Abstract
T cells and B cells are crucial in the initiation and maintenance of multiple sclerosis (MS), and the activation of these cells is believed to be mediated through specific recognition of antigens by the T‐ and B‐cell receptors. The antigen receptors are highly polymorphic due to recombination (T‐ and B‐cell receptors) and mutation (B‐cell receptors) of the encoding genes, which can therefore be used as fingerprints to track individual T‐ and B‐cell clones. Such studies can shed light on mechanisms driving the immune responses and provide new insights into the pathogenesis. Here, we summarize studies that have explored the T‐ and B‐cell receptor repertoires using earlier methodological approaches, and we focus on how high‐throughput sequencing has provided new knowledge by surveying the immune repertoires in MS in even greater detail and with unprecedented depth.
Introduction
Multiple sclerosis (MS) is believed to be mediated by an immunological attack on the central nervous system (CNS), orchestrated by T cells and B cells of the adaptive immune system. Already half a century ago, a local synthesis of immunoglobulin G (IgG) was identified in the cerebrospinal fluid (CSF) of MS patients.1, 2 It was later shown that this IgG is produced by B cells in the CSF and CNS.3, 4 Other studies have demonstrated clonal expansions of T and B cells,5, 6 and deposition of immunoglobulins,7 in active demyelinating lesions. Tertiary lymphoid structures, which could be sites of B‐cell differentiation and affinity maturation, are present in the meninges of some patients with long‐standing disease, and have been linked to cortical pathology.8, 9 In further support of the idea that T and B cells mediate CNS damage in MS, specifically killing them or hindering their recruitment to the CNS efficiently suppresses disease activity.10 Finally, genetic studies indicate that adaptive immunity may play a role also in the initiation of the disease.11, 12
T and B cells recognize specific antigens through their antigen receptors.13 The T‐cell receptor (TCR) binds peptides presented on human leukocyte antigen (HLA) molecules, whereas the B‐cell immunoglobulin (herein referred to as B‐cell receptor, BCR) binds linear or conformational epitopes on native antigens (Fig. 1A). If the lymphocyte receives appropriate co‐stimulatory signals, antigen recognition leads to activation and proliferation known as clonal expansion. Although immunization with myelin antigens induces an MS‐like disease in rodents, the target antigens of the T‐ and B‐cell responses in MS have not been identified. It is a particular paradox that the specificity of oligoclonal IgG within the CSF of patients with MS remains unknown, whereas it was proven more than four decades ago that oligoclonal IgG in CNS infection target the causative agent.14 Since then, the target antigens of oligoclonal CSF IgG have also been identified in patients with noninfectious immune‐mediated diseases, such as Yo antigens in paraneoplastic cerebellar degeneration.15 This could either suggest that we need more refined methods to identify MS antigens still hiding, or that there are no particular target antigens in MS. Importantly, the first alternative implies that MS could be treated by specific immune intervention strategies. In this review, we summarize studies that have surveyed the immune repertoires in MS using earlier techniques. We discuss how the introduction of high‐throughput sequencing has provided new knowledge, and anticipate how it may continue to unravel important aspects of the adaptive immune responses in MS.
Figure 1.
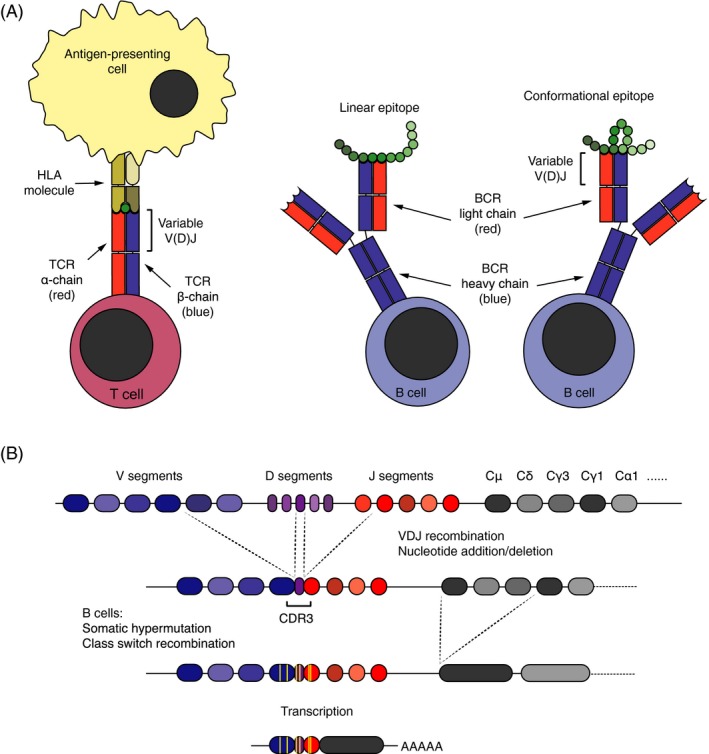
Structure, function, and diversification of antigen receptors. (A) The T‐cell receptor (TCR) binds to linear epitopes presented on HLA molecules by antigen‐presenting cells. The B‐cell receptor (BCR), in contrast, recognizes linear or conformational epitopes on native antigens. Both antigen receptors are composed of distinct pair of chains: The TCR of an α‐ and a β‐chain, and the BCR of two heavy and two light chains. The variable part of the receptor chains, encoded by V, J, and D (TCR β and BCR heavy) gene segments, constitutes their antigen‐binding surface. (B) During T‐ and B‐cell development, the V, J, and D (TCR β and BCR heavy) gene segments are stochastically recombined, and nucleotides may also be randomly added and deleted between them. The diversity of the receptor genes concentrates in the third complementarity determining region (CDR3), which encodes the center of the antigen‐binding surface. In the course of antigen‐driven immune responses, B cells may go through an additional round of diversification in germinal centers, where they undergo somatic hypermutation and clonal selection. During this process, they also switch the isotype of the constant chain. Class switch recombination leading to isotype switching from IgM (encoded by Cμ gene segments) to immunoglobulin G (IgG)1 (encoded by Cγ1 gene segments) is depicted.
Immune Receptors and Repertoires
The TCR and the BCR share structural similarities (Fig. 1A). Both comprise distinct pair of chains, one α‐ and one β‐chain for the αβ TCR and two heavy‐ and two light chains for the BCR, with variable domains mediating binding to antigens.13 The genes encoding the variable domains undergo somatic diversification during lymphocyte development. In this process, the variable (V), joining (J), and diversity (D; for the TCR β and BCR heavy chain) gene segments are rearranged (Fig. 1B). In addition, nucleotides may be randomly added or removed between the segments. The greatest diversity is found within the complementarity determining regions (CDRs), in particular the CDR3, which contributes most to the specificity of the receptors. Pairing of the receptor chains leads to further divergence of the repertoire, and the potential diversity has been estimated to 1018 αβ TCRs and 5 × 1013 BCRs.13 Upon encountering an antigen, B cells undergo an additional round of diversification in lymphoid germinal centers. This involves the enzyme activity‐induced cytidine deaminase and is known as somatic hypermutation. The variable domains of TCRs and BCRs can thus be used as molecular fingerprints to track lymphocytes of similar specificities. The totality of different antigen receptors with distinct variable domains in a given individual is here denoted the immune repertoire.
