Abstract
Yap1 is a transcriptional co‐activator of the Hippo pathway. The importance of Yap1 in early cell fate decision during embryogenesis has been well established, though its role in embryonic stem (ES) cells remains elusive. Here, we report that Yap1 plays crucial roles in normal differentiation rather than self‐renewal of ES cells. Yap1‐depleted ES cells maintain undifferentiated state with a typical colony morphology as well as robust alkaline phosphatase activity. These cells also retain comparable levels of the core pluripotent factors, such as Pou5f1 and Sox2, to the levels in wild‐type ES cells without significant alteration of lineage‐specific marker genes. Conversely, overexpression of Yap1 in ES cells promotes nuclear translocation of Yap1, resulting in disruption of self‐renewal and triggering differentiation by up‐regulating lineage‐specific genes. Moreover, Yap1‐deficient ES cells show impaired induction of lineage markers during differentiation. Collectively, our data demonstrate that Yap1 is a required factor for proper differentiation of mouse ES cells, while remaining dispensable for self‐renewal.
Keywords: embryonic stem cells, Yap1, differentiation, Hippo pathway, self‐renewal
Subject Categories: Signal Transduction, Stem Cells
Introduction
The Hippo signaling pathway, modulated by cell density and cell–cell contact, is implicated in diverse cellular processes including cell proliferation 1, 2, 3, 4, 5, apoptosis 5, 6, and organ size control 7, 8. Yap1 is a transcriptional co‐activator of the Hippo pathway and is known to play a crucial role in the segregation of inner cell mass (ICM) and trophectoderm (TE) during early embryogenesis 9, 10, 11, 12, 13. While Yap1 resides in the nucleus of trophectodermal cells and functions as a critical co‐activator for TE development, it is sequestered mainly in the cytoplasm of ICM as a phosphorylated inactive form due to active Hippo signaling 9. However, the role of Yap1 in ICM is still elusive 9, 12. In addition to Yap1, Taz and Tead family members are also crucial players in the Hippo pathway. A homologue of Yap1, Taz, shares redundant functions such as controlling cell proliferation and sensing mechanical stress 14, 15. Furthermore, Tead proteins—important in TE differentiation during early embryogenesis, form a complex with Yap1 and are known to activate their downstream target genes 16, 17.
Two different observations on the roles of Yap1 in embryonic stem (ES) cells are of note. Recent studies suggested that Yap1 plays an important role in the maintenance of mouse ES cells as an active factor in the nucleus 18, 19. These works showed knockdown (KD) of Yap1 promotes differentiation of ES cells while overexpression (OE) of Yap1 not only enhances self‐renewal but also inhibits differentiation of ES cells, even under neuronal differentiation conditions 18. However, the study showing nuclear localization of Yap1 in ES cells is somewhat contradictory to the function of the Hippo signaling, since mouse ES cells grow as tightly packed colonies. It has been suggested that high cell density or cell–cell contact activates the Hippo signaling and subsequent sequestration of Yap1 in the cytoplasm of various cell lines such as HaCaT and NIH‐3T3 1, 3, 7, 20. Accordingly, another recent study has claimed that both Yap1 and Taz are dispensable for self‐renewal of ES cells in 2i (Gsk3β and Mek inhibitors) culture condition 21. In this case, Yap1‐ and Taz‐depleted ES cells maintained undifferentiated state under differentiation‐promoting culture conditions 21. Consistent with this observation, studies of neuronal differentiation from ES cells have shown that high cell density, which activates the Hippo signaling and sequesters Yap1 in the cytoplasm, blocks differentiation of ES cells 22, 23, suggesting that nuclear localization of Yap1 might be important in normal differentiation of ES cells.
In the current study, we show that Yap1 is dispensable for the maintenance of ES cells but critical in their differentiation. Additional testing of Yap1‐associated factors including Tead family proteins and Taz also supports the dispensability of Yap1 for the self‐renewal of ES cells. In line with gradual up‐regulation of Yap1 level upon differentiation of ES cells, OE of Yap1 in ES cells enhances nuclear abundance of Yap1 accompanied by induction of various lineage‐specific marker genes. On the contrary, Yap1‐depleted ES cells showed impaired differentiation. Taken together, our data demonstrate a critical role of Yap1 in normal differentiation rather than self‐renewal of ES cells.
Results and Discussion
Yap1 is dispensable for self‐renewal of mouse ES cells
Previous studies reported that Yap1 is required in the maintenance of mouse ES cells by showing that KD of Yap1 promotes differentiation of ES cells 18, 19. Conversely, another study claimed that double KD of Yap1 and Taz in 2i media does not disrupt self‐renewal of ES cells 21. To decipher the roles of Yap1 in self‐renewal and pluripotency of ES cells, we first performed KD of Yap1 using lentivirus‐delivered shRNAs in J1 mouse ES cells (Fig EV1A and Dataset EV1). In contrast to the previous reports 18, 19, we found that even with > 85% of Yap1 KD, ES cells maintain normal colony morphology with high alkaline phosphatase (AP) activity, whereas ES cells with KD of Pou5f1 undergo differentiation accompanied by loss of AP activity, as expected (Fig 1A). We additionally found that these Yap1‐depleted ES cells show comparable proliferation rate to that of control ES cells (Fig EV1B). In agreement with these observations, overall expression levels of ES cell core pluripotency factors, such as Pou5f1 and Nanog, as well as several lineage‐specific regulators were not significantly altered upon KD of Yap1 (Figs 1B–D and EV1C). We validated our observation by testing two additional ES cell lines (E14 and CJ7) and confirmed that KD of Yap1 does not significantly affect features of normal self‐renewing ES cells (Fig EV2A–F).
Figure EV1. Yap1 is dispensable for self‐renewal of J1 mouse ES cells (related to Fig 1).
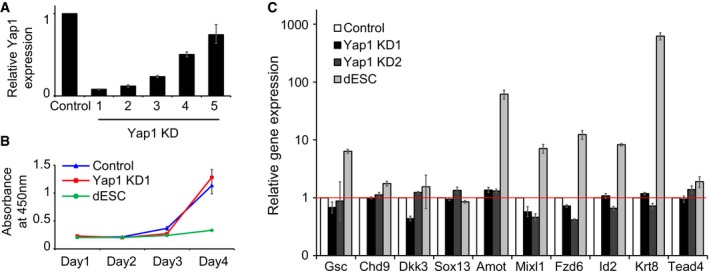
- Yap1 mRNA levels measured by RT–qPCR upon shRNA‐based KD. Five different shRNA sequences were tested, and shRNAs 1 and 2 (KD1 and KD2) were used for further studies. Data are represented as mean ± SD.
- Cell proliferation rates of Yap1 KD cells and control cells.
- mRNA levels of lineage‐specific marker genes upon KD of Yap1. Differentiating ES cells (dESC) were used as control cells. Data are represented as mean ± SD.
Figure 1. Yap1 is dispensable for self‐renewal of J1 mouse ES cells (see also Figs EV1, EV2, EV3).
