Abstract
Purpose
To identify new therapeutic targets for non-small cell lung cancer (NSCLC), we systematically searched 2 cancer cell line databases for sensitivity data on a broad range of drugs. We identified polo-like kinase 1 (PLK1) as the most promising target for further investigation based on a subset of sensitive NSCLC cell lines and inhibitors that were in advanced clinical development.
Experimental Design
To identify potential biomarkers of response of NSCLC to PLK1 inhibition and mechanisms of PLK1 inhibitor-induced apoptosis, integrated analysis of gene and protein expression, gene mutations, and drug sensitivity was performed using 3 PLK1 inhibitors (volasertib, BI2536, and GSK461364) with a large panel of NSCLC cell lines.
Results
The NSCLC cell lines had different sensitivities to PLK1 inhibition, with a minority demonstrating sensitivity to all 3 inhibitors. PLK1 inhibition led to G2/M arrest, but only treatment-sensitive cell lines underwent substantial apoptosis following PLK1 inhibition. NSCLC lines with high epithelial-mesenchymal transition gene signature scores (mesenchymal cell lines) were more sensitive to PLK1 inhibition than were epithelial lines (P < 0.02). Likewise, proteomic profiling demonstrated that E-cadherin expression was higher in the resistant cell lines than in the sensitive ones (P < 0.01). Induction of an epithelial phenotype by expression of the microRNA miR-200 increased cellular resistance to PLK1 inhibition. Also, KRAS mutation and alterations in the tight-junction, ErbB, and Rho signaling pathways correlated with drug response of NSCLC.
Conclusions
In this first reported large-scale integrated analysis of PLK1 inhibitor sensitivity, we demonstrated that epithelial-mesenchymal transition leads to PLK1 inhibition sensitivity of NSCLC cells. Our findings have important clinical implications for mesenchymal NSCLC, a significant subtype of the disease that is associated with resistance to currently approved targeted therapies.
Keywords: polo-like kinase 1, non-small cell lung cancer, targeted therapy, epithelial-mesenchymal transition
Introduction
Polo-like kinases (PLKs) are 5 highly conserved serine/threonine kinases. PLK1 is the best characterized member of this kinase family, and its overexpression transforms 3T3 cells. PLK1 regulates centrosomes at the G2/M transition and orchestrates cell-cycle progression and response to DNA damage (1). Whereas PLK1 is scarcely detectable in most normal adult tissues, it is overexpressed in many tumors, and its overexpression is associated with poor prognosis for non-small cell lung cancer (NSCLC) (2). A subset of patients with refractory NSCLC have responses to PLK1 inhibition, and authors have experienced long-term disease control (3, 4). A predictive biomarker of response to treatment with PLK1 inhibitors has yet to be identified.
Researchers have identified several potential candidate biomarkers to predict PLK1 inhibitor sensitivity, however. In a genome-wide RNA interference screen to identify synthetic lethal interactions of kinases with the KRAS oncogene in a pair of isogenic colon cancer cell lines, a short hairpin RNA against PLK1 was more toxic in KRAS-mutant cells than in cells wild-type for KRAS. Another study confirmed this effect of the PLK1 inhibitor BI2536 (5). Also, PLK1 overexpression was significantly associated with the presence of TP53 mutations in breast and lung tumors (6, 7). Cell line screening with the PLK1 inhibitor GSK461364 demonstrated that cells with loss of p53 expression were more sensitive to treatment with GSK461364 than were cells with intact p53 expression. The more sensitive lines also had higher levels of chromosome instability than did the resistant cells (8). In contrast, loss of TP53 expression in isogenic colon cancer lines did not affect PLK1 inhibitor sensitivity unless the cells were exposed to ionizing radiation (9). Finally, PLK1 inhibitors were particularly toxic to glioblastoma and breast cancer stem cells (10, 11).
To enhance future clinical translation of our research, we chose to examine 3 PLK1 inhibitors that were the most advanced in clinical development and for which we had safety and pharmacokinetic data: BI2536, volasertib, and GSK461364. All 3 are ATP-competitive kinase inhibitors. BI2536 and volasertib (Boehringer Ingelheim) are dihydropteridinone derivatives. Characterization of these inhibitors using in vitro kinase assays demonstrated that BI2536 inhibited PLK1, PLK2, and PLK3 activity at IC50s of 0.83, 3.50, and 9.00 nM, respectively, and exhibited 1000-fold greater selectivity for PLK1 than for a panel of 63 other kinases (12). In comparison, volasertib inhibited PLK1, PLK2, and PLK3 at IC50s of 0.87, 5.00, and 56.00 nM, respectively. In vivo, volasertib has more sustained tumor exposure, a higher volume of distribution, and a longer terminal half-life than does BI2536 (13). GSK461364 (GlaxoSmithKline) has an IC50 of 2.2 nM, with 400-fold greater potency for PLK1 than for PLK2 (IC50, ~860 nM) and PLK3 (IC50, ~1000 nM) and 1000-fold greater selectivity for PLK1 than for a panel of 48 other kinases (14).
In the present study, to identify potential biomarkers of response of NSCLC to treatment with PLK1 inhibitors and mechanisms of PLK1 inhibitor-induced apoptosis in NSCLC cells, we performed an integrated analysis of gene expression, protein expression, gene mutations, and drug sensitivity in a large panel of NSCLC cell lines. In addition to examining correlations with all 3 inhibitors individually, we analyzed a group of cell lines that were consistently sensitive or resistant to them (termed universal cell lines) to reduce the potential for off-target drug effects and confirmed our results using PLK1 knockdown in NSCLC cells. To demonstrate the functional significance of the correlation with EMT, we induced an epithelial or mesenchymal phenotype in NSCLC cells and measured their sensitivity to PLK1 inhibition.
Materials and Methods
Materials
Anti-PLK1 (Invitrogen); anti-phosphorylated Myt1 (Thr495) and Myt1 (Thermo Fisher Scientific); anti-poly(ADP-ribose) polymerase, -cleaved poly(ADP-ribose) polymerase, -phosphorylated translational controlled tumor protein (TCTP; Ser46), -cyclin B1, -E-cadherin, -vimentin, -β-catenin, and -Snail (Cell Signaling Technology); anti-TCTP (Abcam); and anti-β-actin (Sigma) antibodies were used. The PLK1 inhibitors BI2536, volasertib, and GSK461364 were purchased from Selleck Chemicals and prepared as 10-mM stock solutions in dimethyl sulfoxide. Predesigned sets of 4 independent small interfering RNA (siRNA) sequences of the target genes PLK1 and CDH1 (siGENOME SMARTpool; Dharmacon) were used. Human transforming growth factor-β1 was purchased from Cell Signaling Technology.