Studies of immune repertoires in MS are important for several reasons: (1) The composition of the repertoires may indicate whether they are the result of antigen stimulation or other means of activation; (2) If the sequences encoding both antigen receptor chains are available, it is possible to express recombinant receptors to search for target antigens; and (3) Dominant clones could represent potential biomarkers or targets for selective immunotherapy.
Analyses of Immune Repertoires in MS
The TCR repertoire
A summary of the techniques most commonly used to investigate the immune repertoires in MS is given in Table 1. Using Southern blot, early studies explored the diversity of the TCR repertoires in the CSF.16, 17 The methods included cloning and in vitro expansion of individual T cells. Accordingly, only a limited number of CSF T‐cell clones were studied, and this limitation could possibly explain conflicting results.16, 17 Subsequent investigations of T‐cell clones from CSF and blood using gene‐specific primers mapping to the TCR Vβ genes indicated a common TCR Vβ gene usage in some MS patients.18 Combining PCR amplification of TCR Vβ genes and conventional Sanger sequencing, another study found an oligoclonal TCR repertoire in the CSF of patients with MS and also other inflammatory neurological conditions.19 Through PCR amplification of TCR Vβ genes and a subsequent immunoenzymatic approach, Gran and colleagues found a skewed TCR Vβ usage in the blood.20
Table 1.
Principles, outcomes, and challenges with techniques used to study immune repertoires in multiple sclerosis
| Technology | Principle | Outcome | Challenges |
|---|---|---|---|
| Southern blot | DNA is digested by restriction enzymes, separated on agarose gels, and blotted onto nitrocellulose membranes. DNA fragments are hybridized with gene probes. | Clonal composition and diversity |
|
| CDR3 spectratyping | T‐cell receptor cDNA is amplified by PCR across the CDR3 region using primers mapping to different families of variable genes. Since T‐cell clones differ in CDR3 length, the distribution of lengths of the resultant PCR products reflects the overall diversity. Deviations from a bell‐shaped distribution indicate clonal expansions. | Overview of the clonal composition and diversity |
|
| Flow cytometry based T‐cell receptor (TCR) Vβ repertoire analysis | A cell sample is stained with antibodies against different TCR‐Vβ‐families and analyzed on a flow cytometer. | Overview of the clonal composition |
|
| Sanger sequencing | The sequencing technique is based on selective incorporation of chain‐terminating dideoxynucleotides. The resulting DNA fragments are separated by electrophoresis. | Nucleic acid sequence, up to 700 bp |
|
| High‐throughput sequencing technologies | |||
| Roche 454 | “Sequencing by synthesis,” based on the release of pyrophosphate on nucleotide incorporation. | Nucleic acid sequence, 400 bp (recently upgraded to 1000 bp) |
|
| Illumina (HiSeq and MiSeq) | “Sequencing by synthesis,” based on cyclic reversible termination, which is an adaption of Sanger sequencing. “Paired end” sequencing makes it possible to sequence both ends of a fragment, and subsequently align the reads to cover longer sequences. | Nucleic acid sequence, 2 × 150 bp for HiSeq and 2 × 300 bp for MiSeq |
|
In CDR3 spectratyping, the lengths of the CDR3 sequences are investigated. This technique showed an overrepresentation of certain Vβ genes in the blood,21 clonal T cell expansions in the CSF,22 a diversification of the TCR repertoire after autologous stem cell transplantation,23 and more recently that the drugs natalizumab and fingolimod might influence the peripheral TCR repertoire.24, 25 It is also possible to explore the TCR Vβ repertoire by flow cytometry using fluorochrome‐labeled antibodies against different Vβ families.26 A study using this technique demonstrated clonal expansions mainly among CD8+ T cells in the CSF.27
Oksenberg and colleagues were the first to explore the TCR repertoires expressed in brain lesions.28 Through specific PCR amplification, they found a restricted TCR Vα usage. In contrast, another early study using PCR amplification and Southern blot reported a polyclonal TCR repertoire in MS brains.29 Later studies combining CDR3 spectratyping with Sanger sequencing of candidate T‐cell clones have indicated that the TCR repertoire in MS brains is diverse, but nevertheless dominated by expanded clones.30, 31 Many of these are CD8+ T cells, and at least some can also be detected in the CSF and blood at different time points.30 Interestingly, some expanded clones within MS lesions express TCR β sequences that are similar on the amino acid level, but encoded by different nucleotide combinations.6, 31 The existence of such shared TCR sequences is suggested to be due to “convergent recombination,” which makes some TCRs to be produced more frequently than others.32 Since similar TCRs are believed to share specificity, the presence of such sequences within an MS brain argues that they have been recruited by the same antigen.
The repertoire of myelin basic protein (MBP)‐specific T cells in blood has been studied by PCR amplification of TCR Vβ genes followed by Southern blotting or conventional Sanger sequencing in combination with antibodies against different Vβ families.33, 34, 35, 36, 37 While some of these studies demonstrated a restricted TCR repertoire,33, 34, 35 other research groups found a broader and more diverse composition.36, 37 Later studies focusing on the change in the MBP‐specific T‐cell repertoire over time showed some cases of preserved specificities,38 but also instances of diversification of the repertoire due to epitope spreading,39 which may have implications for antigen‐specific tolerization strategies.40
Previous studies using restriction fragment length polymorphisms (RFLP) as genetic markers have reported conflicting results regarding the influence of TCR gene polymorphisms on MS risk.41, 42, 43, 44 A more recent study used single‐nucleotide polymorphism markers to revisit this and identified three potential loci of interest in TCR alpha V and constant gene regions.45
The BCR repertoire
PCR amplification, cloning, and Sanger sequencing of immunoglobulin heavy‐chain V (IGHV) genes in CSF B cells from MS patients have demonstrated clonal expansion and somatic hypermutation.46, 47, 48 Using similar techniques, clonal expansions of B cells have also been identified in the brain and meninges.5, 49, 50 Moreover, antigen‐experienced B cells within the CSF and CNS tend to preferentially use VH4 germline segments, which is indicative of an antigen‐driven response,5, 46, 49, 50 and it has been suggested that certain patterns of somatic mutations within these VH4 segments could be used as a diagnostic tool.51
PCR amplification of IGHV transcripts from single sorted B cells allows faithful pairing of the heavy and light chains, and has enabled researchers to express expanded CSF and CNS B‐cell clones as recombinant antibodies to identify target antigens. This approach has proven successful for aquaporin‐4‐specific B cells from the CSF in neuromyelitis optica and measles‐specific B cells from the brain in subacute sclerosing panencephalitis.52, 53 Initial attempts to identify the specificity of recombinant antibodies from expanded CSF B cells in MS, in contrast, was not conclusive.54 However, some of these antibodies has recently been found to bind to astrocytes and neuronal antigens, and to cause complement‐mediated tissue destruction in spinal cord explants.55 Other studies have found evidence of myelin reactivity of recombinant Fab fragments and antibodies from the CSF.56, 57 A very recent study investigating recombinant antibodies from expanded B cells in MS brains did not succeed in identifying specificities unique to the disease.58 Taken together, the evidence so far show that BCRs from MS patients have a mutation pattern compatible with an antigen‐driven immune response, but does not unambiguously point to a particular target antigen.