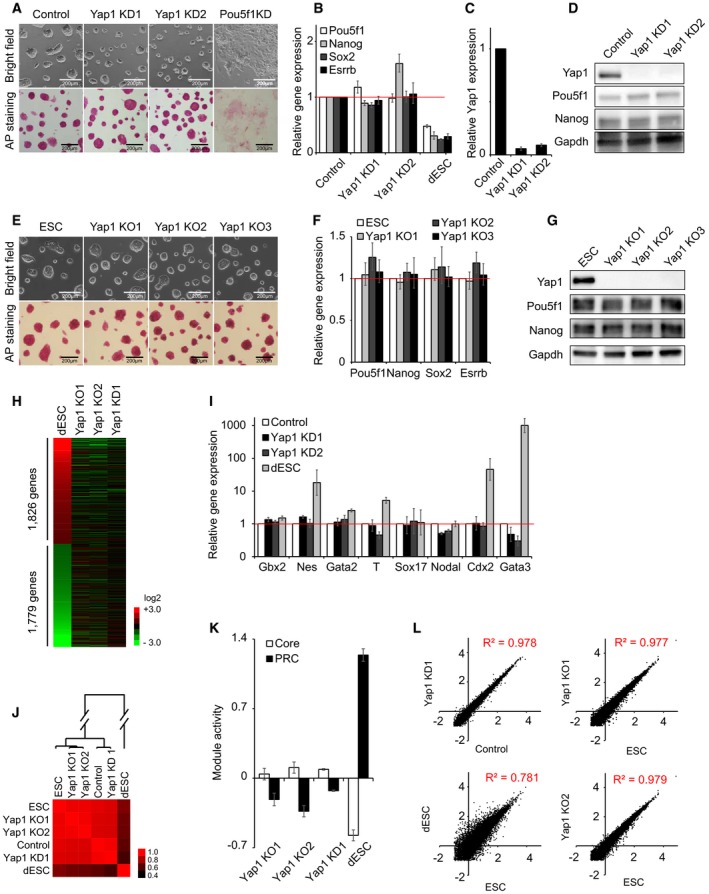
-
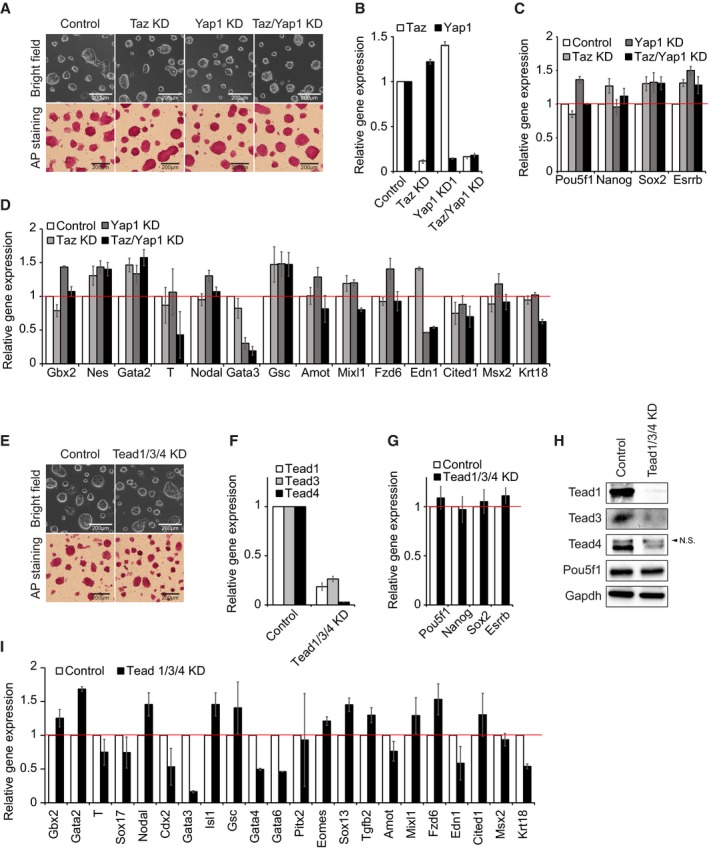
AColony morphology and alkaline phosphatase (AP) activity of ES cells upon KD of Yap1 and Pou5f1. KD1 and KD2 indicate two different shRNA sequences tested. All the following cell morphology and AP staining pictures were taken two passages (4 days) after lentivirus infection unless otherwise stated.
-
B, CmRNA expression levels of Pou5f1, Nanog, Sox2, Esrrb (B), and Yap1 (C) upon KD of Yap1. All the following mRNA samples were harvested 4 days after lentivirus infection while passaged every 2 days unless otherwise stated. Data are represented as mean ± SD.
-
DProtein levels of Yap1, Pou5f1, and Nanog upon KD of Yap1. All the following protein samples were harvested 4 days after lentivirus infection while passaged every 2 days unless otherwise stated.
-
EColony morphology and AP activity of mouse embryonic stem cells (ESC) and three Yap1 KO clones (KO1‐KO3).
-
FmRNA levels of Pou5f1, Nanog, Sox2, and Esrrb upon KO of Yap1. Data are represented as mean ± SD.
-
GProtein levels of Yap1, Pou5f1, and Nanog in Yap1 KO clones.
-
HA heatmap showing relative mRNA expression levels of 3,605 genes differentially expressed (> twofold) between ES cells and differentiating ES cells (dESC). Genes were sorted by the fold changes of gene expression between dESC and ES cells. Corresponding gene expression profiles obtained from Yap1 KO1, Yap1 KO2, and Yap1 KD cells are also shown.
-
ImRNA expression levels of lineage‐specific marker genes upon KD of Yap1. dESC were used as control cells.
-
JA heatmap showing Pearson's correlation coefficients of gene expression profiles obtained from ESC, control virus‐infected ES cells (Control), dESC, Yap1 KD cells, and Yap1 KO cells.
-
KRelative average module activities (Core and PRC) in Yap1 KD1 cells, KO cells, and dESC. Module activities were normalized by the data obtained in ES cells. Data are represented as mean ± SEM.
-
LScatter plots showing log10 (FPKM) values of genes in Yap1 KD1 cells and Control (upper left panel), dESC and ESC (bottom left panel), and Yap1 KO cells and ES cells (right two panels). Pearson's correlation coefficients (R 2) are indicated. “FPKM” indicates fragments per kilobase of transcript per million fragments mapped.
Figure EV2. Yap1 is dispensable for self‐renewal of E14 and CJ7 mouse ES cells (related to Fig 1).