Cell culture and characterization
Sixty-three human NSCLC cell lines were authenticated via DNA fingerprinting, routinely tested for the presence of Mycoplasma species, and maintained as described previously (15). The cell line Cal-12T was purchased from The Leibniz Institute DSMZ. The cell lines’ mutational profiles for 264 genes (Supplementary Table S1) were obtained from COSMIC (version 67; http://cancer.sanger.ac.uk/cosmic) and the Cancer Cell line Encyclopedia (16). Baseline mRNA (48,804 probe sets) and protein (193 proteins and phosphoproteins) expression levels were determined using Illumina and reverse-phase protein arrays, respectively, as described previously (17, 18).
Cell viability assays
Fifty NSCLC cell lines were incubated with dimethyl sulfoxide (vehicle control), BI2536, or volasertib for 120 hours at 9 distinct concentrations, with the maximum dose being the peak drug concentration in humans (Cmax): 1.6 μM for BI2536 and 1.2 μM for volasertib (4, 19). Cell viability was measured using an MTT assay as described previously (20). In addition, 63 NSCLC cell lines were incubated with dimethyl sulfoxide or GSK461364 for 72 hours at 7 distinct concentrations, with the maximum dose being the Cmax (1 μM) (21). A CellTiter-Glo luminescent cell viability assay (Promega) was performed as per the manufacturer's specifications. For both assays, 6 replicates were tested at each concentration, and each test was completed at least twice on different days.
For colony formation assays, cells were treated for 24 hours with dimethyl sulfoxide or volasertib and then incubated in a drug-free medium for 14-21 days. Plates were stained with crystal violet, and the total colony area per well was estimated using the ImageJ software program (National Institutes of Health) as described previously (22).
Statistical analysis
IC50 and IC70 values were estimated using the best-fit dose-response model selected by calculating the residual standard error using the R software program drexplorer (23).
Gene expression data were available for 43 of the 50 cell lines treated with BI2536 or volasertib and 50 of the 63 lines treated with GSK461364. Gene expression and drug sensitivity data were available for 41 cell lines treated with all 3 drugs. Reverse-phase protein array data were obtained as described previously (17, 18, 24) and available for 44 lines treated with BI2536 or volasertib and 45 lines treated with GSK461364. Among the cell lines that were consistently sensitive or resistant to treatment with all 3 drugs (universal group), gene expression data were available for 10 sensitive and 11 resistant lines, and protein expression data were available for 10 sensitive and 13 resistant lines.
To compare gene and protein expression patterns in cell lines resistant and sensitive to the 3 drugs, 2-sample t-tests were performed on a gene-by-gene or protein-by-protein basis. To adjust for multiple testing, the beta-uniform mixture model was applied to modeling P values to select an appropriate false-discovery rate cutoff (25). For correlations between drug sensitivity and gene mutations, we performed the Fisher exact test. Additionally, associations between drug sensitivity and epithelial-mesenchymal transition (EMT) score (17, 26) were evaluated using 2-sample t-tests.
To evaluate pre-established signatures for KRAS dependency (27) and chromosomal instability (28), a 2-way hierarchical clustering technique (17) was applied to the data set to produce heatmaps. Correlation of the sensitivities to the 3 study drugs was analyzed using the chi-square test. The Fisher exact test was performed for assessment of associations between pairs of drugs. All statistical analyses were performed using the R programming language (29).
Pathway analysis was performed with the Ingenuity Pathway Analysis tool (QIAGEN) using gene sets as described in the Results section.
The animal data had repeated tumor size measurements at different time points. Therefore we considered the correlations between the measurements on the same subject. We fit this linear model (with treatment, time effect and its interaction effect) using the generalized least squares (GLS) method. The parameters were estimated by maximizing restricted log-likelihood (REML) method. We used the Akaike information criterion (AIC) for selecting the best correlation structure. The analysis was performed using nlme package in R (version 3.1.3).
Cell-cycle analysis and apoptosis assays
For cell-cycle analysis, cells were harvested, fixed, incorporated with bromodeoxyuridine (BrdU), and stained with 7-aminoactinomycin D (BrdU Flow kit; BD Biosciences). The DNA content was analyzed using a cytofluorimeter, a fluorescence-activated cell sorter (FACScan; Becton Dickinson), and the ModFit software program (Verity Software House) (20). Terminal deoxynucleotidyl transferase dUTP nick end labeling staining (APO-BrdU kit; BD Biosciences) was performed to measure apoptosis, and bromodeoxyuridine incorporation was quantitated using fluorescence-activated cell sorting (BrdU Flow kits; BD Biosciences) according to the manufacturer's protocols.
Western blotting and immunoprecipitation
NSCLC cells were lysed on ice, and the lysates were centrifuged at 20,000 g for 5 minutes at 4°C as described previously (30). For immunoprecipitation, lysates were precleared, incubated with an anti-TCTP antibody, and washed as described previously (30). Whole-celllysates containing 30-50 μg of proteins (Western blot analysis) or immunoprecipitates were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotted with the indicated primary antibodies, and detected using a horseradish peroxidase-conjugated secondary antibody (Bio-Rad) and an enhanced chemiluminescence reagent (Amersham Biosciences).
Transfections
NSCLC cells were transduced using Lipofectamine LTX or lentiviral delivery of pTRIPz–miR-200, pc-DNA3.1-ZEB1, or a control vector. Lentiviruses were produced by co-transfecting HEK-293 cells with 1.5 μg of the viral packaging vector psPAX2, 0.5 μg of the viral envelope vector pMD2.G, and 2 μg of pTRIPz–miR-200 or the control vector using Lipofectamine LTX. The HEK-293 cell medium was changed 24 hours after transfection, and the cells were incubated at 37°C for 48 hours to allow for virus production. After 48 hours, HEK-293 media containing viral particles were transferred onto NSCLC cell culture plates and incubated at 37°C for 48 hours. After transduction, fresh RPMI 1640 medium with 10% fetal bovine serum was added to the cell culture plates, and the cells were allowed to recover for 24 hours. Cells were selected using 3 μg/mL puromycin in miR-200 expressing cells and 500 μg/mL G418 in ZEB1-expressing cells, and 2 μg/mL doxycycline was used for induction. Induced RFP expression was used to visually verify successfully infected cells. H1299 cells expressing miR-200 were sorted using flow cytometry for RFP positivity to obtain a transfected cell population.
For siRNA transfection, NSCLC cells were harvested, washed, and suspended (106 cells/100 μL) in Nucleofector Solution V (Amaxa), and siRNA (200 pmol/100 μL) was added to the cells. The cells were then electroporated using Nucleofector program U24 (Amaxa) as described previously (30).
NSCLC xenograft models
All animal research was conducted in accordance with The University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee policies as described previously (31). Briefly, 40 female 6-week-old Swiss nu/nu mice were injected subcutaneously with 4 million H1975 or Calu6 cells. When visible tumors developed, (32)the mice were stratified according to tumor size into 2 groups and then randomly assigned intravenous treatment with a vehicle control or volasertib at 30 mg/kg weekly for 3 weeks or sooner if tumor burden was excessive. Following euthanasia, E-Cadherin and Ki67 immunohistochemistry was performed and scored as previously described (33).