Some earlier genetic association studies have explored BCR gene polymorphisms. The use of DNA probes mapping to different V gene segments and RFLP showed significant associations.59, 60 On the other hand, the use of microsatellite markers within the IGH cluster produced negative results.61, 62 One study using gene‐specific primers to explore an IGHV4‐39 germline deletion did not detect any association linking the polymorphism to disease susceptibility or progression.63
High‐Throughput Sequencing of Immune Repertoires
As described above, molecular methods such as Sanger sequencing and CDR3 spectratyping have provided important insights on the clonality of intrathecal T‐ and B‐cell repertoires, and have provided promising tools for studies of the specificity of CSF and CNS B cells. Cloning and conventional Sanger sequencing, however, only allows interrogation of a small fraction of the immune repertoires. CDR3 spectratyping, on the other hand, gives an “eagle eye” perspective, but no information about the sequence determining receptor specificity. These constraints have hampered an unbiased characterization of the complete immune repertoires within each immunological or anatomic compartment. High‐throughput sequencing overcomes these limitations and offers the best of both worlds. It enables sequencing of millions of short templates in parallel, capturing even infrequent clones. The diversity is accurately determined by estimating the contribution of single clones to the total repertoire.
Although the theoretical diversity of the αβ TCR repertoire in the human body is assumed to be 1018,13 the number of different TCRs present in a given individual was estimated in an earlier study only to 106 different β chains, each potentially pairing with one of 25 different α chains.64 High‐throughput sequencing has recently been used to revisit this, and revealed that the true number of different β chains present is higher. By combining high‐throughput sequencing with a computational approach, Robins and colleagues found that the TCR β diversity was at least fourfold greater than previous estimates.65 Qi and colleagues found an even higher estimate of 108 different TCR β chain genes in young adults.66 It has been claimed that these approximations could be biased by sample size, and that the true diversity of the TCR repertoire might still be greater.67
Although well suited to study immune repertoires, high‐throughput sequencing poses new experimental and computational challenges.68 For T cells, one has to decide whether to sequence TCR α or β chain genes. Up to 10% of T cells may express two functional α chains, whereas it has been estimated that less than 1% express two functional β chains.69 Consequently, the distribution of TCR β genes more accurately reflects the repertoire on a cellular level in a given sample, and high‐throughput sequencing of TCR β genes would often be preferable to TCR α genes to estimate the number of T cells expressing a given TCR. Another important question is whether one should use complementary DNA (cDNA) that are reverse transcribed from mRNA, or genomic DNA (gDNA) for the investigation of immune repertoires. Due to multiple copies of the same RNA transcript per cell, the use of cDNA is much more sensitive than gDNA. This is advantageous if cell numbers are low, as for CSF B cells.70, 71 The use of cDNA also excludes introns and simplifies the sequencing strategy, and it reduces the number of unproductive sequences.72 However, RNA copy number depends on the activation status, and may consequently not be proportional to the number of cells. Activation of T cells, for instance, modifies the expression of TCR genes.73 Moreover, antibody‐secreting effector cells, such as plasmablasts in the CSF, show increased levels of BCR transcripts.74 The amount of gDNA, on the other hand, has been shown to correlate well with cell numbers if PCR bias is eliminated.75
There are different options for sequencing technologies, and a few have been used in MS (Table 1). The Roche 454 technology has been utilized to study the BCR repertoires in CSF and blood.70, 76, 77, 78 This technology provides longer sequence reads than other platforms, but has a high rate of artifactual insertions and deletions, which might be impossible to distinguish from true insertions and deletions introduced during receptor gene recombination. We and others have applied Illumina sequencing technology to study the TCR79, 80, 81, 82, 83 and the BCR71, 84 repertoires. Artifacts are rarer, but the sequence lengths on the HiSeq platform are shorter, and consequently not covering all possible somatic mutations within IGHV genes.
Strategies to process and analyze high‐throughput sequencing data of antigen receptors have recently been reviewed.68, 85 The raw sequence reads must first be filtered and clustered in order to correct for sequencing and PCR errors,86 and some software solutions that perform these algorithms have been developed.87, 88 Next, the sequences are classified according to their V(D)J germline segment, which can be achieved using web‐based tools such as IMGT/HighV‐QUEST or IgBLAST,89, 90 or available software.91, 92 Several software packages and web‐based analysis tools, such as Adaptive Biotechnologies ImmunSEQ platform, is accessible for further analysis of sequencing data.93, 94, 95
The need for publicly available databases dedicated to rearranged TCR and BCR sequences are becoming increasingly clear.96 This would simplify the search for previously published identical or highly similar CDR3 sequences that may represent public TCRs or BCRs. Moreover, there is also a need for standardization of sequencing protocols and data handling, which would enable analyses of meta data.97 For the time being, the National Center for Biotechnology Information (NCBI) BLAST search engine is a valuable tool to identify sequences in the nonredundant protein database obtained with previously available sequencing technology.98 For high‐throughput sequencing data deposited in the NCBI Sequence Read Archive, it is possible to perform a BLAST search within files of published projects.99
High‐Throughput Sequencing of TCRs in Multiple Sclerosis
Autologous stem cell transplantation and the monoclonal antibody alemtuzumab are assumed to “reset” the immune system. To test this hypothesis in MS patients treated with stem cell transplantation, Muraro, Robins and colleagues followed the renewal of the T‐cell repertoire applying high‐throughput TCR β sequencing.80 This allowed a comprehensive assessment of the regenerated CD4+ and CD8+ T‐cell repertoires, including the impact of individual surviving clones on the new repertoire. Interestingly, whereas the CD4+ T‐cell repertoire after transplantation were largely composed of newly generated clones, the CD8+ T‐cell repertoire were reestablished from clones already present before treatment.80 The latter mechanism, known as homeostatic proliferation, was shown by assessment of thymic output by TCR excision circles also to be prominent after treatment with alemtuzumab.79 Less diverse TCR repertoires were associated with a poor response to stem cell transplantation and with the development of secondary autoimmunity after treatment with alemtuzumab.79, 80 These results show that high‐throughput sequencing may enable monitoring of disease‐relevant T‐cell clones following therapies with fundamental effects on the immune system.