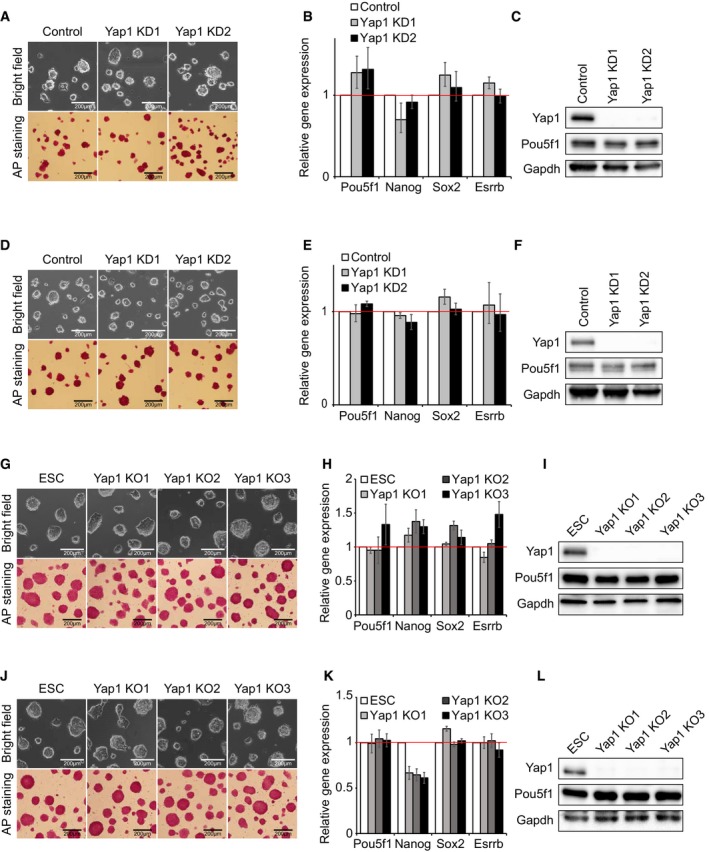
-
A–CData from CJ7 ES cells. Colony morphology and AP activity of Yap1 KD cells (A), mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb in Yap1 KD cells (B), and protein expression levels of Yap1, Pou5f1, and Gapdh were measured in Yap1 KD cells (C). mRNA level data are represented as mean ± SD.
-
D–FData from E14 ES cells. Colony morphology and AP activity of Yap1 KD cells (D), mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb in Yap1 KD cells (E), and protein expression levels of Yap1, Pou5f1, and Gapdh were measured in Yap1 KD cells (F). mRNA level data are represented as mean ± SD.
-
G–IData from CJ7 ES cells (ESC). Colony morphology and AP activity of Yap1 KO clones (G), mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb in Yap1 KO clones (H), and protein expression levels of Yap1, Pou5f1, and Gapdh were measured in Yap1 KO clones (I). mRNA level data are represented as mean ± SD.
-
J–LData from E14 ES cells. Colony morphology and AP activity of Yap1 KO clones (J), mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb in Yap1 KO clones (K), and protein expression levels of Yap1, Pou5f1, and Gapdh were measured in Yap1 KO clones (L). mRNA level data are represented as mean ± SD.
To rule out the possibility of off‐target effects and incomplete depletion due to shRNA‐mediated KD strategies, we additionally established Yap1 knockout (KO) ES cell lines harboring premature stop codons on both alleles by CRISPR‐Cas9‐based genome editing strategies (Dataset EV1) 24, 25. Consistent with the KD results, Yap1 KO ES cells sustained self‐renewing status and showed normal ES colony morphology and high AP activity, and levels of pluripotency‐related genes were comparable to those of wild‐type ES cells (Figs 1E–G and EV2G–L). Yap1 KO ES cells were able to maintain self‐renewal for more than 1 month in culture (Fig EV3A–C). Taken together, these results indicate that Yap1 is dispensable for self‐renewal of mouse ES cells.
Figure EV3. Yap1‐depleted ES cells can maintain self‐renewal for more than a month in culture (related to Fig 1).
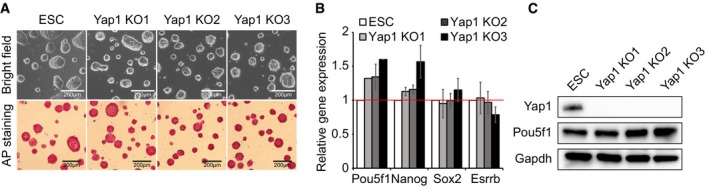
- Colony morphology and AP activity of Yap1 KO clones cultured for more than a month.
- mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb in Yap1 KO clones shown in (A). Data are represented as mean ± SD.
- Protein expression levels of Yap1, Pou5f1, and Gapdh were measured in Yap1 KO clones shown in (A).
To further validate the dispensability of Yap1 in self‐renewal of ES cells, we sought to monitor the global gene expression profiles of Yap1 KD and Yap1 KO ES cells using RNA‐seq approaches. As expected, comparison of expression profiles between ES and differentiating ES cells revealed many differentially expressed genes (DEGs) (Fig 1H and Dataset EV2). However, expression levels of these genes were not altered significantly upon KD or KO of Yap1 (Fig 1H) which was further confirmed by RT–qPCR (Fig 1I). Overall, these results indicate that the depletion of Yap1 does not trigger differentiation of ES cells.
Unlike differentiating ES cells, a hierarchical clustering of global expression data revealed that Yap1 KD and Yap1 KO ES cells were clustered together with normal and control ES cells, indicating that Yap1‐deficient ES cells have similar expression profiles to those of normal ES cells (Fig 1J). We also investigated the activity of previously defined functional modules in ES cells (Core and PRC) 26. Module activity is defined as an averaged expression of all genes in each module. Briefly, the Core module includes core pluripotency factors such as Pou5f1, Nanog, and Sox2, most of which are highly expressed in self‐renewing ES cells. On the other hand, the PRC module includes many lineage‐specific regulators, such as Fgf5, Bmp4, and Hand1, most of which are repressed in ES cells. Since differentiation of ES cells decreases the activity of Core module but increases the activity of PRC module 26, we sought to examine module activities upon KD or KO of Yap1 to test whether cells maintain self‐renewal. As shown in Fig 1K, ES cells with depleted Yap1 did not show down‐regulation of Core module activity or up‐regulation of PRC module activity, suggesting that Yap1‐depleted ES cells largely maintain self‐renewing and undifferentiated states. Further correlation analyses verified that the global expression patterns of Yap1‐depleted ES cells showed higher correlation with those of control ES cells (R 2 = 0.978 for Yap1 KD, R 2 = 0.977 for Yap1 KO1, and R 2 = 0.979 for Yap1 KO2) than differentiating ES cells (R 2 = 0.781) (Fig 1L). Collectively, these data provide strong evidence that the depletion of Yap1 does not significantly alter the self‐renewal of ES cells.
Taz and Tead family proteins are not required for self‐renewal of ES cells and do not compensate Yap1 functions in Yap1‐depleted ES cells
Taz is homologous to Yap1 and has similar functions to Yap1, such as regulation of proliferation and activation of TE lineage markers 14, 27. To rule out the possibility of compensation by Taz in Yap1 KD ES cells, we performed both single and double KD of Yap1 and Taz using shRNAs under drug selections (blasticidin and puromycin, respectively). J1 ES cells with depletion of both Yap1 and Taz (> 85% of KD for each) maintained typical colony morphology as well as high AP activity (Fig 2A and B). Additionally, the levels of pluripotency markers, such as Pou5f1 and Nanog, as well as various lineage markers were not significantly affected by either single or double KD of Yap1 and Taz (Fig 2C and D), indicating that the dispensability of Yap1 in self‐renewing ES cells is not due to the compensatory effect of Taz.
Figure 2. Taz and Tead family proteins are not required for the self‐renewal of J1 mouse ES cells (see also Fig EV4).
-
AColony morphology and AP activity of ES cells upon KD of Yap1 and Taz.
