Abstract
Mis18 is a key regulator responsible for the centromere localization of the CENP‐A chaperone Scm3 in Schizosaccharomyces pombe and HJURP in humans, which establishes CENP‐A chromatin that defines centromeres. The molecular and structural determinants of Mis18 centromere targeting remain elusive. Here, by combining structural, biochemical, and yeast genetic studies, we show that the oligomerization of S. pombe Mis18, mediated via its conserved N‐terminal Yippee‐like domain, is crucial for its centromere localization and function. The crystal structure of the N‐terminal Yippee‐like domain reveals a fold containing a cradle‐shaped pocket that is implicated in protein/nucleic acid binding, which we show is required for Mis18 function. While the N‐terminal Yippee‐like domain forms a homodimer in vitro and in vivo, full‐length Mis18, including the C‐terminal α‐helical domain, forms a homotetramer in vitro. We also show that the Yippee‐like domains of human Mis18α/Mis18β interact to form a heterodimer, implying a conserved structural theme for Mis18 regulation.
Keywords: CENP‐A, centromere, epigenetics, Mis18, Yippee
Subject Categories: Cell Cycle; DNA Replication, Repair & Recombination; Structural Biology
Introduction
The accurate distribution of genetic information to daughter cells during cell division relies on the physical attachment of chromosomes to spindle microtubules mediated by kinetochores. Kinetochores are large protein assemblies deposited at specific chromosomal loci known as centromeres 1, 2, 3. Defective centromere function results in chromosome segregation errors that can contribute to genomic instability implicated in cancer 4. Hence, understanding the molecular mechanisms that promote kinetochore establishment and maintenance at centromeres is of prime importance.
The location of most eukaryotic centromeres is determined by the assembly of specialized chromatin composed of nucleosomes in which canonical histone H3 is replaced by the centromere‐specific H3 variant CENP‐A in vertebrates and Cnp1 (CENP‐ACnp1) in Schizosaccharomyces pombe 3, 5. Thus, the establishment and maintenance of kinetochores requires CENP‐A to be recruited to and deposited at centromeres. In S. pombe, CENP‐ACnp1 is specifically incorporated into chromatin over the central domain of endogenous centromeres where it is flanked by heterochromatin formed on outer repeat elements 6, 7.
During S phase, CENP‐ACnp1 levels at fission yeast centromeres are halved following DNA replication. Subsequently, CENP‐ACnp1 levels are replenished during G2 phase of the cell cycle 8. In early mitosis, the Mis18 complex comprising Mis18, Mis16, Eic1/Mis19/Kis1, and Eic2/Mis20, along with the CENP‐ACnp1 chaperone Scm3 (counterpart of vertebrate HJURP; HJURPScm3), dissociates from centromeres and re‐associates in mid/late anaphase following chromosome segregation 9, 10, 11, 12, 13, 14, 15. Exclusion of the Mis18 complex and HJURPScm3 from centromeres during mitosis likely provides an opportunity for the CENP‐ACnp1 loading cycle to reset and thereby prevent continual CENP‐ACnp1 deposition 16. HJURPScm3 directly interacts with CENP‐ACnp1 and is essential for the deposition of new CENP‐ACnp1 at centromeres 14. Genetic studies have shown that Mis16, Mis18, and Eic1/Mis19/Kis1 are essential genes that are required for the localization of HJURPScm3 to centromeres and hence CENP‐ACnp1 maintenance at centromeres 10, 11, 13, 14, 17.
Most of the components of the CENP‐A assembly pathway are conserved among eukaryotes, but with a few striking differences. Humans possess two isoforms of Mis18 (Mis18α and Mis18β) and Mis16 (RbAp46 and RbAp48) 10. During telophase, the human Mis18 complex comprising Mis18α, Mis18β, and Mis18BP1/KNL2, along with RbAp46 and RbAp48, associates with centromeres 17. Mis18BP1/KNL2 has no detectable S. pombe homolog, but Eic1/Mis19/Kis1 appears to perform an analogous function 11, 13. As in S. pombe, the human Mis18 complex is required for HJURP recruitment to centromeres, where CENP‐A is deposited during early G1 rather than G2 9. In addition, the stable incorporation of CENP‐A at human centromeres requires the small GTPase activity of Cdc42 regulated by MgcRacGap/ECT2 18.