We have recently used high‐throughput sequencing to explore the TCR β repertoires of MS patients and controls with other types of neuroinflammation.81 Compared to previously available technology, high‐throughput sequencing allowed a better estimate of the diversity in the CSF and blood, and a precise quantitation of the overlap between the two compartments. We found highly diverse repertoires both in CSF and blood, and a significant overlap between the compartments, indicating that most CSF T cells have entered from blood. The most frequent clones within the CSF were, however, infrequent in blood, indicating that they had expanded locally.81 Such dominant clones only made up a small proportion of the total TCR β repertoire in CSF, but were remarkably stable for at least one year. Applying high‐throughput TCR sequencing in peripheral blood, another recent study demonstrated that children with MS were characterized by a long VJ junction region, indicating a self‐reactive TCR repertoire, and a skewed TCR Vβ family usage.95
MS is associated with Epstein–Barr virus (EBV).100 In order to track EBV‐reactive T cells and determine if they accumulate in the CSF, we created reference TCR β libraries by sorting and sequencing EBV‐reactive CD4+ and CD8+ T cells in blood.81 Because the TCR β sequence is unique for each memory clone, the sequences of EBV‐reactive T cells from blood could be used to track T cells from the same clone in CSF. By these means, we found an enrichment of EBV‐reactive CD8+ T cells in the CSF of MS patients, but not the controls.81 Among the EBV‐reactive CD8+ T cells in the CSF, we identified several public TCR β sequences. These sequences are known to be shared across individuals, and to be carried by TCRs that recognize common viral antigens in the context of particular HLA molecules. To our surprise, one of the public EBV‐specific sequences, and another almost identical sequence, had previously been identified in MS lesions.31 This confirms that EBV‐specific T cells gain entry to the crime scene in MS, and that high‐throughput sequencing can catch them in flagrante.
Combining high‐throughput sequencing and CDR3 spectratyping, a recent study compared the TCR β repertoires in MS lesions, CSF, and blood from the same individuals.82 The repertoire in the CNS appeared to be closer to the repertoire in the CSF than to that in blood, and closer to the CD8+ than to the CD4+ peripheral T‐cell compartment. Characterization of clonally expanded CD8+ T cells in blood by flow cytometry showed that these cells were biased toward a memory phenotype with increased expression of CCR5, CD11a, and Granzyme B.82 Another study used high‐throughput sequencing to explore the TCR repertoire present in so‐called pattern II MS lesions, which are believed to be associated with antibody‐mediated pathology.83 The authors identified expanded T‐cell clones and were subsequently able to isolate them from the CSF of the same patient and show that they secreted Th2 cytokines and were able to provide B cell help.83 They did not observe identical TCR sequences encoded by different nucleotide combinations, a phenomenon observed by others and taken in support of an antigen‐driven T cell response.6, 31 However, several sequences had identical CDR3 sequence but slightly different TCRβ V gene segments.83 The CDR1 and CDR2 sequences in these TCRs were highly similar, which might indicate that the TCRs recognize the same antigen/HLA combination.
These examples demonstrate the ability of high‐throughput sequencing to track disease‐relevant T cells in different compartments, and the possibility of combining the technique with functional characterization of T‐cell clones.
High‐Throughput Sequencing of BCRs in Multiple Sclerosis
While previously available technology enabled expression of recombinant antibodies from a limited number of CSF B cells, high‐throughput sequencing made it possible to sequence previously unattainable numbers of IGHV genes from CSF and blood. This paved the way for identification of related B‐cell clones in CSF and blood, first shown by von Büdingen and colleagues and later by us.70, 71 Figure 2 shows data from two MS patients, based on high‐throughput sequencing of IGHV genes as described elsewhere.71 By comparing the BCR transcriptome with the IgG proteome characterized by mass spectroscopy, we were able to ensure that the sequence data are correct and reflect B cells producing IgG within the CSF. Somatic hypermutation of the IGHV genes makes it possible to track the maturation of B‐cell clones in CSF and blood. This can be visualized as “trees” where distinct B‐cell clones that have acquired somatic mutations are depicted as different “branches” emerging from a hypothetical germline clone. In the present data, we detected related B‐cell clones in CSF and blood of the patients investigated (Fig. 2). Previous publications have shown that some related clones present in both compartments match CSF IgG.71, 76 One of the B‐cell lineages presented here has members matching both CSF and serum IgG (Fig. 2, MS‐1, rightmost tree).
Figure 2.
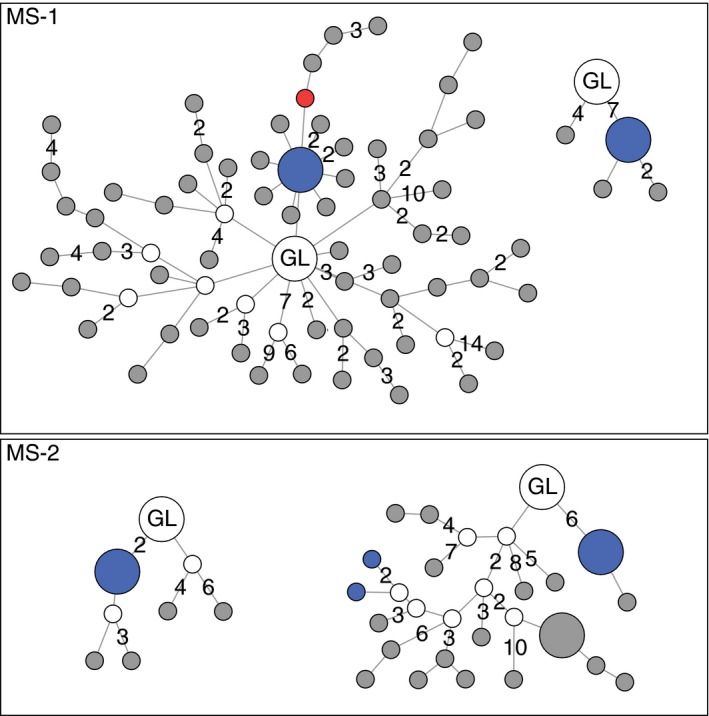
Maturation of four immunoglobulin G (IgG)‐producing B‐cell clones from two multiple sclerosis (MS) patients. High‐throughput sequencing of immunoglobulin heavy‐chain variable (IGHV) transcripts from cerebrospinal fluid (CSF) and blood was performed, and IgG from CSF and serum was analyzed by mass spectrometry. Each node represents a single IGHV sequence. The hypothetical germline (GL) sequence is set as origo, and the connecting lines depict somatic mutations. Lines without numbers denotes a single‐nucleotide exchange, “2” denotes two mutations, and so on. Gray nodes represent sequences only detected in CSF, the red node is a sequence only detected in blood, and blue nodes are identical sequences detected in both CSF and blood, whereas white nodes represent hypothetical intermediates. Larger nodes represent the most abundant transcripts. The CDR3 of all lineage trees matched CSF IgG. The rightmost tree of MS‐1 also matched IgG from serum.