Results
BI2536, volasertib, and GSK461364 inhibit PLK1 substrates in NSCLC cell lines
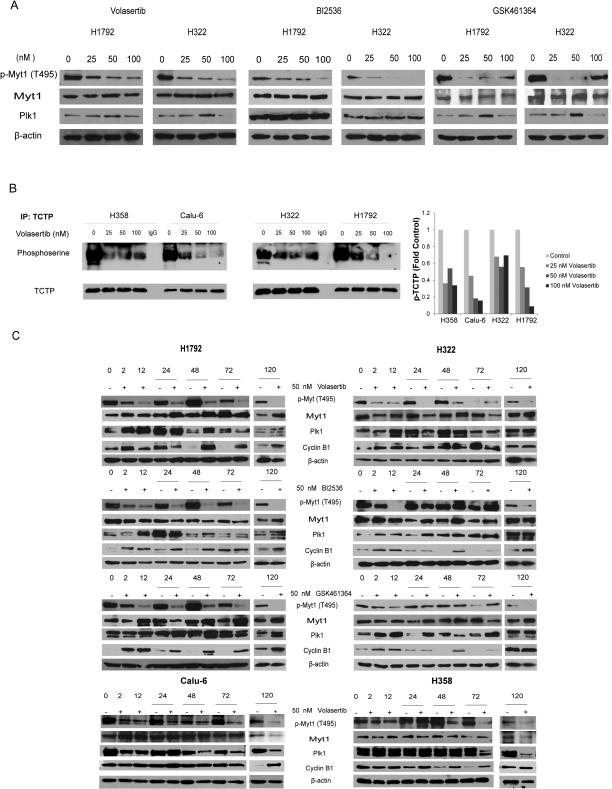
To confirm that treatment with BI2536, volasertib, and GSK461364 inhibits the activation of PLK1 in NSCLC cell lines, we measured the levels of PLK1 substrates, including phosphorylated TCTP and phosphorylated Myt1, in NSCLC cells treated with PLK1 inhibitors at different time points and concentrations (Fig. 1). We tested both PLK1 inhibitor-sensitive (H1792 and Calu-6) and -resistant (H322 and H358) NSCLC cell lines (PLK1 inhibitor sensitivity defined below). As expected, we observed decreases in the levels of phosphorylated TCTP and phosphorylated Myt1 following treatment with the PLK1 inhibitors that were dose-dependent with the exception of GSK461364 at high doses (Fig. 1A and B). Substrate inhibition persisted for 120 hours after administration of 1 dose; this was the maximal period the cells were exposed to drugs in the viability assays, confirming durable target inhibition. GSK461364 induced ABCB1, ABCG2, and ABCC1 expression at high concentrations, which likely led to a loss of PLK1 inhibition as suggested by the recovery of phosphorylated Myt1 (Supplementary Fig. S1). We also measured cyclin B1 expression in the cells, as proteasomal degradation of cyclin B1 is PLK1-dependent. As expected, at time points later, cyclin B1 expression increased (Fig. 1C).
Figure 1.
PLK1 inhibitors inhibit the activation of PLK1 substrates in NSCLC cell lines. NSCLC cell lines were incubated with dimethyl sulfoxide (vehicle control), BI2536, volasertib, or GSK461364 at the indicated concentrations for 2 hours (A and B) or at 50 nM for the indicated periods (in hours; C). Cells were then lysed and subjected to Western blotting with the indicated primary antibodies (A and C) or immunoprecipitated with an anti-TCTP antibody (B). Quantitation of the p-TCTP bands is shown in B.
NSCLC cell lines have differing sensitivity to treatment with PLK1 inhibitors
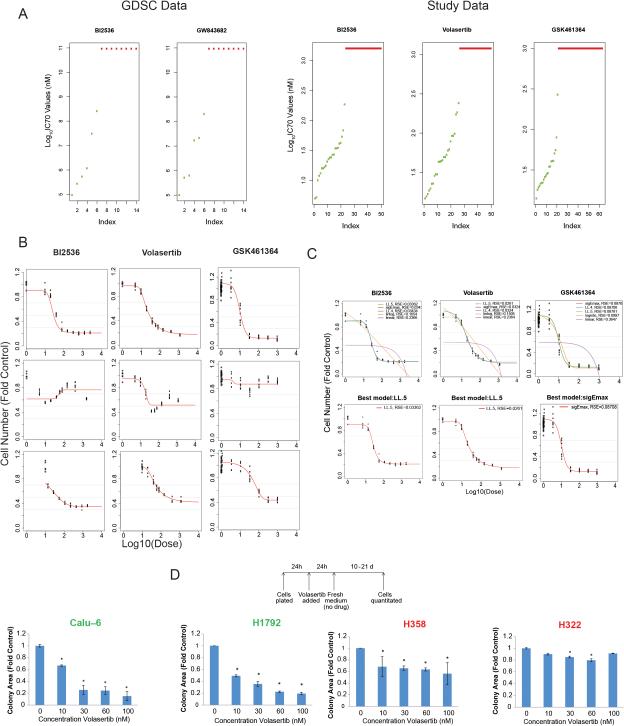
The Genomics of Drug Sensitivity in Cancer project (http://www.cancerrxgene.org/) (16, 34) has drug sensitivity data on 14 NSCLC cell lines treated with 2 PLK1 inhibitors (Fig. 2A). We tested 50 NSCLC cell lines to determine their sensitivity to treatment with BI2536 (Cmax, 1.6 μM) and volasertib (Cmax, 1.2 μM). To validate our findings, we also tested 63 NSCLC lines to determine their sensitivity to GSK461364 (Cmax, 1 μM) (Fig. 2A-C). For many cell lines, the dose-response curve plateaued at or near the IC50 (representative data shown in Fig. 1B and C), so we used IC70 values to distinguish treatment-sensitive and -resistant cell lines. We considered cell lines with IC70 values greater than the Cmax to be resistant and those with IC70 values less than or equal to the Cmax to be sensitive. We termed the cell lines that were consistently sensitive or resistant (10 and 13 cell lines, respectively) to treatment with the 3 PLK1 inhibitors universal cell lines.
Figure 2.
NSCLC cell lines have diverse sensitivities to PLK1 inhibition. A, the sensitivity to 2 PLK1 inhibitors in 14 NSCLC cell lines according to data obtained from the Genomics of Drug Sensitivity in Cancer (GDSC) database is shown at left. Each data point indicates the IC75 for 1 cell line. NSCLC cell lines were treated with BI2536, volasertib, or GSK461364 for 72-120 hours, and their viability was estimated using an MTT or CellTiter-Glo assay. Each data point on the right indicates the IC70 for 1 cell line. B and C, replicate data were graphed for each concentration, and the best dose-response model was selected using the residual standard error (RSE) method (representative data). The best model of 5 tested— Sigmoid Emax model (sigEmax), 4-parameter log-logistic model (LL.4), 5-parameter log-logistic model (LL.5), log linear model (linlog), and linear model (linear)—was then used to calculate the IC70 for each cell line. D, NSCLC cell lines were incubated with volasertib at the indicated concentrations for 24 hours; the medium was then changed to remove the drug, and colony formation was measured 14 days later. The names of sensitive cell lines (D) and data for sensitive cell lines (A) are shown in green, whereas those of and for resistant cell lines are shown in red.
