Abstract
Upstream open reading frames (uORFs) are known to regulate a few specific transcripts, and recent computational and experimental studies have suggested candidate uORF regulation across the genome. In this issue, Johnstone et al (2016) use ribosome profiling to identify translated uORFs and measure their effects on downstream translation. Furthermore, they show that regulatory uORFs are conserved across species and subject to selective constraint. Recognizing the potential of uORFs in regulating translation expands our understanding of the dynamic regulation of gene expression.
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology, Systems & Computational Biology
Differences in the translation level of distinct mRNAs play an important role in controlling the production of the encoded protein. Changes in translation drive posttranscriptional gene expression programs that play critical roles in diverse processes ranging from cellular stress responses to memory formation. Despite the importance of differential translation, we have a limited understanding of the cis‐acting mRNA features that determine the stability or translation state of an mRNA (Brar et al, 2012; Arribere & Gilbert, 2013; Calvo et al, 2009). Johnstone et al (2016) now provide evidence for a global impact of short upstream open reading frames (uORFs) on translation.
In eukaryotes, the small subunit of the ribosome typically scans from the 5′ end of an mRNA until it recognizes a start codon. The transcript leader sequence (TLS), also known as the 5′ untranslated region, scanned by the ribosome can thus modulate translation initiation in order to control protein synthesis. In particular, the presence of upstream start codons in the transcript leader can recruit scanning ribosomes to an alternate reading frame, reducing the fraction that reach the start codon for the major protein (Johnstone et al, 2016; Arribere & Gilbert, 2013). Regions that show no overlap with the CDS are termed upstream open reading frames (uORFs), while those that start prior to CDS AUG but finish within the CDS are called overlapping open reading frames (oORFs; Johnstone et al, 2016). Calvo et al (2009) provided direct evidence that the presence of a uORF represses downstream CDS translation in the context of reporters bearing endogenous, uORF‐containing TLSs. Furthermore, uORFs can specify dynamic regulation. In the stress‐specific transcription factors ATF4 (in mammals) and GCN4 (in yeast), upstream initiation sites repress protein production under normal cellular conditions (Sonenberg & Hinnebusch, 2009). Under stress, global translation levels decrease; however, ATF4 and GCN4 synthesis is paradoxically upregulated in an upstream initiation site‐dependent manner.
While it is possible to computationally identify upstream TLS AUG codons, these potential initiation sites may not actually be engaged by ribosomes. The advent of ribosome profiling now allows direct interrogation of ribosome occupancy in TLSs (Fig 1A) and has identified translated uORFs (Ingolia et al, 2009; Arribere & Gilbert, 2013) that in some cases produce peptides detectable by mass spectrometry (Bazzini et al, 2014; Saghatelian & Couso, 2015). Ribosome profiling provides information about ribosome occupancy at single nucleotide precision, and several metrics now exist to calculate the likelihood of a ribosome profiling signal corresponding to active translation of a given reading frame (Ingolia et al, 2014; Chew et al, 2013). Bazzini et al (2014) utilized the triplet periodicity from such nucleotide level profiles to classify translation status on a genomewide level across zebrafish developmental stages, using a metric termed ORF score (Fig 1B). Johnstone et al (2016) now ask how empirically verified uORF translation acts across the transcriptome.
Figure 1. Ribosome profiling detects uORF translation and downstream repression.