Stern and colleagues applied high‐throughput sequencing on B cells in paired samples from cervical lymph nodes and brains of MS patients.84 They found considerable overlap between the B‐cell repertoire in the CNS and the lymph nodes. On the other hand, some clones were only present in a single compartment, and the repertoire tended to be more focused in the CNS compared to the periphery. Most importantly, they found that a greater proportion of clonal founders within cervical lymph nodes, suggesting that initiation of the B‐cell maturation occur in secondary lymphoid tissue outside the CNS.84
The findings of clonally related T cells and B cells on both sides of the blood–brain barrier are particularly interesting in the light of the discovery of the drainage of the interstitial fluid of the brain through the so‐called “glymphatic” pathway,101 and more recently of a lymphatic drainage system from the brain to deep cervical lymph nodes.102, 103 These findings corroborate early studies suggesting that the afferent arm of the immune system is operating in the brain.104 The existence of such lymphatic vessels also offers a path for the egression of immune cells from the CNS, and could thus contribute to explain the finding of identical and clonally related T cells and B cells, as well as IgG, in blood and CSF.
Prospects and Promise
High‐throughput sequencing of immune repertoires in MS has so far been limited to genes encoding one of the two antigen receptor chains. Paired sequencing of both chains will significantly increase the resolution of the analysis. For BCR sequencing, paired data will make it possible to explore somatic mutations also in the V region of the light chain, capturing a more accurate picture of the affinity maturation. Importantly, paired‐chain sequencing will also enable recombinant expression of TCRs and BCRs for studies of receptor specificities.
For high‐frequency B‐cell clones, it is possible to pair heavy and light chains based on their relative frequencies,105 and a combinatorial approach has recently been developed to match the α‐ and β‐chains in high‐throughput TCR sequencing.106 Recent technological advances have also made it possible to increase the throughput of single‐cell analyses.107 By barcoding transcripts from the same cell or by linking sequences during cDNA synthesis, it is possible to perform high‐throughput sequencing on paired templates in heterogeneous samples.108, 109 Moreover, in order to link TCR and BCR sequences to functional T‐ and B‐cell phenotypes, it is possible to simultaneously assess the receptor sequences and the transcript expression from single cells.110
It has been shown that immunoglobulin V region polymorphisms may have a profound impact on immune responses,111 and high‐throughput sequencing has recently revealed that there is a plethora of undiscovered immunoglobulin V gene segment alleles.112 By detailed mapping of germline immunoglobulin segments in large cohorts of MS patients and controls, high‐throughput sequencing technology may reveal novel disease‐associated V gene polymorphisms.
As our knowledge of the MS pathogenesis expands, new applications of high‐throughput sequencing of immune repertoires will likely emerge. Novel strategies for antigen‐specific tolerization have recently been suggested.40 If selective immunotherapy were to become a reality, high‐throughput sequencing would be well suited to monitor pathogenic clones posttreatment. This would parallel the tracking of minimal residual disease, which shows great promise in the follow‐up of lymphoid malignancies.113
Conflict of Interest
None declared.
References
- 1. Kabat EA, Moore DH, Landow H. An electrophoretic study of the protein components in cerebrospinal fluid and their relationship to the serum proteins. J Clin Invest 1942;21:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lowenthal A, Vansande M, Karcher D. The differential diagnosis of neurological diseases by fractionating electrophoretically the CSF gamma‐globulins. J Neurochem 1960;6:51–56. [DOI] [PubMed] [Google Scholar]
- 3. Obermeier B, Mentele R, Malotka J, et al. Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat Med 2008;14:688–693. [DOI] [PubMed] [Google Scholar]
- 4. Obermeier B, Lovato L, Mentele R, et al. Related B cell clones that populate the CSF and CNS of patients with multiple sclerosis produce CSF immunoglobulin. J Neuroimmunol 2011;233:245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baranzini SE, Jeong MC, Butunoi C, et al. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J Immunol 1999;163:5133–5144. [PubMed] [Google Scholar]
- 6. Babbe H, Roers A, Waisman A, et al. Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J Exp Med 2000;192:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breij EC, Brink BP, Veerhuis R, et al. Homogeneity of active demyelinating lesions in established multiple sclerosis. Ann Neurol 2008;63:16–25. [DOI] [PubMed] [Google Scholar]
- 8. Magliozzi R, Howell OW, Reeves C, et al. A Gradient of neuronal loss and meningeal inflammation in multiple sclerosis. Ann Neurol 2010;68:477–493. [DOI] [PubMed] [Google Scholar]
- 9. Howell OW, Reeves CA, Nicholas R, et al. Meningeal inflammation is widespread and linked to cortical pathology in multiple sclerosis. Brain 2011;134:2755–2771. [DOI] [PubMed] [Google Scholar]
- 10. McPherson RC, Anderton SM. Adaptive immune responses in CNS autoimmune disease: mechanisms and therapeutic opportunities. J Neuroimmune Pharmacol 2013;8:774–790. [DOI] [PubMed] [Google Scholar]
- 11. International Multiple Sclerosis Genetics C, Wellcome Trust Case Control C, Sawcer S, et al. . Genetic risk and a primary role for cell‐mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease‐associated variation in regulatory DNA. Science 2012;337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murphy K. Janeway's Immunobiology. New York: Garland Science, 2012. [Google Scholar]
- 14. Vandvik B, Norrby E. Oligoclonal IgG antibody response in the central nervous system to different measles virus antigens in subacute sclerosing panencephalitis. Proc Natl Acad Sci USA 1973;70:1060–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Storstein A, Monstad SE, Honnorat J, Vedeler CA. Paraneoplastic antibodies detected by isoelectric focusing of cerebrospinal fluid and serum. J Neuroimmunol 2004;155:150–154. [DOI] [PubMed] [Google Scholar]
- 16. Rotteveel FT, Kokkelink I, van Walbeek HK, et al. Analysis of T cell receptor‐gene rearrangement in T cells from the cerebrospinal fluid of patients with multiple sclerosis. J Neuroimmunol 1987;15:243–249. [DOI] [PubMed] [Google Scholar]