The sensitivities to the 3 drugs correlated at a P value of 1.4e-06 (chi-square test). When we compared the drug sensitivities pair-wise using the Fisher exact test, the correlation between the sensitivities to BI2536 and volasertib (P = 1.1e-06) was stronger than that between GSK461364 and volasertib (P = 0.186) and that between GSK461364 and BI2536 (P = 0.020). We expected this result given these drugs’ similarities. To ensure that the observed sensitivity differences did not result from technical differences between the MTT and CellTiter-Glo assays, we tested the universal cell lines to determine their sensitivity to all 3 drugs under identical conditions. Although the IC70 values were not identical using the 2 assays, all of the universal cell lines remained sensitive or resistant to the treatment regardless of the assay used (Supplementary Table S2).
To independently measure drug sensitivity, we incubated 2 PLK1 inhibitor-sensitive and 2 PLK1 inhibitor-resistant NSCLC cell lines with 50 nM volasertib for 24 hours and then measured colony formation 10-21 days later. In agreement with the results of the MTT assays, formation of sensitive cell colonies but not of resistant ones was markedly impaired (Fig. 2D). This result demonstrated the irreversibility of the effect of PLK1 inhibition on cell viability and is consistent with our subsequent experiments demonstrating that PLK1 inhibition causes apoptosis of inhibitor-sensitive cell lines.
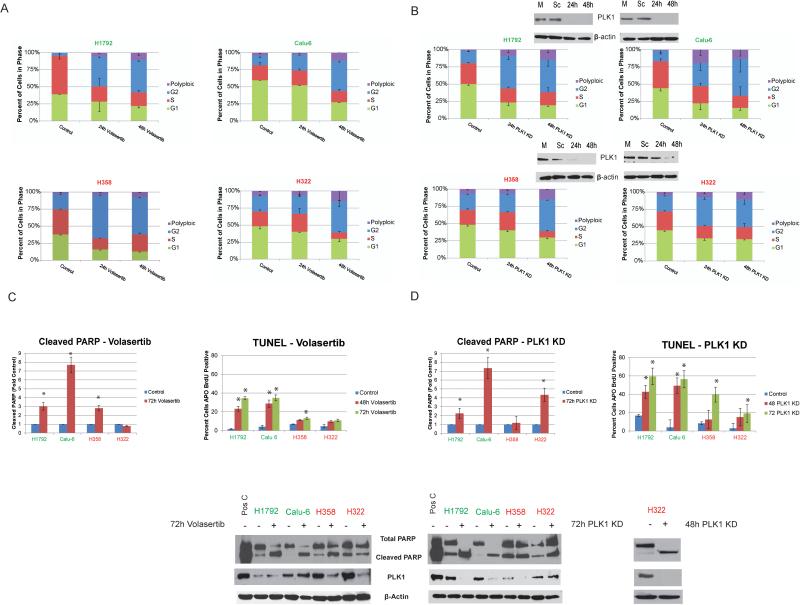
PLK1 inhibition and knockdown lead to cell-cycle arrest and apoptosis in NSCLC cell lines
The effect of PLK1 inhibition on the cell cycle is well characterized, and it typically leads to prometaphase arrest. We observed an increase in the number of NSCLC cells in G2/M and in the number of cells with greater than 4N DNA content (polyploid) in both volasertib-sensitive and -resistant lines treated with this drug (Fig. 3A). In contrast, treatment with volasertib induced substantial apoptosis only in sensitive cell lines (Fig. 3C and D). To demonstrate the specificity of PLK1 inhibitors, we knocked down PLK1 expression in NSCLC cell lines using siRNA and observed G2/M arrest in all cell lines, with more apoptosis in the PLK1 inhibitor-sensitive lines than in the resistant ones (Fig. 3B-D).
Figure 3.
PLK1 inhibition or knockdown leads to cell-cycle arrest and apoptosis in NSCLC cell lines.NSCLC cells with different sensitivities to treatment with PLK1 inhibitors were treated with 50 nM volasertib (A and C) or transfected with siRNA against PLK1 (B and D). A and B, cell-cycle stages determined using 7-aminoactinomycin D and BrdU incorporation. C and D, apoptosis measurement using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining and Western blotting of NSCLC cells for cleaved poly(ADP-ribose) polymerase (PARP). *P ≤ 0.05. PLK1 inhibition-sensitive and -resistant cell lines are indicated in green and red, respectively.
Mesenchymal NSCLC cell lines are more sensitive to PLK1 inhibition than are epithelial NSCLC cell lines
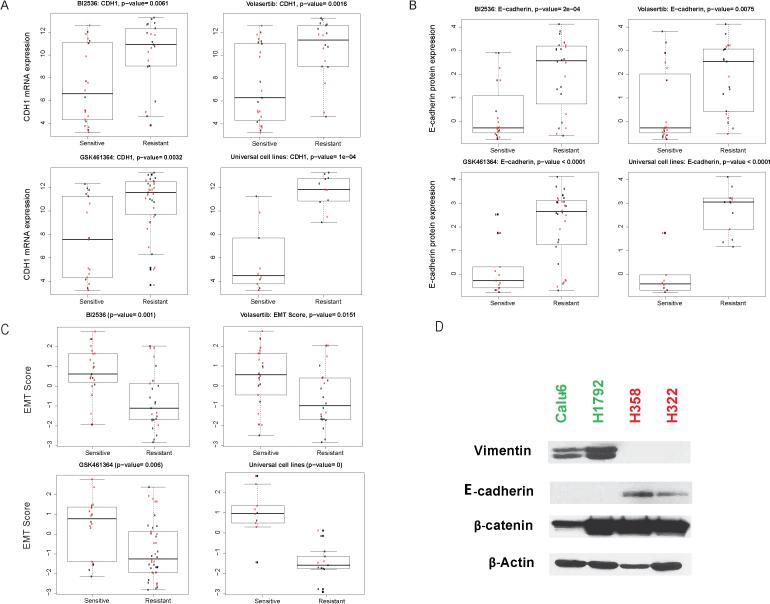
To determine why the NSCLC cell lines had diverse sensitivities to PLK1 inhibition, we examined the relationship between drug sensitivity and basal gene/protein expression in cell lines resistant and sensitive to BI2536, volasertib, or GSK461364 and the universal cell lines. We discovered that the expression of several genes and proteins correlated with drug sensitivity (Supplementary Fig. S2, Supplementary Table S3 and S4). Also, E-cadherin and β-catenin (CTNNB1) were consistently expressed at higher levels in the resistant lines than in the sensitive ones, and DNA-dependent protein kinase catalytic subunit and thymidylate synthase were consistently overexpressed in the sensitive lines.