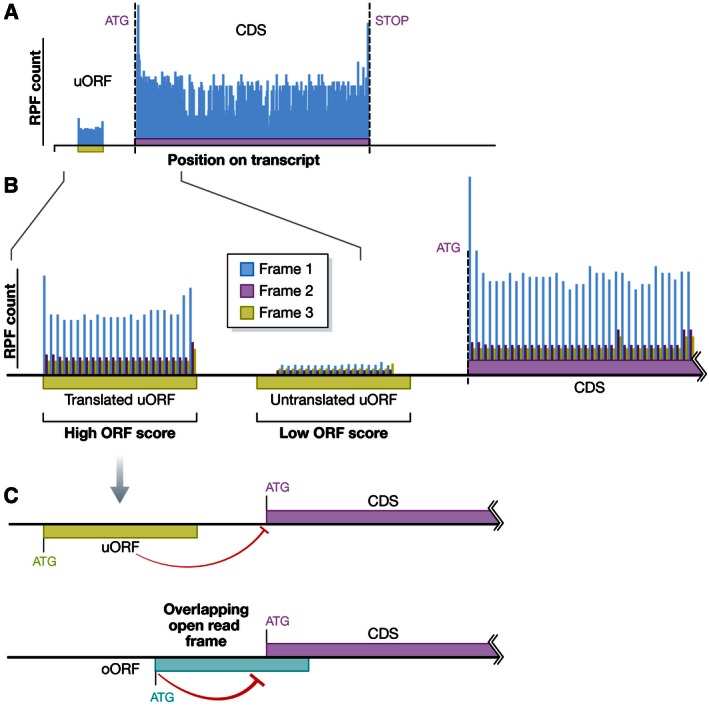
A) Ribosome profiling maps the location of ribosomes across a transcript using deep sequencing of ribosome protected fragments (RPFs) (Ingolia et al, 2009). In addition to the major protein coding sequence (CDS), RPFs can be seen in upstream open reading frames (uORF). B) RPFs offer nucleotide level resolution that can map to any of the three potential reading frames. Bazzini et al (2014) utilized this triplet periodicity to generate an ORFscore. This ORFscore can be used to classify actively translated regions. In this issue, Johnstone et al (2016) apply ORFscore classification to separate computationally defined uORFs by their translation status. C) Johnstone et al (2016) find that confidently translated uORFs correlate with repression of the downstream CDS translation. Moreover, overlapping open reading frames (oORFs) act as stronger repressors of CDS translation.
Using ORF score, Johnstone et al (2016) identify over a thousand zebrafish transcripts with confidently translated uORFs, including several developmentally important genes. Previous studies suggest uORFs generally repress the downstream CDS (Calvo et al, 2009; Arribere & Gilbert, 2013), and using matched RNA sequencing, Johnstone et al (2016) calculate a score of translation efficiency (TE) for the major transcript product as well. They show that on a genomewide scale, the presence of a translated uORF correlates with decreased downstream CDS translation and transcript stability relative to non‐uORF‐containing transcripts (Fig 1C). Moreover, the repression of the downstream CDS is greater in the presence of overlap with the upstream region (oORFs) or stronger uORF translation.
uORFs provide functionally important repression, mediated by the titration of initiating ribosomes away from downstream genes. Johnstone et al (2016) demonstrate conservation of the presence of uORFs and selective constraints on sequence features that confer strong translation initiation. Furthermore, these sequence features positively correlate with the strength of downstream translation repression across zebrafish genes. In contrast, the peptide sequences of regulatory uORFs do not show selective constraint as seen in truly functional micropeptides (Saghatelian & Couso, 2015). This suggests that the functional importance of uORFs is dependent on features that drive uORF translation rather than the specific peptide produced.
The next challenge for the field will be understanding the mechanism by which these TLS reading frames can provided dynamic regulation. In the case of ATF4 and GCN4, translation of the CDS is only accomplished under cases of cellular stress, and loss of this stress‐induced translation is linked to diabetes and neurodegenerative disorders (Sidrauski et al, 2015; Sonenberg & Hinnebusch, 2009). This regulation depends on phosphorylation of translation initiation factor eIF2α, which is thought to promote the bypass of repressive uORFs.
Because eIF2α is required for essentially all translation initiation events, this acts as a global method of regulation. Dynamic regulation of specific transcripts can result from the interaction between repressive uORFs and sequence‐specific RNA‐binding proteins, as seen in Drosophila SXL2 control of msl2 translation (Medenbach et al, 2011). Direct detection and measurement of uORF translation by ribosome profiling, as demonstrated by Johnstone et al (2016), promises further insights into the dynamic process of uORF‐mediated translational control.
See also: TG Johnstone et al (April 2016)
References
- Arribere JA, Gilbert WV (2013) Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23: 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ (2014) Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33: 937–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS (2012) High resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E (2013) Ribosome profiling reveals resemblance between long noncoding RNAs and 5' leaders of coding RNAs. Development 140: 2828–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern‐Ginossar N, Harris MS, Talhouarne GJS, Jackson SE, Wills MR, Weissman JS (2014) Ribosome profiling reveals pervasive translation outside of annotated protein coding genes. Cell Rep 8: 1365–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ (2016) Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35: 706–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW (2011) Translational control via protein‐regulated upstream open reading frames. Cell 145: 902–913 [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Couso JP (2015) Discovery and characterization of smORF‐encoded bioactive polypeptides. Nat Chem Biol 11: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P (2015). The small molecule ISRIB reverses the effects of eIF2a phosphorylation on translation and stress granule assembly. eLife 4: e05033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]