- 17. Hafler DA, Duby AD, Lee SJ, et al. Oligoclonal T lymphocytes in the cerebrospinal fluid of patients with multiple sclerosis. J Exp Med 1988;167:1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee SJ, Wucherpfennig KW, Brod SA, et al. Common T‐cell receptor V beta usage in oligoclonal T lymphocytes derived from cerebrospinal fluid and blood of patients with multiple sclerosis. Ann Neurol 1991;29:33–40. [DOI] [PubMed] [Google Scholar]
- 19. Gestri D, Baldacci L, Taiuti R, et al. Oligoclonal T cell repertoire in cerebrospinal fluid of patients with inflammatory diseases of the nervous system. J Neurol Neurosurg Psychiatry 2001;70:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gran B, Gestri D, Sottini A, et al. Detection of skewed T‐cell receptor V‐beta gene usage in the peripheral blood of patients with multiple sclerosis. J Neuroimmunol 1998;85:22–32. [DOI] [PubMed] [Google Scholar]
- 21. Matsumoto Y, Yoon WK, Jee Y, et al. Complementarity‐determining region 3 spectratyping analysis of the TCR repertoire in multiple sclerosis. J Immunol 2003;170:4846–4853. [DOI] [PubMed] [Google Scholar]
- 22. Muraro PA, Cassiani‐Ingoni R, Chung K, et al. Clonotypic analysis of cerebrospinal fluid T cells during disease exacerbation and remission in a patient with multiple sclerosis. J Neuroimmunol 2006;171:177–183. [DOI] [PubMed] [Google Scholar]
- 23. Muraro PA, Douek DC, Packer A, et al. Thymic output generates a new and diverse TCR repertoire after autologous stem cell transplantation in multiple sclerosis patients. J Exp Med 2005;201:805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warnke C, Mausberg AK, Stettner M, et al. Natalizumab affects the T‐cell receptor repertoire in patients with multiple sclerosis. Neurology 2013;81:1400–1408. [DOI] [PubMed] [Google Scholar]
- 25. Chiarini M, Sottini A, Bertoli D, et al. Newly produced T and B lymphocytes and T‐cell receptor repertoire diversity are reduced in peripheral blood of fingolimod‐treated multiple sclerosis patients. Mult Scler 2015;21:726–734. [DOI] [PubMed] [Google Scholar]
- 26. Muraro PA, Jacobsen M, Necker A, et al. Rapid identification of local T cell expansion in inflammatory organ diseases by flow cytometric T cell receptor Vbeta analysis. J Immunol Methods 2000;246:131–143. [DOI] [PubMed] [Google Scholar]
- 27. Jacobsen M, Cepok S, Quak E, et al. Oligoclonal expansion of memory CD8 + T cells in cerebrospinal fluid from multiple sclerosis patients. Brain 2002;125:538–550. [DOI] [PubMed] [Google Scholar]
- 28. Oksenberg JR, Stuart S, Begovich AB, et al. Limited heterogeneity of rearranged T‐cell receptor V alpha transcripts in brains of multiple sclerosis patients. Nature 1990;345:344–346. [DOI] [PubMed] [Google Scholar]
- 29. Wucherpfennig KW, Newcombe J, Li H, et al. T cell receptor V alpha‐V beta repertoire and cytokine gene expression in active multiple sclerosis lesions. J Exp Med 1992;175:993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skulina C, Schmidt S, Dornmair K, et al. Multiple sclerosis: brain‐infiltrating CD8 + T cells persist as clonal expansions in the cerebrospinal fluid and blood. Proc Natl Acad Sci USA 2004;101:2428–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Junker A, Ivanidze J, Malotka J, et al. Multiple sclerosis: T‐cell receptor expression in distinct brain regions. Brain 2007;130:2789–2799. [DOI] [PubMed] [Google Scholar]
- 32. Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T‐cell responses? Nat Rev Immunol 2008;8:231–238. [DOI] [PubMed] [Google Scholar]
- 33. Wucherpfennig KW, Ota K, Endo N, et al. Shared human T cell receptor V beta usage to immunodominant regions of myelin basic protein. Science 1990;248:1016–1019. [DOI] [PubMed] [Google Scholar]
- 34. Kotzin BL, Karuturi S, Chou YK, et al. Preferential T‐cell receptor beta‐chain variable gene use in myelin basic protein‐reactive T‐cell clones from patients with multiple sclerosis. Proc Natl Acad Sci USA 1991;88:9161–9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ben‐Nun A, Liblau RS, Cohen L, et al. Restricted T‐cell receptor V beta gene usage by myelin basic protein‐specific T‐cell clones in multiple sclerosis: predominant genes vary in individuals. Proc Natl Acad Sci USA 1991;88:2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin R, Utz U, Coligan JE, et al. Diversity in fine specificity and T cell receptor usage of the human CD4+ cytotoxic T cell response specific for the immunodominant myelin basic protein peptide 87‐106. J Immunol 1992;148:1359–1366. [PubMed] [Google Scholar]
- 37. Meinl E, Weber F, Drexler K, et al. Myelin basic protein‐specific T lymphocyte repertoire in multiple sclerosis. Complexity of the response and dominance of nested epitopes due to recruitment of multiple T cell clones. J Clin Invest 1993;92:2633–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovett‐Racke AE, Martin R, McFarland HF, et al. Longitudinal study of myelin basic protein‐specific T‐cell receptors during the course of multiple sclerosis. J Neuroimmunol 1997;78:162–171. [DOI] [PubMed] [Google Scholar]
- 39. Goebels N, Hofstetter H, Schmidt S, et al. Repertoire dynamics of autoreactive T cells in multiple sclerosis patients and healthy subjects: epitope spreading versus clonal persistence. Brain 2000;123(Pt 3):508–518. [DOI] [PubMed] [Google Scholar]
- 40. Lutterotti A, Martin R. Antigen‐specific tolerization approaches in multiple sclerosis. Expert Opin Investig Drugs 2014;23:9–20. [DOI] [PubMed] [Google Scholar]
- 41. Seboun E, Robinson MA, Doolittle TH, et al. A susceptibility locus for multiple sclerosis is linked to the T cell receptor beta chain complex. Cell 1989;57:1095–1100. [DOI] [PubMed] [Google Scholar]
- 42. Oksenberg JR, Sherritt M, Begovich AB, et al. T‐cell receptor V alpha and C alpha alleles associated with multiple and myasthenia gravis. Proc Natl Acad Sci USA 1989;86:988–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lynch SG, Rose JW, Petajan JH, Leppert M. Discordance of the T‐cell receptor alpha‐chain gene in familial multiple sclerosis. Neurology 1992;42:839–844. [DOI] [PubMed] [Google Scholar]
- 44. Dyment DA, Steckley JL, Morrison K, et al. TCR beta polymorphisms and multiple sclerosis. Genes Immun 2004;5:337–342. [DOI] [PubMed] [Google Scholar]
- 45. Watson CT, Para AE, Lincoln MR, et al. Revisiting the T‐cell receptor alpha/delta locus and possible associations with multiple sclerosis. Genes Immun 2011;12:59–66. [DOI] [PubMed] [Google Scholar]