-
B–DmRNA expression levels of Yap1 and Taz (B), Pou5f1, Nanog, Sox2, and Esrrb (C), and lineage‐specific marker genes (D) upon KD of Yap1 and Taz. Data are represented as mean ± SD.
-
EColony morphology and AP activity of ES cells upon KD of Tead1/Tead3/Tead4.
-
F, GmRNA expression levels of Tead1, Tead3, and Tead4 (F) and Pou5f1, Nanog, Sox2, and Esrrb (G) upon KD of Tead 1/3/4. Data are represented as mean ± SD.
-
HProtein levels of Tead1, Tead3, Tead4, and Pou5f1 upon KD of Tead1/Tead3/Tead4. N.S., non‐specific.
-
ImRNA expression levels of lineage‐specific marker genes upon KD of Tead1/Tead3/Tead4. Data are represented as mean ± SD.
Since Yap1 is known to require Tead family proteins to activate its downstream target genes in NIH‐3T3 and MCF10A cell lines 17, 28, 29, we investigated whether Tead proteins are also dispensable for the maintenance of ES cells. To do so, we first performed KD of Tead2 in ES cells. In contrast to the previous report 19, we did not observe any significant alteration of cell morphology or reduced AP activity upon down‐regulation of Tead2 (at least > 90% of KD in mRNA levels) (Fig EV4A and B). In accordance with the colony morphology, Tead2 KD ES cells expressed similar levels of pluripotency genes compared to wild‐type ES cells (Fig EV4C and D). These results were confirmed by generation of three independent Tead2 KO ES cell clones using CRISPR‐Cas9 strategies (Dataset EV1). These Tead2 KO clones also maintained self‐renewal without differentiation (Fig EV4E–G). We additionally conducted triple KD of Tead1/3/4 with triple drug selection (at least > 80% of KD for each), and did not observe any significant alteration of cell morphology or AP activity which is in contrast to the previous report 18 (Fig 2E and F). Similar to the results obtained from the KD of Yap1, triple Tead KD ES cells expressed comparable levels of pluripotency genes shown in wild‐type ES cells without any significant activation of lineage‐specific regulators (Fig 2G–I). The results were further validated by double KD of Tead1/3 in Tead4 KO cells. ES cells with Tead4 KO and Tead1/3 KD also maintained self‐renewal (Fig EV4H–J). Collectively, our data suggest that both Yap1 and Yap1‐associated proteins such as Taz and Tead are not required for self‐renewal of ES cells.
Figure EV4. Tead family proteins are not required for the self‐renewal of ES cells (related Fig 2).
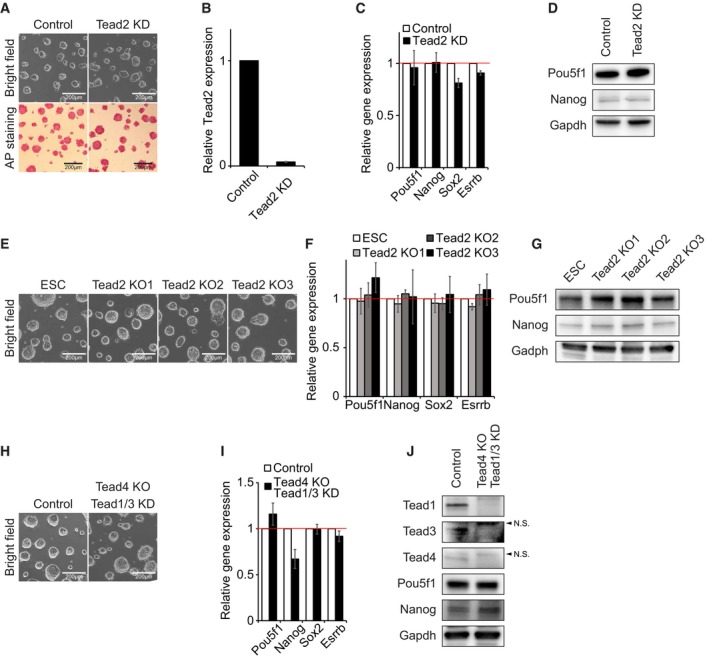
- Colony morphology and AP activity of Tead2 KD ES cells.
- mRNA levels of Tead2 measured by RT–qPCR upon shRNA‐based KD.
- mRNA expression levels of Pou5f1, Nanog, Sox2, and Esrrb upon KD of Tead2. Data are represented as mean ± SD.
- Protein levels of Pou5f1 and Nanog upon KD of Tead2.
- Colony morphology of three Tead2 KO clones (KO1‐KO3) and control ES cells.
- mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb upon KO of Tead2. Data are represented as mean ± SD.
- Protein levels of Pou5f1 and Nanog in Tead2 KO clones.
- Colony morphology of ES cells upon KO of Tead4 and KD of Tead1/3.
- mRNA levels of Pou5f1, Nanog, Sox2, and Esrrb upon KO of Tead4 and KD of Tead1/3. Data are represented as mean ± SD.
- Protein levels of Pou5f1 and Nanog in Tead4 KO and Tead1/3 KD ES cells. N.S., non‐specific bands.
Yap1 is induced and translocated into the nucleus upon differentiation of ES cells
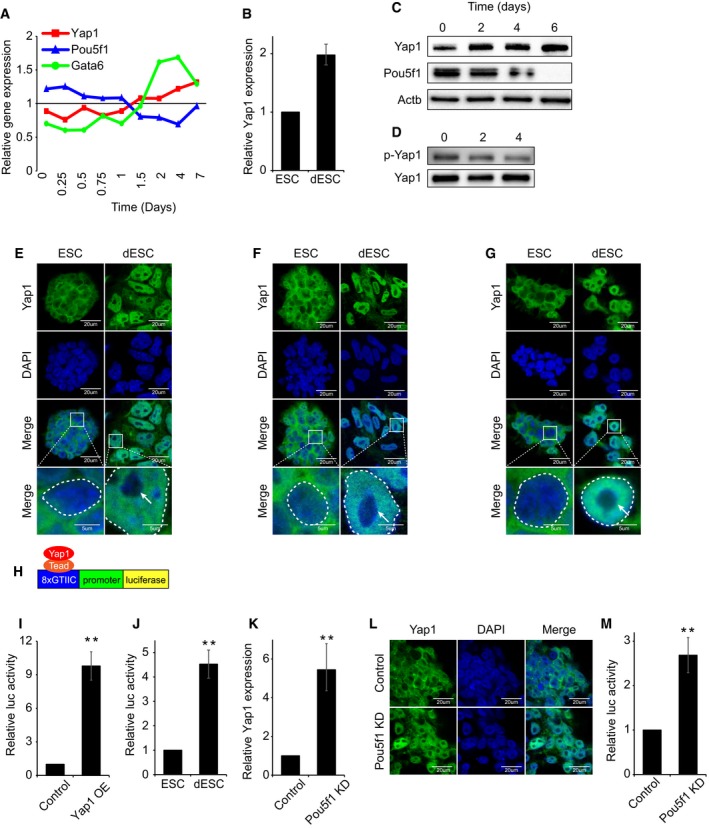
In order to investigate the roles of Yap1 in differentiation of ES cells, we examined the expression level of Yap1 in self‐renewing mouse ES cells as well as upon differentiation of ES cells. Analysis of published mRNA expression data obtained upon time‐course differentiation of embryoid body (EB) 30 revealed that Yap1 is moderately expressed in ES cells while its expression gradually increases upon differentiation (Fig 3A). We differentiated mouse J1 ES cells by the withdrawal of leukemia inhibitory factor (LIF) in the culture media and examined the level of Yap1. Consistent with the results from the EB differentiation, both mRNA and protein levels of Yap1 were moderately increased upon spontaneous differentiation (Fig 3B and C).