We hypothesized that the genes that were expressed in all 4 groups of cell lines and that had the greatest differences in gene and protein expression between resistant and sensitive cell lines are the genes most likely to be involved in sensitivity—specifically, to PLK1 inhibition—rather than off-target drug effects. After correction for multiple comparisons, none of the genes’ expression correlated with sensitivity to BI2536 or volasertib. As an exploratory analysis, we examined probe sets with at least a 2-fold difference in mean gene expression between sensitive and resistant lines and corresponding P values less than 0.05 for this difference. Using these criteria, we identified 39 probe sets (37 genes) that overlapped in the 4 groups (Supplementary Table S5).
We next performed pathway analysis of cell lines with expression of genes that correlated with sensitivity to GSK461364 and the universal cell lines as well as the 39 overlapping probe sets described above. We observed that the tight-junction, ErbB, and Rho signaling pathways were altered significantly in multiple gene sets (Supplementary Table S6).
Based on the differential expression of E-cadherin (Fig. 4A and B) and the several genes involved in EMT as well as our observations that NSCLC cells with mesenchymal morphologies were more sensitive to PLK1 inhibition than were epithelial cells, we hypothesized that mesenchymal NSCLC cells are more sensitive than epithelial cells to PLK1 inhibition. To test this hypothesis, we examined the ability of a 76-gene EMT gene signature score developed by our group to distinguish PLK1 inhibition-sensitive and -resistant cell lines and validated it using multiple data sets (17, 24, 26). EMT scores were highly correlated with sensitivity to PLK1 inhibition in cell lines sensitive to any of the 3 study drugs, particularly in universal cell lines (Fig. 4C). Cell lines with higher EMT scores (mesenchymal) were more sensitive to treatment with PLK1 inhibitors than were cells with lower scores.
Figure 4.
NSCLC cell lines with mesenchymal gene expression patterns are more sensitive to PLK1 inhibition than are those with epithelial gene expression patterns. E-cadherin mRNA (CDH1; A) and protein (B) expression levels were significantly higher in PLK1 inhibition-resistant cell lines than in sensitive ones. C, EMT scores for PLK1 inhibition-sensitive and -resistant cell lines determined using a published signature (17). Data on KRAS-mutant cell lines are shown in red. D, E-cadherin and vimentin protein expression levels in 2 representative cell lines according to Western blotting. PLK1 inhibition-sensitive and -resistant cell lines are indicated in green and red, respectively.
Induction of an epithelial or mesenchymal phenotype affects sensitivity of NSCLC cell lines to PLK inhibition
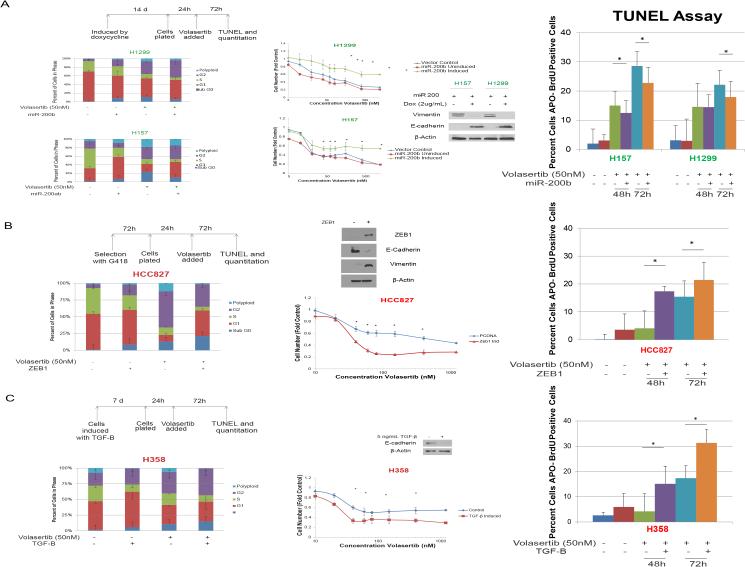
To determine whether the correlation between EMT score and drug sensitivity was functionally significant, we manipulated NSCLC cells and found that PLK1 inhibition predominantly led to apoptosis in mesenchymal cells and cell-cycle arrest in epithelial ones. Specifically, we transfected NSCLC cell lines with miR-200b and miR-200ab, which induce an epithelial protein expression pattern and phenotype (35). In H1299 and H157 cells, transfection with miR-200b and miR-200ab, respectively, led to increased expression of E-cadherin and decreased expression of vimentin. In agreement with the finding that epithelial NSCLC cell lines were more resistant than mesenchymal cells to PLK1 inhibition, induction of an epithelial phenotype increased the cell lines’ resistance to treatment with volasertib, with IC70 values increasing from 226 nM to greater than 1200 nM in H157 cells and from 253 nM to greater than 1200 nM in H1299 cells (Fig. 5A). The induction of a mesenchymal phenotype by ZEB1 overexpression (Fig. 5B) or transforming growth factor-β (Fig. 5C, Supplementary Fig. S3) led to an increase in volasertib-induced apoptosis and sensitivity to treatment with volasertib.
Figure 5.
Mesenchymal cells are more sensitive to PLK1 inhibition than are epithelial cells in isogenic human NSCLC models. A, forced expression of miR-200 led to increased E-cadherin expression and decreased vimentin expression according to Western blotting and to volasertib resistance according to an MTT assay. Induction of a mesenchymal phenotype by ZEB1 expression (B) or 5 ng/mL transforming growth factor-β (C) led to volasertib sensitivity and increased volasertib-induced apoptosis. *P ≤ 0.05 compared with a control or as indicated. PLK1 inhibition-sensitive and -resistant cell lines are indicated in green and red, respectively.
Owing to the striking correlation of PLK1 inhibitor sensitivity with E-cadherin expression, we examined the effect of knocking down E-cadherin expression on volasertib sensitivity in NSCLC cells. E-cadherin knockdown alone was not sufficient to change the biological effects of PLK1 inhibition on these cells (Supplementary Fig. S4).
PLK1 inhibition leads to decreased NSCLC tumor size in vivo
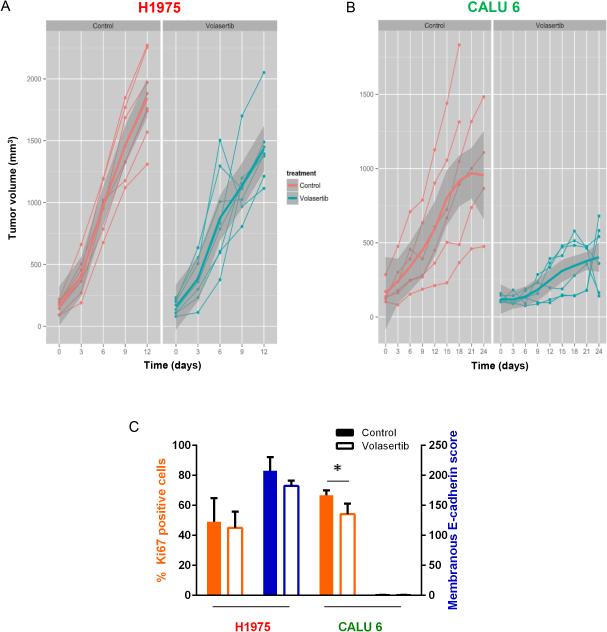
We examined the effect of PLK1 inhibition via treatment with volasertib on mouse xenograft models of NSCLC using mesenchymal (Calu6) and epithelial (H1975) cell lines. In the epithelial tumors, we observed a nonsignificant trend of decreased tumor growth with this treatment (P = 0.56). Nonparallel mean curves diverged progressively over time, suggesting that a significant difference with treatment may have occurred if volasertib could have been administered longer (Fig. 6A). In contrast, treatment with volasertib resulted in decreased mesenchymal NSCLC tumor size in vivo (P = 0.008) (Fig. 6B).