- 46. Qin Y, Duquette P, Zhang Y, et al. Clonal expansion and somatic hypermutation of V(H) genes of B cells from cerebrospinal fluid in multiple sclerosis. J Clin Invest 1998;102:1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Colombo M, Dono M, Gazzola P, et al. Accumulation of clonally related B lymphocytes in the cerebrospinal fluid of multiple sclerosis patients. J Immunol 2000;164:2782–2789. [DOI] [PubMed] [Google Scholar]
- 48. Owens GP, Ritchie AM, Burgoon MP, et al. Single‐cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the B cell response in multiple sclerosis cerebrospinal fluid. J Immunol 2003;171:2725–2733. [DOI] [PubMed] [Google Scholar]
- 49. Owens GP, Kraus H, Burgoon MP, et al. Restricted use of VH4 germline segments in an acute multiple sclerosis brain. Ann Neurol 1998;43:236–243. [DOI] [PubMed] [Google Scholar]
- 50. Lovato L, Willis SN, Rodig SJ, et al. Related B cell clones populate the meninges and parenchyma of patients with multiple sclerosis. Brain 2011;134:534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cameron EM, Spencer S, Lazarini J, et al. Potential of a unique antibody gene signature to predict conversion to clinically definite multiple sclerosis. J Neuroimmunol 2009;213:123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Owens GP, Shearer AJ, Yu X, et al. Screening random peptide libraries with subacute sclerosing panencephalitis brain‐derived recombinant antibodies identifies multiple epitopes in the C‐terminal region of the measles virus nucleocapsid protein. J Virol 2006;80:12121–12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bennett JL, Lam C, Kalluri SR, et al. Intrathecal pathogenic anti‐aquaporin‐4 antibodies in early neuromyelitis optica. Ann Neurol 2009;66:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Owens GP, Bennett JL, Lassmann H, et al. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Ann Neurol 2009;65:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blauth K, Soltys J, Matschulat A, et al. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid cause demyelination of spinal cord explants. Acta Neuropathol 2015;130:765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lambracht‐Washington D, O'Connor KC, Cameron EM, et al. Antigen specificity of clonally expanded and receptor edited cerebrospinal fluid B cells from patients with relapsing remitting MS. J Neuroimmunol 2007;186:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. von Budingen HC, Harrer MD, Kuenzle S, et al. Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin‐specific antibodies. Eur J Immunol 2008;38:2014–2023. [DOI] [PubMed] [Google Scholar]
- 58. Willis SN, Stathopoulos P, Chastre A, et al. Investigating the Antigen Specificity of Multiple Sclerosis Central Nervous System‐Derived Immunoglobulins. Front Immunol 2015;6:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walter MA, Gibson WT, Ebers GC, Cox DW. Susceptibility to multiple sclerosis is associated with the proximal immunoglobulin heavy chain variable region. J Clin Invest 1991;87:1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hashimoto LL, Walter MA, Cox DW, Ebers GC. Immunoglobulin heavy chain variable region polymorphisms and multiple sclerosis susceptibility. J Neuroimmunol 1993;44:77–83. [DOI] [PubMed] [Google Scholar]
- 61. Feakes R, Chataway J, Sawcer S, et al. Susceptibility to multiple sclerosis and the immunoglobulin heavy chain gene cluster. Ann Neurol 1998;44:984. [DOI] [PubMed] [Google Scholar]
- 62. Feakes R, Sawcer S, Smillie B, et al. No evidence for the involvement of interleukin 2 or the immunoglobulin heavy chain gene cluster in determining genetic susceptibility to multiple sclerosis. J Neurol Neurosurg Psychiatry 2000;68:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Watson CT, Ramagopalan SV, Morrison KM, et al. IGHV4‐39 deletion polymorphism does not associate with risk or outcome of multiple sclerosis. J Neuroimmunol 2010;225:164–166. [DOI] [PubMed] [Google Scholar]
- 64. Arstila TP, Casrouge A, Baron V, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science 1999;286:958–961. [DOI] [PubMed] [Google Scholar]
- 65. Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T‐cell receptor beta‐chain diversity in alphabeta T cells. Blood 2009;114:4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Qi Q, Liu Y, Cheng Y, et al. Diversity and clonal selection in the human T‐cell repertoire. Proc Natl Acad Sci USA 2014;111:13139–13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Laydon DJ, Bangham CR, Asquith B. Estimating T‐cell repertoire diversity: limitations of classical estimators and a new approach. Philos Trans R Soc Lond B Biol Sci 2015;370. pii: 20140291. doi: 10.1098/rstb.2014.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Calis JJ, Rosenberg BR. Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends Immunol 2014;35:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Balakrishnan A, Morris GP. The highly alloreactive nature of dual TCR T cells. Curr Opin Organ Transplant 2015. Nov 9. doi: 10.1097/MOT.0000000000000261. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. von Budingen HC, Kuo TC, Sirota M, et al. B cell exchange across the blood‐brain barrier in multiple sclerosis. J Clin Invest 2012;122:4533–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Johansen JN, Vartdal F, Desmarais C, et al. Intrathecal BCR transcriptome in multiple sclerosis versus other neuroinflammation: equally diverse and compartmentalized, but more mutated, biased and overlapping with the proteome. Clin Immunol 2015;160:211–225. [DOI] [PubMed] [Google Scholar]
- 72. Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity 1998;8:135–141. [DOI] [PubMed] [Google Scholar]
- 73. Paillard F, Sterkers G, Bismuth G, et al. Lymphokine mRNA and T cell multireceptor mRNA of the Ig super gene family are reciprocally modulated during human T cell activation. Eur J Immunol 1988;18:1643–1646. [DOI] [PubMed] [Google Scholar]
- 74. Jack HM, Wabl M. Immunoglobulin mRNA stability varies during B lymphocyte differentiation. EMBO J 1988;7:1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Carlson CS, Emerson RO, Sherwood AM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013;4:2680. [DOI] [PubMed] [Google Scholar]