Figure 3. Yap1 is up‐regulated and translocated into nucleus during ES cell differentiation (see also Fig EV5).
-
ARelative mRNA levels of Yap1, Pou5f1, and Gata6 during time‐course embryoid body (EB) differentiation. Gene expression data were obtained from GSE3749. Pou5f1 and Gata6 serve as representative ES cell marker and lineage‐specific marker, respectively.
-
BRelative Yap1 mRNA levels in ES cells (ESC) and differentiating ES cells (dESC) (LIF withdrawal for 4 days) and data are represented as mean ± SD. To differentiate ES cells, cells were incubated in LIF‐withdrawn medium for 4 days. Both ESC and dESC were passaged every 2 days.
-
CProtein levels of Yap1 and Pou5f1 during time‐course differentiation upon LIF withdrawal.
-
DPhospho‐Yap1 levels during time‐course differentiation. Samples were normalized by total Yap1 level.
-
E–GImmunofluorescence (IF) images depicting localization of Yap1 in J1 (E), CJ7 (F), and E14 (G) mouse ESC (top) and dESC (bottom). The white arrow indicates nucleolus. Bottom panels represent higher magnification of the above panels. Dashed circle indicates nucleus border.
-
HA schematic diagram depicting a Yap1‐responsive luciferase reporter (8xGTIIC) construct.
-
ILuciferase reporter assay using Yap1‐responsive luciferase reporter (8xGTIIC) upon transient overexpression (OE) Yap1 in ES cells. P‐values were calculated using Student's t‐test. Data are represented as mean ± SD. **P < 0.01. “Control” indicates ES cells infected with control virus not expressing any specific shRNA sequence.
-
JRelative activity of Yap1‐responsive luciferase reporter gene in ESC and dESC. P‐values were calculated using Student's t‐test. Data are represented as mean ± SD. **P < 0.01.
-
KRelative Yap1 mRNA levels in Control and Pou5f1 KD ES cells. Data are represented as mean ± SD. **P < 0.01. “Control” indicates ES cells infected with control virus not expressing any specific shRNA sequence.
-
LIF images depicting localization of Yap1 in Control and Pou5f1 KD ES cells.
-
MRelative activity of Yap1‐responsive luciferase reporter gene upon Pou5f1 KD in ES cells. P‐values were calculated using Student's t‐test. Data are represented as mean ± SD. **P < 0.01.
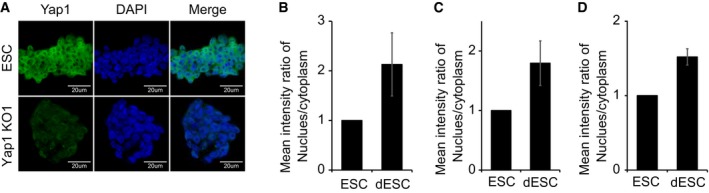
Since active Hippo signaling leads to phosphorylation and cytoplasmic sequestration of Yap1, blocking Yap1's function as a transcriptional coactivator 7, 9, 12, we examined the levels of phospho‐Yap1 and its subsequent localization in both self‐renewing and differentiating ES cells. Western blot analysis showed that Yap1 is highly phosphorylated in self‐renewing ES cells, but the level of phospho‐Yap1 is reduced in differentiating ES cells (Fig 3D). Given the fact that phospho‐Yap1 is sequestered in the cytoplasm 7, 9, 12, we examined Yap1 localization by immunofluorescence (IF). Consistent with hyper‐phosphorylation of Yap1 in ES cells, IF results revealed that Yap1 resides primarily in the cytoplasm of self‐renewing ES cells (Fig 3E–G). However, upon differentiation of multiple mouse ES cell lines we tested (J1, CJ7, and E14), Yap1 was translocated into the nucleus (Figs 3E–G and EV5A–D). Cytoplasmic Yap1 in ES cells could be attributed to compact ES cell colonies with active Hippo signaling 7, while lower cell density of differentiating ES cells growing in a monolayer leads to inactive Hippo signaling, resulting in the nuclear localization of Yap1.
Figure EV5. Yap1 is translocated into the nucleus upon differentiation of ES cells (related to Fig 3).
-
AImmunofluorescence (IF) images depicting Yap1 signals in J1 ES cells (ESC) and Yap1 KO clone.
-
B–DQuantification of relative Yap1 localization between ESC and differentiating ES cells (dESC) from three different cell lines: J1 (B), CJ7 (C), and E14 (D). See Appendix Supplementary Methods for detailed quantification method. Data are represented as mean ± SD.
We further investigated the activity of nuclear Yap1 using a synthetic Yap1‐responsive luciferase (8xGTIIC) construct as previously designed for the measurement of Yap1 transcriptional activity in mechanical stress condition (Fig 3H) 15, 19, 31, 32. The luciferase construct contains repeated Yap1‐Tead binding motifs (eight times) in front of the minimal cTNT promoter followed by a luciferase reporter gene 15, 32, 33. As shown in Fig 3I and J, we observed a significant increase in luciferase activity in both ES cells with transient OE of Yap1 and in differentiating ES cells compared to the reporter activity in self‐renewing ES cells, thereby indicating the increased level of nuclear Yap1 either by OE of Yap1 or by ES cell differentiation promotes transcription of the reporter gene. An induced level and nuclear localization of Yap1 were also confirmed, along with the increased Yap1 activity in Pou5f1 KD ES cells undergoing TE differentiation (Fig 3K–M).
Yap1 is required for normal differentiation of ES cells
As we observed increased expression levels and nuclear localization of Yap1 in differentiating ES cells (Fig 3), we hypothesized that Yap1 may have critical roles in differentiation of ES cells. To address this, we tested differentiation potential of Yap1 KD ES cells. Upon 3 days of differentiation, completely differentiated and mono‐layered cellular morphology with reduced AP activity were observed in control ES cells. However, Yap1 KD cells maintained typical colony morphology with high AP activity comparable to that of self‐renewing ES cells even after 2–3 days of differentiation (Fig 4A). We further found that expression levels of some pluripotency factors such as Sox2 and Esrrb were relatively highly maintained in Yap1‐depleted cells upon differentiation, although the expression of other core factors, Pou5f1 and Nanog, was decreased similarly to their levels in control cells upon differentiation (Fig EV6A). Moreover, up‐regulation of various lineage‐specific markers, such as Nes, T, Gsc, Gata6, Cdx2, and Gata3, was significantly impaired during differentiation of Yap1‐depleted cells (Fig EV6B), suggesting that the depletion of Yap1 affects differentiation potential of ES cells.