Figure 6.
PLK1 inhibition leads to decreased mesenchymal NSCLC tumor growth in vivo. Mice bearing epithelial (H1975; A) or mesenchymal (Calu6; B) NSCLC tumors were given 30 mg/kg volasertib intravenously each week or a vehicle control. Tumors were measured every 3 days. Individual tumors are graphed as thin lines with markers. The mean tumor size is indicated by the thick solid line, with the standard deviation indicated in dark gray. (C) Tumors were subjected to immunohistochemical staining with Aperio digital analysis. The average percent of tumor cells positive for Ki67 (orange bars) and the average score for E-cadherin (blue bars) were calculated with error bars representing standard deviation. Open bars, volasertib. Solid bars, control. *P ≤ 0.05 compared with control (no volasertib).
NSCLC cell lines with KRAS mutations are more sensitive to PLK1 inhibition than are cell lines wild-type for KRAS
We examined the correlation between mutations of 264 genes that are commonly mutated in cancer (Supplementary Table S1) and the drug sensitivity of all NSCLC cell lines treated with BI2536 or volasertib and for 63 cell lines treated with GSK461364. More NSCLC cell lines with KRAS mutations than cell lines wild-type for KRAS were sensitive to PLK1 inhibition (Supplementary Table S7). No gene mutations other than that of KRAS correlated with sensitivity to more than 1 drug. We also examined the co-occurrence of other gene mutations in the KRAS-mutant population because such co-mutations may define subgroups of NSCLC with distinct clinical outcomes and drug sensitivities. We did not find any statistical correlations between PLK1 inhibitor sensitivity and the presence of LKB1 or TP53 mutations in KRAS-mutant tumors, possibly because the absolute numbers of cell lines in these groups were small (Supplementary Table S8).
To further characterize NSCLC cell lines with KRAS mutations we applied an established KRAS dependency signature to KRAS-mutant NSCLC cell lines (27). KRAS-dependent NSCLC cell lines, defined as those that undergo apoptosis when KRAS expression in them is knocked down, exhibited a classical epithelial morphology, whereas KRAS-independent cells had a mesenchymal phenotype. In an unsupervised analysis, KRAS-independent cell lines were sensitive to treatment with GSK461364 (P = 0.0052), but the other 2 drugs and the universal lines were not (Supplementary Fig. S5A).
Chromosomal instability markers, TP53 mutation, PLK1 expression, histology, and basal ABC transporter expression do not correlate with response to PLK1 inhibition in NSCLC cell lines
Given the relationship between PLK1 function and chromosomal stability (36), we evaluated the association between markers of chromosomal instability and PLK1 inhibitor sensitivity using a signature of 70 genes (59 available in our data set) (28). In an unsupervised clustering analysis, we did not observe enrichment of PLK1 inhibition-sensitive or -resistant NSCLC cell lines in any clusters (Supplementary Fig. S5B). To validate candidate biomarkers of response according to the available literature (6, 7) (8), we examined the correlation of TP53 mutational status (Fisher exact test), PLK1 gene expression (2-sample t-test), NSCLC histology (Fisher exact test), and expression of the ABC transporters ABCB1 and ABCG2 (2-sample t-test) with sensitivity to treatment with the 3 PLK1 inhibitors. We did not find any statistically significant correlations (P > 0.05).
Discussion
Our study is the first large-scale integrated analysis of gene expression, protein expression, gene mutations, and PLK1 inhibitor sensitivity that we conducted to identify potential mechanisms of PLK1 inhibitor-induced apoptosis of NSCLC cells. We demonstrated that NSCLC cell lines have differing sensitivity to PLK1 inhibition, with a minority of them exhibiting sensitivity to multiple PLK1 inhibitors. This finding is consistent with results of clinical trials of PLK1 inhibitors in treatment of solid tumors that demonstrated low response rates (4-14%) with stable disease rates of 26-42% in unselected patients (3, 4, 37, 38). Although both PLK1 inhibitor-sensitive and -resistant cell lines underwent G2/M arrest following PLK1 inhibition, only the sensitive cell lines underwent substantial apoptosis. E-cadherin expression was higher in the resistant cell lines than in the sensitive ones. We discovered that NSCLC cell lines with high EMT scores (mesenchymal lines) were more sensitive to all 3 PLK1 inhibitors than were cell lines with low scores. In addition, forced induction of an epithelial phenotype led to drug resistance in the cells, and forced induction of a mesenchymal phenotype increased their drug sensitivity.
EMT is a dynamic and reversible process in which epithelial cells undergo morphological transition and acquire fibroblast-like characteristics through loss of adhesion and gain of a migratory, invasive phenotype. The tight-junction and Rho signaling pathways are related to EMT, and these pathways were enriched in the sensitive NSCLC cells in our study as evinced by differential gene expression. Tight junctions are crucial components of signaling pathways that regulate epithelial proliferation and differentiation, and the Rho family GTPases are essential for epithelial cell polarity, tight-junction assembly, and actin cytoskeleton regulation (39). In agreement with our results, previous studies demonstrated that in breast cancer and glioblastoma cell lines, cancer stem cells, which are usually mesenchymal, are more sensitive to PLK1 inhibition than non-stem cells are (11, 40). To the best of our knowledge, no published studies have demonstrated a role for PLK family members in EMT. Potential mechanistic links with EMT are forkhead transcription factors, which, upon regulation by PLK1 (41), can affect mitotic progression (42) and EMT (43).
A limitation of our study was the poor correlation of the effects of treatment with BI2536 and volasertib with those of treatment with GSK461364. The difference in the drugs’ effects may be attributed to differences in the drugs’ selectivity, as GSK461364 is more selective for PLK1 than are the other 2 drugs. Authors reported similar inconsistencies in 2 large-scale pharmacogenomic studies demonstrating good correlation of genomic data between the studies but highly discordant responses to the drugs (44). Additionally, we noted that in some NSCLC cell lines, higher drug concentrations had less of an effect on cell viability than did intermediate concentrations (Fig. 2B, cell line H358). Recently, Raab et al. (45) demonstrated a similar pattern in HeLa cells, with less apoptosis at higher drug concentrations, and discovered that treatment with BI2536 inhibits death-associated protein kinase activity. In intact cells, this kinase was inhibited at BI2536 concentrations greater than 500 nM. Thus, in that study, the drugs may have inhibited tumor suppressors such as death-associated protein kinase at high concentrations.