- 76. Bankoti J, Apeltsin L, Hauser SL, et al. In multiple sclerosis, oligoclonal bands connect to peripheral B‐cell responses. Ann Neurol 2014;75:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Palanichamy A, Apeltsin L, Kuo TC, et al. Immunoglobulin class‐switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci Transl Med 2014;6:248ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rounds WH, Salinas EA, Wilks TB 2nd, et al. MSPrecise: A molecular diagnostic test for multiple sclerosis using next generation sequencing. Gene 2015;572:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jones JL, Thompson SA, Loh P, et al. Human autoimmunity after lymphocyte depletion is caused by homeostatic T‐cell proliferation. Proc Natl Acad Sci USA 2013;110:20200–20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Muraro PA, Robins H, Malhotra S, et al. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. J Clin Invest 2014;124:1168–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lossius A, Johansen JN, Vartdal F, et al. High‐throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV‐reactive CD8 + T cells. Eur J Immunol 2014;44:3439–3452. [DOI] [PubMed] [Google Scholar]
- 82. Salou M, Garcia A, Michel L, et al. Expanded CD8 T‐cell sharing between periphery and CNS in multiple sclerosis. Ann Clin Transl Neurol 2015;2:609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Planas R, Metz I, Ortiz Y, et al. Central role of Th2/Tc2 lymphocytes in pattern II multiple sclerosis lesions. Ann Clin Transl Neurol 2015;2:875–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stern JN, Yaari G, Vander Heiden JA, et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med 2014;6:248ra107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol 2013;25:646–652. [DOI] [PubMed] [Google Scholar]
- 86. Bolotin DA, Mamedov IZ, Britanova OV, et al. Next generation sequencing for TCR repertoire profiling: platform‐specific features and correction algorithms. Eur J Immunol 2012;42:3073–3083. [DOI] [PubMed] [Google Scholar]
- 87. Vander Heiden JA, Yaari G, Uduman M, et al. pRESTO: a toolkit for processing high‐throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 2014;30:1930–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shugay M, Britanova OV, Merzlyak EM, et al. Towards error‐free profiling of immune repertoires. Nat Methods 2014;11:653–655. [DOI] [PubMed] [Google Scholar]
- 89. Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V‐(D)‐J repertoires, polymorphisms, and IG mutations: IMGT/V‐QUEST and IMGT/HighV‐QUEST for NGS. Methods Mol Biol 2012;882:569–604. [DOI] [PubMed] [Google Scholar]
- 90. Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 2013;41:W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Thomas N, Heather J, Ndifon W, et al. Decombinator: a tool for fast, efficient gene assignment in T‐cell receptor sequences using a finite state machine. Bioinformatics 2013;29:542–550. [DOI] [PubMed] [Google Scholar]
- 92. Giraud M, Salson M, Duez M, et al. Fast multiclonal clusterization of V(D)J recombinations from high‐throughput sequencing. BMC Genom 2014;15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Adaptive Biotechnologies ImmunoSeq platform. Available at: http://www.adaptivebiotech.com/immunoseq. Last accessed on December 4, 2015.
- 94. Nazarov VI, Pogorelyy MV, Komech EA, et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics 2015;16:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shugay M, Bagaev DV, Turchaninova MA, et al. VDJtools: Unifying Post‐analysis of T Cell Receptor Repertoires. PLoS Comput Biol 2015;11:e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Georgiou G, Ippolito GC, Beausang J, et al. The promise and challenge of high‐throughput sequencing of the antibody repertoire. Nat Biotechnol 2014;32:158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Brusic V, Gottardo R, Kleinstein SH, et al. Computational resources for high‐dimensional immune analysis from the Human Immunology Project Consortium. Nat Biotechnol 2014;32:146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. NCBI Basic Local Alignment Search Tool. Available at: http://blast.ncbi.nlm.nih.gov. Last accessed on December 5, 2015.
- 99. Leinonen R, Sugawara H, Shumway M. International Nucleotide Sequence Database C. The sequence read archive. Nucleic Acids Res 2011;39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lossius A, Johansen JN, Torkildsen O, et al. Epstein‐Barr virus in systemic lupus erythematosus, rheumatoid arthritis and multiple sclerosis‐association and causation. Viruses 2012;4:3701–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Iliff JJ, Wang M, Liao Y, et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med 2012;4:147ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Louveau A, Smirnov I, Keyes TJ, et al. Structural and functional features of central nervous system lymphatic vessels. Nature 2015;523:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Aspelund A, Antila S, Proulx ST, et al. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J Exp Med 2015;212:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cserr HF, Harling‐Berg CJ, Knopf PM. Drainage of brain extracellular fluid into blood and deep cervical lymph and its immunological significance. Brain Pathol 1992;2:269–276. [DOI] [PubMed] [Google Scholar]
- 105. Reddy ST, Ge X, Miklos AE, et al. Monoclonal antibodies isolated without screening by analyzing the variable‐gene repertoire of plasma cells. Nat Biotechnol 2010;28:965–969. [DOI] [PubMed] [Google Scholar]
- 106. Howie B, Sherwood AM, Berkebile AD, et al. High‐throughput pairing of T cell receptor alpha and beta sequences.Sci Transl Med 2015;7:301ra131. [DOI] [PubMed] [Google Scholar]
- 107. Shapiro E, Biezuner T, Linnarsson S. Single‐cell sequencing‐based technologies will revolutionize whole‐organism science. Nat Rev Genet 2013;14:618–630. [DOI] [PubMed] [Google Scholar]
- 108. DeKosky BJ, Ippolito GC, Deschner RP, et al. High‐throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol 2013;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Busse CE, Czogiel I, Braun P, et al. Single‐cell based high‐throughput sequencing of full‐length immunoglobulin heavy and light chain genes. Eur J Immunol 2014;44:597–603. [DOI] [PubMed] [Google Scholar]
- 110. Han A, Glanville J, Hansmann L, Davis MM. Linking T‐cell receptor sequence to functional phenotype at the single‐cell level. Nat Biotechnol 2014;32:684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Raposo B, Dobritzsch D, Ge C, et al. Epitope‐specific antibody response is controlled by immunoglobulin V(H) polymorphisms. J Exp Med 2014;211:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gadala‐Maria D, Yaari G, Uduman M, Kleinstein SH. Automated analysis of high‐throughput B‐cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc Natl Acad Sci USA 2015;112:E862–E870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wu D, Sherwood A, Fromm JR, et al. High‐throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med 2012;4:134ra163. [DOI] [PubMed] [Google Scholar]