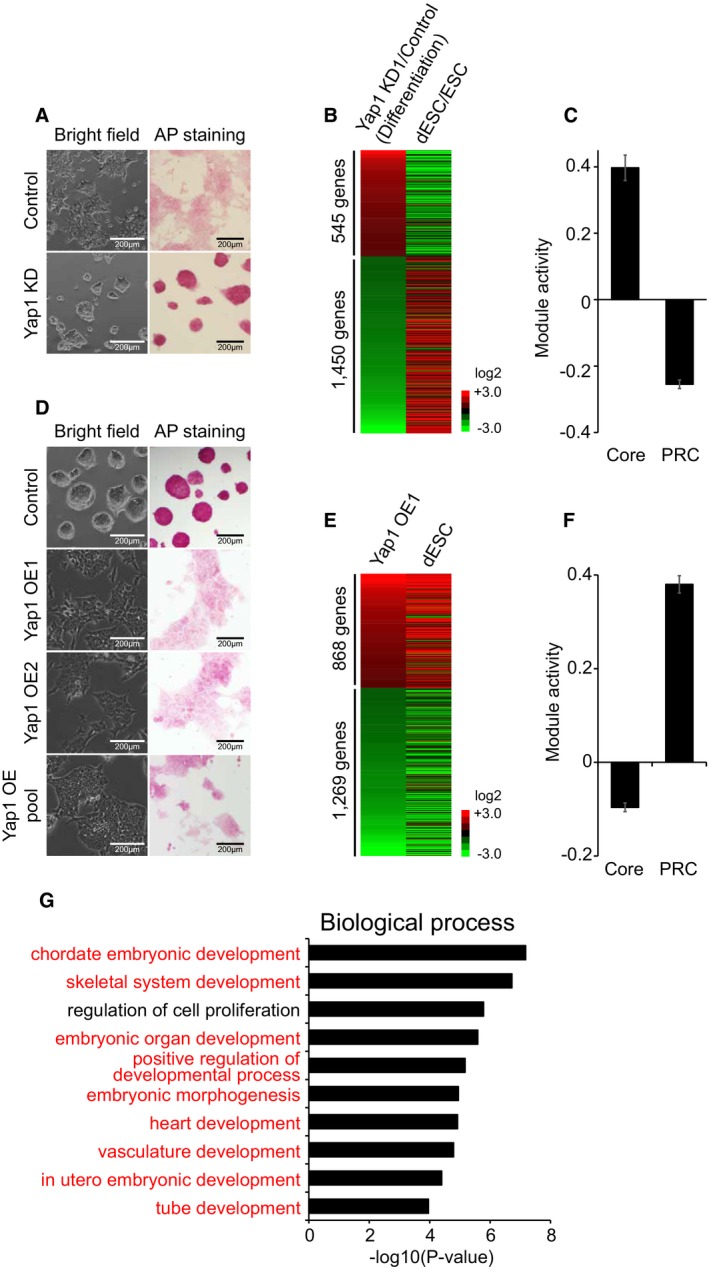
Figure 4. Yap1 is required for differentiation of ES cells (see also Fig EV6).
-
AColony morphology and AP activity of Control and Yap1 KD ES cells upon differentiation. Morphology and AP staining pictures were taken 2 days after differentiation.
-
BA heatmap showing relative mRNA expression levels of 1,995 genes differentially expressed (> twofold) between Yap1 KD ES cells and Control upon 4 days of differentiation. Genes were sorted by the fold changes of gene expression between Yap1 KD ES cells and Control (first column). Corresponding gene expression changes between ES cells (ESC) and differentiating ES cells (dESC) are shown in the second column.
-
CRelative average module activities (Core and PRC modules) between Yap1 KD ES cells and Control cells upon differentiation. Data are represented as mean ± SEM.
-
DColony morphology and AP activity in Yap1 OE cells. Two different Yap1 OE clones (OE1 and OE2) and pool of Yap1 OE (OE pool) were used. Cell morphology and AP staining pictures were taken 3 weeks after electroporation.
-
EA heatmap showing relative mRNA expression levels of 2,137 genes differentially expressed (> twofold) between Yap1 OE ES cells and control ES cells. Genes were sorted by the fold changes of gene expression between Yap1 OE ES cells and control ES cells (first column) and corresponding gene expression profiles obtained from dESC are shown.
-
FRelative average module activities (Core and PRC modules) between Yap1 OE cells and control cells are shown. Data are represented as mean ± SEM.
-
GGenes up‐regulated in Yap1 OE cells were tested using David 6.7. Significantly enriched gene ontology (GO) terms (biological functions) are shown. Developmental process‐related GO terms are highlighted in red.
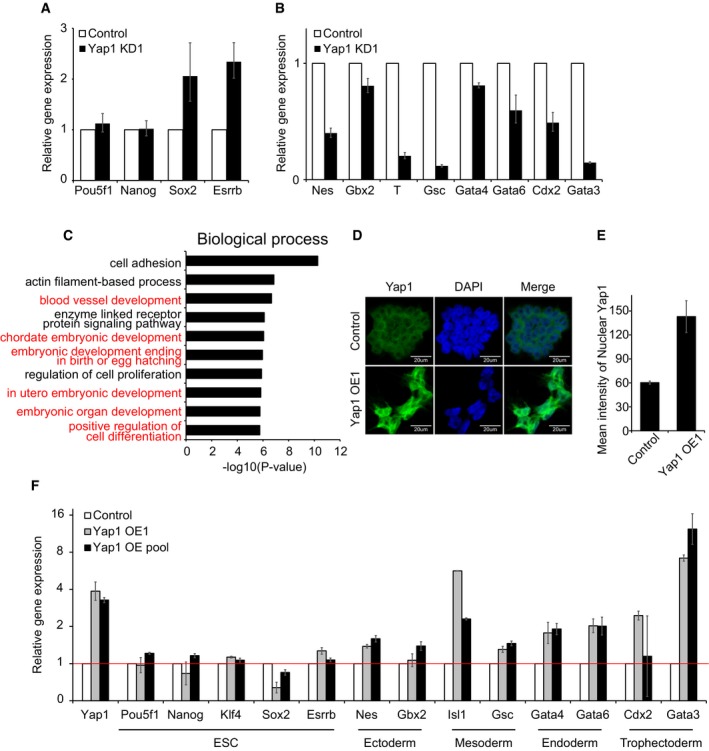
Figure EV6. Alteration of Yap1 affects differentiation of ES cells (related to Fig 4).
-
A, BmRNA levels of ES cell core factors (A) and lineage‐specific markers (B) in Yap1 KD1 cells upon 4 days of differentiation. White bars indicate the levels of genes tested in control virus‐infected ES cells (Control) upon differentiation. Data are represented as mean ± SD.
-
CBar graphs showing significantly enriched gene ontology (GO) terms (biological functions). GO analysis of genes down‐regulated in Yap1 KD cells upon differentiation was performed using David 6.7 tools. Developmental process‐related terms are highlighted in red.
-
DIF images showing localization of Yap1 in control and Yap1 OE cells.
-
EQuantification of nuclear Yap1 in control and Yap1 OE cells. Data are represented as mean ± SD.
-
FmRNA levels of ES cell (ESC) core factors and lineage‐specific marker genes in Yap1 OE cells and Yap1 OE pool. Data are represented as mean ± SD.