We tested several potential biomarkers that were previously demonstrated to correlate with cancer cell line sensitivity to PLK1 inhibition, including PLK1 expression, TP53 mutation (6, 8), chromosomal instability (8), and ABCB1 or ABCG2 overexpression (46). Our findings were consistent with low rates of response of NSCLC to treatment with PLK1 inhibitors in clinical trials despite a high prevalence of PLK1 overexpression in tumors (3, 4, 21). Our findings did not preclude a role for ABCB1 or ABCG2 in drug resistance, as expression of these proteins can be induced following drug exposure. We also cannot exclude a role for chromosomal instability, as we tested only 59 of the 70 genes in the study signature, and that signature has yet to be validated in our NSCLC cell lines.
Our findings have important clinical implications and warrant further investigation in the clinical setting. For example, about 20% of NSCLC tumors have a mesenchymal phenotype (24, 26), and patients with such tumors may benefit from treatment with PLK1 inhibitors, one of which, volasertib, has received breakthrough designation for treatment of leukemia and has a favorable safety profile. Unfortunately, no tumor specimens from prior clinical trials are available for testing of our hypothesis, and published clinical data demonstrating that more aggressive solid tumors are more or less responsive to PLK1 inhibition than less aggressive ones are lacking. In addition to single-agent activity of PLK1 inhibitors, researchers have demonstrated synergism between PLK1 inhibitors and 2 chemotherapeutic agents commonly used for lung cancer: paclitaxel and gemcitabine (47). Additionally, EMT is one of the mechanisms that lead to loss of oncogene addiction (48). Specifically, NSCLC cell lines with mesenchymal gene signatures were markedly more resistant to epidermal growth factor receptor and phosphoinositide 3-kinase inhibitors than were those with an epithelial gene signature (17). A body of evidence also suggests the existence of phenotypic and molecular associations between chemoresistance and EMT, highlighting the importance of targeting cancer cells with mesenchymal phenotypes (49, 50). Another potential therapeutic application of PLK1 inhibition is in the adjuvant setting, when the population of cancer stem cells with mesenchymal phenotypes may be increased.
In conclusion, we identified a subset of NSCLC cell lines that are exquisitely sensitive to PLK1 inhibitors and strong correlations among mesenchymal gene expression patterns, tight-junction signaling, and sensitivity of NSCLC cell lines to treatment with PLK1 inhibitors. Our findings support the use of the EMT score or E-cadherin expression to prospectively select NSCLC patients most likely to benefit from use of PLK1 inhibitors. This is the first published study directly linking PLK1 inhibition with EMT. We will aim our future studies at dissecting the mechanistic pathways that mediate PLK1 inhibitor-induced apoptosis. We also intend to test PLK1 inhibitors using NSCLC cell lines with acquired resistance to epidermal growth factor receptor inhibitor-based treatment and cytotoxic chemotherapy.
Supplementary Material
Translational Relevance.
Our finding that mesenchymal non-small cell lung cancer (NSCLC) is sensitive to polo-like kinase 1 (PLK1) inhibition reported herein has several potential clinical applications. Epithelial-mesenchymal transition occurs in a considerable subset of NSCLC cases (≥20%) and can mediate loss of oncogene addiction, epidermal growth factor receptor inhibitor resistance, and chemotherapy resistance. Thus, PLK1 inhibitors may be particularly active in NSCLC patients in whom current therapies are ineffective. Additionally, PLK1 inhibitors may enhance the efficacy of chemotherapeutic agents commonly used for NSCLC. Given the availability of the PLK1 inhibitor volasertib, which has breakthrough status for leukemia and an favorable safety profile, as well as additional PLK1 inhibitors in clinical trials, these results have the potential to be translated into the clinic for the treatment of NSCLC and provide a strong rationale for using markers of epithelial-mesenchymal transition as candidate predictive markers for selecting NSCLC patients for treatment.
Acknowledgments
We thank Arthur Gelmis and Donald Norwood of the Department of Scientific Publications at MD Anderson for scientific editing of the manuscript.
Grant Support
This work was supported by The Lung Cancer Research Foundation (F.M. Johnson) and The University of Texas MD Anderson Cancer Center Lung Cancer Moon Shot (J.V. Heymach). Reverse-phase protein arrays and flow cytometry were supported by the National Institutes of Health/National Cancer Institute under award number P30CA016672 (to MD Anderson). The gene expression and mutational profiles of the NSCLC lines were previously generated as part of the UT SPORE in Lung Cancer (5-P50-CA090907-15). D.H. Peng was supported by Cancer Prevention and Research Institute of Texas Training Grant RP140106. D.L. Gibbons was supported by National Cancer Institute grant K08-CA151651, the MD Anderson Physician Scientist Award, and Rexanna's Foundation for Fighting Lung Cancer and is an R. Lee Clark Fellow of MD Anderson supported by the Jeanne F. Shelby Scholarship Fund. Bioinformatic analysis also was supported under award number P30CA016672.
Footnotes
Disclosure of Potential Conflicts of Interest
Research funding from GlaxoSmithKline (J.V. Heymach), Astra Zeneca (J.V. Heymach), and PIQUR Pharmaceuticals (F.M. Johnson). Membership on scientific advisory boards for Astra Zeneca (J.V. Heymach), GlaxoSmithKline (J.V. Heymach), Genentech (J.V. Heymach), Novartis (F.M. Johnson), and Bristol-Myers Squibb (F.M. Johnson). All others declare no conflict of interest.
