infant. Arch. Pathol. 1973;95:145–150. [PubMed] [Google Scholar]
- 101.Cali A, et al. Brachiola vesicularum, n. g., n. sp., a new microsporidium associated with AIDS and myositis. J. Eukaryot. Microbiol. 1998;45:240–251. doi: 10.1111/j.1550-7408.1998.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 102.Hocevar SN, et al. Microsporidiosis acquired through solid organ transplantation: a public health investigation. Ann. Intern. Med. 2014;160:213–220. doi: 10.7326/M13-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Norhayati M, et al. A preliminary study on the prevalence of intestinal microsporidiosis in patients with and without gastrointestinal symptoms in Malaysia. Trans. R. Soc. Trop. Med. Hyg. 2008;102:1274–1278. doi: 10.1016/j.trstmh.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 104.Orenstein JM, et al. Fatal pulmonary microsporidiosis due to Encephalitozoon cuniculi following allogeneic bone marrow transplantation for acute myelogenous leukemia. Ultrastruct. Pathol. 2005;29:269–276. doi: 10.1080/01913120590951257. [DOI] [PubMed] [Google Scholar]
- 105.Sharma S, et al. Microsporidial keratitis: need for increased awareness. Surv. Ophthalmol. 2011;56:1–22. doi: 10.1016/j.survophthal.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 106.Chabchoub N, et al. Genetic identification of intestinal microsporidia species in immunocompromised patients in Tunisia. Am. J. Trop. Med. Hyg. 2009;80:24–27. [PubMed] [Google Scholar]
- 107.Didier ES, et al. Isolation and characterization of a new human microsporidian, Encephalitozoon hellem (n. sp.), from three AIDS patients with keratoconjunctivitis. J. Infect. Dis. 1991;163:617–621. doi: 10.1093/infdis/163.3.617. [DOI] [PubMed] [Google Scholar]
- 108.Sharma S, et al. Ocular microsporidiosis. In: Weiss L, Becnel JJ, editors. In Microspor-idia: Pathogens of Opportunity. John Wiley & Sons; 2014. pp. 403–419. [Google Scholar]
- 109.Cali A, et al. Septata intestinalis n. g., n. sp., an intestinal microsporidian associated with chronic diarrhea and dissemination in AIDS patients. J. Eukaryot. Microbiol. 1993;40:101–112. doi: 10.1111/j.1550-7408.1993.tb04889.x. [DOI] [PubMed] [Google Scholar]
- 110.Hamamci B, et al. Prevalence of Encephalitozoon intestinalis and Enterocytozoon bieneusi in cancer patients under chemotherapy. Mikrobiyol. Bul. 2015;49:105–113. doi: 10.5578/mb.8787. [DOI] [PubMed] [Google Scholar]
- 111.Jimenez-Gonzalez GB, et al. Microsporidia in pediatric patients with leukemia or limphoma. Rev. Invest. Clin. 2012;64:25–31. [PubMed] [Google Scholar]
- 112.Orenstein JM, et al. Systemic dissemination by a newly recognized intestinal microsporidia species in AIDS. AIDS. 1992;6:1143–1150. doi: 10.1097/00002030-199210000-00013. [DOI] [PubMed] [Google Scholar]
- 113.Sivgin S, et al. Encephalitozoon intestinalis: a rare cause of diarrhea in an allogeneic hematopoietic stem cell transplantation (HSCT) recipient complicated by albendazole-related hepatotoxicity. Turk. J. Hematol. 2013;30:204–208. doi: 10.4274/Tjh.90692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teachey DT, et al. Pulmonary infection with microsporidia after allogeneic bone marrow transplantation. Bone Marrow Transpl. 2004;33:299–302. doi: 10.1038/sj.bmt.1704327. [DOI] [PubMed] [Google Scholar]
- 115.Carlson JR, et al. Disseminated microsporidiosis in a pancreas/kidney transplant recipient. Arch. Pathol. Lab. Med. 2004;128:e41–e43. doi: 10.5858/2004-128-e41-DMIAKT. [DOI] [PubMed] [Google Scholar]
- 116.Galvan AL, et al. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 2011;49:1301–1306. doi: 10.1128/JCM.01833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rabodonirina M, et al. Enterocytozoon bieneusi as a cause of chronic diarrhea in a heart-lung transplant recipient who was seronegative for human immunodeficiency virus. Clin. Infect. Dis. 1996;23:114–117. doi: 10.1093/clinids/23.1.114. [DOI] [PubMed] [Google Scholar]
- 118.Cali A, et al. Corneal microsporidioses: characterization and identification. J. Protozool. 1991;38:215S–217S. [PubMed] [Google Scholar]
- 119.Pariyakanok L, Jongwutiwes S. Keratitis caused by Trachipleistophora anthropopthera. J. Infect. 2005;51:325–328. doi: 10.1016/j.jinf.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 120.Choudhary MM, et al. Tubulinosema sp. microsporidian myositis in immunosuppressed patient. Emerg. Infect. Dis. 2011;17:1727–1730. doi: 10.3201/eid1709.101926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shadduck JA, et al. Isolation of a microsporidian from a human patient. J. Infect. Dis. 1990;162:773–776. doi: 10.1093/infdis/162.3.773. [DOI] [PubMed] [Google Scholar]
- 122.Fernandes M, Sharma S. Polymicrobial and microsporidial keratitis in a pathien using Boston scleral contact lens for Sjögren’s syndrome and ocular pemphigoid. Cont. Lens Anterior Eye. 2013;36:95–97. doi: 10.1016/j.clae.2012.10.082. [DOI] [PubMed] [Google Scholar]
- 123.Hartskeerl RA, et al. Genetic evidence for the occurrence of extra-intestinal Enterocytozoon bieneusi infections. Nucleic Acids Res. 1993;21:4150. doi: 10.1093/nar/21.17.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Del Aguila C, et al. Identification of Enterocytozoon bieneusi spores in respiratory samples from an AIDS patient with a 2-year history of intestinal microsporidiosis. J. Clin. Microbiol. 1997;35:1862–1866. doi: 10.1128/jcm.35.7.1862-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rutrecht ST, Brown MJF. Differential virulence in a multiple-host parasite of bumble bees: resolving the paradox of parasite survival? Oikos. 2009;118:941–949. [Google Scholar]
- 126.Mayack C, Naug D. Energetic stressin the honeybee Apis mellifera from Nosema ceranae infection. J. Invert. Pathol. 2009;100:185–188. doi: 10.1016/j.jip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 127.Huang W-F, et al. Nosema ceranae escapes fumagillin control in honey bees. PLOS Pathog. 2013;9:e1003185. doi: 10.1371/journal.ppat.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]