In order to get further insight into the roles of Yap1 in global transcriptional regulation during differentiation, we analyzed gene expression profiles obtained from RNA‐seq of normal ES cells and Yap1‐depleted ES cells before and after differentiation. As shown in Fig 4B, gene expression patterns of DEGs (Yap1 KD ES cells/wild‐type ES cells) upon differentiation showed an inverse correlation with the expression patterns of wild‐type differentiating cells over self‐renewing ES cells (Dataset EV3). The heatmap results clearly revealed that Yap1‐depleted ES cells are not properly differentiated. Additional analyses of the Core and PRC module activity consistently indicated that Yap1 depletion causes stronger Core module activity with weaker PRC module activity during differentiation, indicating that KD of Yap1 delayed or impaired proper differentiation of ES cells (Fig 4C). Gene ontology (GO) term analysis using the genes that are not properly induced in Yap1‐depleted cells compared to wild‐type ES cells upon differentiation also revealed that these genes are implicated in various development‐related processes, such as blood vessel development, chordate embryonic development, and in utero embryonic development (Fig EV6C). These collectively demonstrate that adequate levels of Yap1 are critical in normal differentiation of ES cells.
Ectopic expression of Yap1 in ES cells is sufficient to induce up‐regulation of lineage marker genes
To further test roles of Yap1 in ES cell differentiation, we performed OE of Yap1 in ES cells. Yap1 mainly resides in the cytoplasm of self‐renewing ES cells. While OE of Yap1 increases both nuclear and cytoplasmic Yap1 levels, we detected more nuclear Yap1 in Yap1 OE cells, indicating that exogenous Yap1 can translocate into the nucleus and act on its target genes (Fig EV6D and E). Yap1 OE cells also showed flattened morphology similar to that of differentiating ES cells with reduced AP activity even in the presence of LIF (Fig 4D). We further examined global gene expression profiles of Yap1 OE cells, and a clustering analysis showed that the DEGs upon OE of Yap1 (Yap1 OE/ES cells) are highly similar to the DEGs of differentiating ES cells over self‐renewing ES cells (Fig 4E and Dataset EV4). Consistently, the activity of the Core module was significantly decreased upon OE of Yap1, while the PRC module activity was dramatically increased (Figs 4F and EV6F). These results suggest that OE of Yap1 is sufficient to trigger ES cell differentiation. Additional GO term analysis revealed that genes up‐regulated upon OE of Yap1 are significantly enriched in developmental processes, such as chordate embryonic development, skeletal system development, and embryonic organ development (Fig 4G), further demonstrating that the ectopic expression of Yap1 promotes differentiation of ES cells.
Unlike the well‐established functions of the Hippo signaling pathway in the first cell fate decision, the roles of Yap1 in ES cells, ICM, and during differentiation of ES cells or ICM are still not well understood. Here, we reveal that Yap1, a transcriptional effector of Hippo pathway, is a crucial factor implicated in differentiation rather than self‐renewal of ES cells. In contrast to the previous reports 18, 19, a depletion of Yap1 does not show any significant effect on the maintenance of multiple ES cells we tested. This is consistent with the nonessential roles of Yap1 in ES cells grown under the 2i condition 21. In agreement with the dispensability of Yap1 in ES cells, other key effectors of the Hippo pathway (Tead family proteins and Taz) do not compensate Yap1, and are also not necessary for the maintenance of ES cells, implying that the transcriptional effectors of the Hippo pathway are at least dispensable for self‐renewal of mouse ES cells. In addition, we show that Yap1 is mainly sequestered in the cytoplasm of self‐renewing ES cells, while it localizes in the nucleus upon differentiation. The predominant nuclear localization of Yap1 in differentiating ES cells may be due to inactive Hippo signaling in the cells growing with lower density. Consistently, OE of Yap1 in ES cells triggers nuclear localization of Yap1 and induces differentiation along with activation of diverse lineage markers. Our global expression analyses further suggest that Yap1 may promote differentiation by activating differentiation‐related genes rather than repressing pluripotency‐related genes, which is consistent with the observation of Yap1 KO embryo, which dies around E10 due to the defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation 34. Notably, a recent study done in human ES cells suggested that YAP1 represses mesendoderm differentiation 35, possibly due to the differences in signaling pathways between human and mouse ES cells 36, 37. In‐depth investigation of the impaired regulation of three lineage‐specific genes as well as TE‐related genes in Yap1 KD ES cells upon differentiation may provide further insights into the functions of Yap1 in early embryogenesis. Together, our data establish that Yap1 is a critical regulator for proper differentiation but dispensable for self‐renewal of ES cells.
Materials and Methods
Cell culture
J1, E14, and CJ7 mouse ES cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco Ref. 11965) supplemented with 18% of fetal bovine serum (FBS), penicillin/streptomycin/L‐glutamine (Gibco Ref. 10378), MEM nonessential amino acid (Gibco Ref. 11140), nucleosides (Millipore Cat. ES‐008‐D), 100 μM β‐mercaptoethanol (Sigma M3148), and 1,000 U/ml recombinant leukemia inhibitory factor (LIF, Millipore Cat. ESG1107). ES cells were cultured on 0.1% gelatin‐coated plates at 37°C and 5% CO2 and passaged every 2 days. HEK 293T cells were maintained in DMEM supplemented with 10% of FBS and penicillin/streptomycin/L‐glutamine. To differentiate ES cells, cells were washed three times with the media without LIF and then incubated for 4 days while passaging every 2 days.
shRNA lentiviral production and infection
HEK 293T cells were plated at ~6 × 106 cells per 100 mm2 and then transfected with 6 μg of pLKO.1 shRNA vector (Sigma) (Dataset EV1), 4 μg of pCMV‐Δ8.9, and 2 μg of VSVG plasmids using 30 μl of Fugene 6 (Promega Ref. 2692), according to the manufacturer's protocol. After 24 h, HEK 293T medium was replaced with ES medium. Two days after transfection, supernatant containing viral particles was collected and filtered through 0.45‐μm Supor® membrane (PALL Ref. 4654). ~2 × 105 ES cells were plated on 12‐well plate with virus containing media supplemented with 10 μg/ml polybrene (Millipore Cat. TR‐1003‐G). After 1 day of infection, cells are selected with appropriate antibiotics and passaged every 2 days. Cell morphology, AP staining, protein and mRNA levels were examined two passages after the infection.
Luciferase reporter gene assay
For the luciferase reporter gene assay, 2.5 × 105 J1 ES cells in each 24 well were co‐transfected with 100 ng of the GTIIC vector, 5 ng of PGL4.75 vector containing a Renilla reporter gene as an internal control reporter using Lipofectamine 3000 (Life Technologies, Cat. L3000008) and then cultivated for 24 h. To measure luciferase reporter gene activity, cells were washed two times with PBS, lysed, and the luciferase activities were measured using the Dual Luciferase® assay kit (Promega, E1910).
Immunofluorescence
~3 × 105/ml ES cells were plated on 0.1% gelatin pre‐coated μ‐Slide VI0.4 (Ibidi Cat. 80606). Slides were fixed with 3.7% paraformaldehyde for 15 min at room temperature and permeabilized with 0.5% Triton X‐100 for 10 min. Slides were then incubated with blocking solution (3% BSA and 1% normal horse serum in PBS) for 1 h at room temperature, Yap1 primary antibody solution (1:200 dilution, Santa Cruz sc‐101199) overnight at 4°C, and secondary antibody solution (1:1,000 dilution) conjugated to Alexa Fluor 488 for 1 h at room temperature. Lastly, slides were mounted with ProLong® Gold antifade reagent with DAPI (Molecular Probes P36935) and imaged on a Zeiss 710 laser scanning confocal and structured illumination microscope.