References
- 1.Takaki T, Trenz K, Costanzo V, Petronczki M. Polo-like kinase 1 reaches beyond mitosis--cytokinesis, DNA damage response, and development. Curr Opin Cell Biol. 2008;20:650–60. doi: 10.1016/j.ceb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Wolf G, Elez R, Doermer A, Holtrich U, Ackermann H, Stutte HJ, et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene. 1997;14:543–9. doi: 10.1038/sj.onc.1200862. [DOI] [PubMed] [Google Scholar]
- 3.Sebastian M, Reck M, Waller CF, Kortsik C, Frickhofen N, Schuler M, et al. The efficacy and safety of BI 2536, a novel Plk-1 inhibitor, in patients with stage IIIB/IV non-small cell lung cancer who had relapsed after, or failed, chemotherapy: results from an open-label, randomized phase II clinical trial. J Thorac Oncol. 2010;5:1060–7. doi: 10.1097/JTO.0b013e3181d95dd4. [DOI] [PubMed] [Google Scholar]
- 4.Schoffski P, Awada A, Dumez H, Gil T, Bartholomeus S, Wolter P, et al. A phase I, dose-escalation study of the novel Polo-like kinase inhibitor volasertib (BI 6727) in patients with advanced solid tumours. Eur J Cancer. 2012;48:179–86. doi: 10.1016/j.ejca.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, et al. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14:R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degenhardt Y, Greshock J, Laquerre S, Gilmartin AG, Jing J, Richter M, et al. Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol Cancer Ther. 2010;9:2079–89. doi: 10.1158/1535-7163.MCT-10-0095. [DOI] [PubMed] [Google Scholar]
- 9.Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Jr., Kinzler KW, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106:3964–9. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danovi D, Folarin A, Gogolok S, Ender C, Elbatsh AM, Engstrom PG, et al. A high- content small molecule screen identifies sensitivity of glioblastoma stem cells to inhibition of polo-like kinase 1. PLoS ONE. 2013;8:e77053. doi: 10.1371/journal.pone.0077053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu K, Law JH, Fotovati A, Dunn SE. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 2012;14:R22. doi: 10.1186/bcr3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–22. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 13.Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J, et al. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res. 2009;15:3094–102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 14.Gilmartin AG, Bleam MR, Richter MC, Erskine SG, Kruger RG, Madden L, et al. Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 2009;69:6969–77. doi: 10.1158/0008-5472.CAN-09-0945. [DOI] [PubMed] [Google Scholar]
- 15.Johnson FM, Saigal B, Talpaz M, Donato NJ. Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses invasion and induces cell cycle arrest and apoptosis of head and neck squamous cell carcinoma and non-small cell lung cancer cells. Clin Cancer Res. 2005;11:6924–32. doi: 10.1158/1078-0432.CCR-05-0757. [DOI] [PubMed] [Google Scholar]
- 16.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An Epithelial- Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clin Cancer Res. 2013;19:279–90. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mross K, Frost A, Steinbild S, Hedbom S, Rentschler J, Kaiser R, et al. Phase I dose escalation and pharmacokinetic study of BI 2536, a novel Polo-like kinase 1 inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2008;26:5511–7. doi: 10.1200/JCO.2008.16.1547. [DOI] [PubMed] [Google Scholar]
- 20.Sen B, Saigal B, Parikh N, Gallick G, Johnson FM. Sustained Src inhibition results in signal transducer and activator of transcription 3 (STAT3) activation and cancer cell survival via altered Janus-activated kinase-STAT3 binding. Cancer Res. 2009;69:1958–65. doi: 10.1158/0008-5472.CAN-08-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olmos D, Barker D, Sharma R, Brunetto AT, Yap TA, Taegtmeyer AB, et al. Phase I study of GSK461364, a specific and competitive Polo-like kinase 1 inhibitor, in patients with advanced solid malignancies. Clin Cancer Res. 2011;17:3420–30. doi: 10.1158/1078-0432.CCR-10-2946. [DOI] [PubMed] [Google Scholar]
- 22.Brannan JM, Sen B, Saigal B, Prudkin L, Behrens C, Solis L, et al. EphA2 in the early pathogenesis and progression of non-small cell lung cancer. Cancer Prev Res (Phila Pa) 2009;2:1039–49. doi: 10.1158/1940-6207.CAPR-09-0212. [DOI] [PubMed] [Google Scholar]
- 23.Tong P, Coombes KR, Johnson FM, Byers LA, Diao L, Liu DD, et al. drexplorer: A tool to explore dose-response relationships and drug-drug interactions. Bioinformatics. 2015;31:1692–4. doi: 10.1093/bioinformatics/btv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–42. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nature communications. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 29.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2012. [Google Scholar]
- 30.Sen B, Peng S, Tang X, Erickson HS, Galindo H, Mazumdar T, et al. Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med. 2012;4:136ra70. doi: 10.1126/scitranslmed.3003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byers LA, Sen B, Saigal B, Diao L, Wang J, Nanjundan M, et al. Reciprocal regulation of c-Src and STAT3 in non-small cell lung cancer. Clin Cancer Res. 2009;15:6852–61. doi: 10.1158/1078-0432.CCR-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng S, Creighton CJ, Zhang Y, Sen B, Mazumdar T, Myers JN, et al. Tumor grafts derived from patients with head and neck squamous carcinoma authentically maintain the molecular and histologic characteristics of human cancers. J Transl Med. 2013;11:198. doi: 10.1186/1479-5876-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, et al. Epithelial-to mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Mod Pathol. 2009;22:668–78. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Ahn YH, Chen Y, Tan X, Guo L, Gibbons DL, et al. ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J Clin Invest. 2014;124:2696–708. doi: 10.1172/JCI72171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li JJ, Li SA. Mitotic kinases: the key to duplication, segregation, and cytokinesis errors, chromosomal instability, and oncogenesis. Pharmacol Ther. 2006;111:974–84. doi: 10.1016/j.pharmthera.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Stadler WM, Vaughn DJ, Sonpavde G, Vogelzang NJ, Tagawa ST, Petrylak DP, et al. An open-label, single-arm, phase 2 trial of the Polo-like kinase inhibitor volasertib (BI 6727) in patients with locally advanced or metastatic urothelial cancer. Cancer. 2014;120:976–82. doi: 10.1002/cncr.28519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoffski P, Blay JY, De Greve J, Brain E, Machiels JP, Soria JC, et al. Multicentric parallel phase II trial of the polo-like kinase 1 inhibitor BI 2536 in patients with advanced head and neck cancer, breast cancer, ovarian cancer, soft tissue sarcoma and melanoma. The first protocol of the European Organization for Research and Treatment of Cancer (EORTC) Network Of Core Institutes (NOCI). Eur J Cancer. 2010;46:2206–15. doi: 10.1016/j.ejca.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 39.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–36. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 40.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan C, Wang L, Zhou L, Fu Z. The function of FOXO1 in the late phases of the cell cycle is suppressed by PLK1-mediated phosphorylation. Cell Cycle. 2014;13:807–19. doi: 10.4161/cc.27727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–82. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Wang Y, Luo J, Fu Z, Ying J, Yu Y, et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 2012;586:3761–5. doi: 10.1016/j.febslet.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 44.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–93. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raab M, Pachl F, Kramer A, Kurunci-Csacsko E, Dotsch C, Knecht R, et al. Quantitative chemical proteomics reveals a Plk1 inhibitor-compromised cell death pathway in human cells. Cell Res. 2014;24:1141–5. doi: 10.1038/cr.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu CP, Sim HM, Huang YH, Liu YC, Hsiao SH, Cheng HW, et al. Overexpression of ATP-binding cassette transporter ABCG2 as a potential mechanism of acquired resistance to vemurafenib in BRAF(V600E) mutant cancer cells. Biochem Pharmacol. 2013;85:325–34. doi: 10.1016/j.bcp.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jimeno A, Rubio-Viqueira B, Rajeshkumar NV, Chan A, Solomon A, Hidalgo M. A fine- needle aspirate-based vulnerability assay identifies polo-like kinase 1 as a mediator of gemcitabine resistance in pancreatic cancer. Mol Cancer Ther. 2010;9:311–8. doi: 10.1158/1535-7163.MCT-09-0693. [DOI] [PubMed] [Google Scholar]
- 48.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 49.Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–8. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.