RNA sequencing and data processing
One μg of RNAs was used to generate Illumina‐compatible sequencing libraries using mRNA isolation kit (NEB, E7490L) and RNA library prep kit (NEB, E7530S) according to the manufacturer's protocol. Adapter ligation was done with sample‐specific barcodes. RNA‐seq libraries were sequenced using an Illumina NextSeq 500 machine. Single‐end reads from RNA‐seq were mapped onto the mouse genome assembly (mm9) using default setting of Tophat2. Transcript‐level expression analysis was performed using Cuffdiff to calculate FPKM (fragments per kilobase of transcript per million mapped reads) 38.
Data deposition
The RNA‐seq data used in this study were deposited at the Gene Expression Omnibus (GEO) under the accession number GSE69669.
Author contributions
HWC, NU, and WS performed the experiments. HWC and B‐KL analyzed RNA‐seq data. HWC, B‐KL, JL, and JK conceived work and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Review Process File
Acknowledgements
We thank all the members of Kim laboratory for their help and support and the ICMB Microscopy and Imaging Facility as well as the Genome Sequencing and Analysis Facility (GSAF) at UT‐Austin. The project is supported by R01GM112722 from NIH/NIGMS and R1106 from the Cancer Prevention Research Institute of Texas (CPRIT) to J.K. J.K. is a CPRIT scholar.
EMBO Reports (2016) 17: 519–529
References
- 1. Kim N, Koh E (2011) E‐cadherin mediates contact inhibition of proliferation through Hippo signaling‐pathway components. Proc Natl Acad Sci USA 2011: 11930–11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR et al (2011) Yap1 acts downstream of α‐catenin to control epidermal proliferation. Cell 144: 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI (2014) Hippo signaling regulates microprocessor and links cell‐density‐dependent miRNA biogenesis to cancer. Cell 156: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silvis MR, Kreger BT, Lien W‐H, Klezovitch O, Rudakova GM, Camargo FD, Lantz DM, Seykora JT, Vasioukhin V (2011) α‐catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal 4: ra33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang J, Wu S, Barrera J, Matthews K, Pan D (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122: 421–434 [DOI] [PubMed] [Google Scholar]
- 6. Lee K‐K, Yonehara S (2012) Identification of mechanism that couples multisite phosphorylation of Yes‐associated protein (YAP) with transcriptional coactivation and regulation of apoptosis. J Biol Chem 287: 9568–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L et al (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17: 2054–2060 [DOI] [PubMed] [Google Scholar]
- 9. Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N et al (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16: 398–410 [DOI] [PubMed] [Google Scholar]
- 10. Hirate Y, Hirahara S, Inoue K‐I, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K et al (2013) Polarity‐dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol 23: 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis A‐K, de la Pompa JL et al (2014) Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 30: 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cockburn K, Biechele S, Garner J, Rossant J (2013) The Hippo pathway member Nf2 is required for inner cell mass specification. Curr Biol 23: 1195–1201 [DOI] [PubMed] [Google Scholar]
- 13. Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, Ralston A (2014) HIPPO pathway members restrict Sox2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 10: e1004618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Imajo M, Ebisuya M, Nishida E (2014) Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol 17: 7–19 [DOI] [PubMed] [Google Scholar]
- 15. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183 [DOI] [PubMed] [Google Scholar]
- 16. Ota M, Sasaki H (2008) Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069 [DOI] [PubMed] [Google Scholar]
- 17. Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y (2010) Structural insights into the YAP and TEAD complex. Genes Dev 24: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R et al (2010) The role of YAP transcription coactivator in regulating stem cell self‐renewal and differentiation. Genes Dev 24: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamm C, Böwer N, Annerén C (2011) Regulation of mouse embryonic stem cell self‐renewal by a Yes‐YAP‐TEAD2 signaling pathway downstream of LIF. J Cell Sci 124: 1136–1144 [DOI] [PubMed] [Google Scholar]
- 20. Varelas X, Samavarchi‐Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL (2010) The Crumbs complex couples cell density sensing to Hippo‐dependent control of the TGF‐β‐SMAD pathway. Dev Cell 19: 831–844 [DOI] [PubMed] [Google Scholar]
- 21. Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V et al (2014) YAP/TAZ incorporation in the β‐catenin destruction complex orchestrates the Wnt response. Cell 158: 157–170 [DOI] [PubMed] [Google Scholar]
- 22. Ying Q‐L, Stavridis M, Griffiths D, Li M, Smith A (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186 [DOI] [PubMed] [Google Scholar]
- 23. Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D (2001) Direct Neural Fate Specification from Embryonic Stem Cells. Neuron 30: 65–78 [DOI] [PubMed] [Google Scholar]
- 24. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA‐guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH (2010) A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Home P, Saha B, Ray S, Dutta D, Gunewardena S, Yoo B, Pal A, Vivian JL, Larson M, Petroff M et al (2012) Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci USA 109: 7362–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang C‐Y, Chinnaiyan AM et al (2008) TEAD mediates YAP‐dependent gene induction and growth control. Genes Dev 22: 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao B, Kim J, Ye X, Lai ZC, Guan KL (2009) Both TEAD‐binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes‐associated protein. Cancer Res 69: 1089–1098 [DOI] [PubMed] [Google Scholar]
- 30. Hailesellasse SK, Porter CJ, Palidwor G, Perez‐Iratxeta C, Muro EM, Campbell PA, Rudnicki MA, Andrade‐Navarro MA (2007) Gene function in early mouse embryonic stem cell differentiation. BMC Genom 8: 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao JH, Davidson I, Matthes H, Garnier J‐M, Chambon P (1991) Cloning, expression, and transcriptional properties of the human enhancer factor TEF‐1. Cell 65: 551–568 [DOI] [PubMed] [Google Scholar]
- 32. Mahoney W, Hong J, Yaffe M, Farrance I (2005) The transcriptional co‐activator TAZ interacts differentially with transcriptional enhancer factor‐1 (TEF‐1) family members. Biochem J 225: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Farrance I, Mar J, Ordahl C (1992) M‐CAT binding factor is related to the SV40 enhancer binding factor, TEF‐1. J Biol Chem 267: 17234–17240 [PubMed] [Google Scholar]
- 34. Morin‐Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL (2006) Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol 26: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL (2013) Switch enhancers interpret TGF‐β and hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5: 1611–1624 [DOI] [PubMed] [Google Scholar]
- 36. Amit M, Carpenter MK, Inokuma MS, Chiu C‐P, Harris CP, Waknitz MA, Itskovitz‐Eldor J, Thomson JA (2000) Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 227: 271–278 [DOI] [PubMed] [Google Scholar]
- 37. Nichols J, Chambers I, Taga T, Smith A (2001) Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128: 2333–2339 [DOI] [PubMed] [Google Scholar]
- 38. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Review Process File