Although key conserved players involved in the assembly and maintenance of CENP‐A chromatin at centromeres have been identified, the molecular mechanisms through which they exert their function remain unclear. Mis18 is critical for the specification of centromeres from fission yeast to humans, however, what allows Mis18 to regulate centromere specification remains largely unknown. To gain insights into the structural features of S. pombe Mis18 that allow it to bind centromeres and promote HJURPScm3 recruitment and CENP‐ACnp1 assembly, we determined the crystal structure of its highly conserved “Yippee‐like” N‐terminal globular domain. Our structural and biochemical analyses reveal that the Mis18 “Yippee‐like” domain possesses a fold that is implicated in protein/nucleic acid binding and that this domain has an innate tendency to homodimerize both in vitro and in vivo. However, full‐length Mis18 forms a homotetramer in vitro, highlighting a role for the C‐terminal α‐helical domain in influencing the overall oligomeric state of the protein. Genetic analyses using structure‐guided mutants demonstrate that dimerization of the “Yippee‐like” domain is essential for the centromere localization and hence the function of Mis18.
Results and Discussion
spMis18 possesses an N‐terminal Yippee‐like globular domain that is implicated in protein/nucleic acid binding
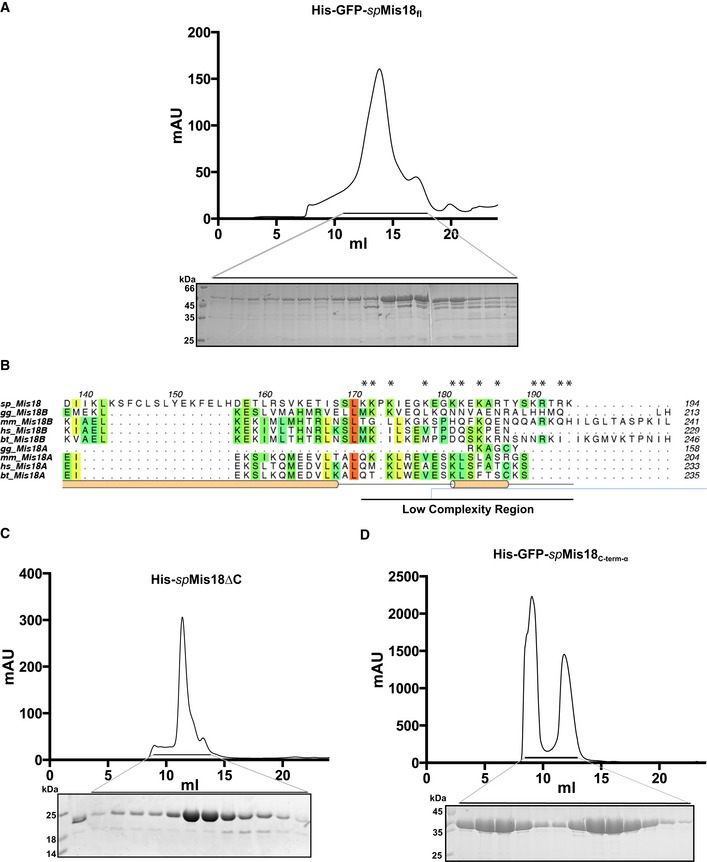
Amino acid sequence and the predicted secondary structure analysis of Schizosaccharomyces pombe Mis18 suggested the presence of a highly conserved N‐terminal globular domain (residues 1–120; spMis181–120) mainly comprised of β‐strands, followed by a moderately conserved C‐terminal α‐helical domain (residues 121–end; spMis18C‐term‐α) (Fig 1A and B). Mis181–120 shares about 20% sequence similarity with a putative Zn2+‐binding protein of unknown function named Yippee, originally identified in Drosophila, that is well conserved from yeast to humans 19. spMis181–120 has two conserved C‐X‐X‐C motifs, which are signature motifs present in metal ion‐binding proteins. Previous analysis showed that mutations within the C‐X‐X‐C motifs of human Mis18α perturb its centromere localization, highlighting the essential role of this domain 17. To structurally characterize spMis181–120, we purified recombinant spMis181–120 as a mono‐disperse sample and obtained crystals that diffracted X‐rays to about 2.6 Å (Fig 1C). X‐ray fluorescence scans of the crystals revealed the presence of bound Zn2+ ions. The structure was determined by single anomalous dispersion (SAD) exploiting the Zn anomalous signal from the bound Zn2+ ions in the crystals. The refined structure has an R factor of 22 and R‐free factor of 26 with good stereochemistry (Table 1 and Fig EV1A). The final model includes amino acid residues 19–118 of spMis181–120 (Fig 1D). The N‐terminal 18 and C‐terminal 2 (119 and 120) residues are presumably disordered and hence could not be modeled.
Figure 1. spMis18 possesses a highly conserved N‐terminal Yippee‐like globular domain and a C‐terminal α‐helical domain.
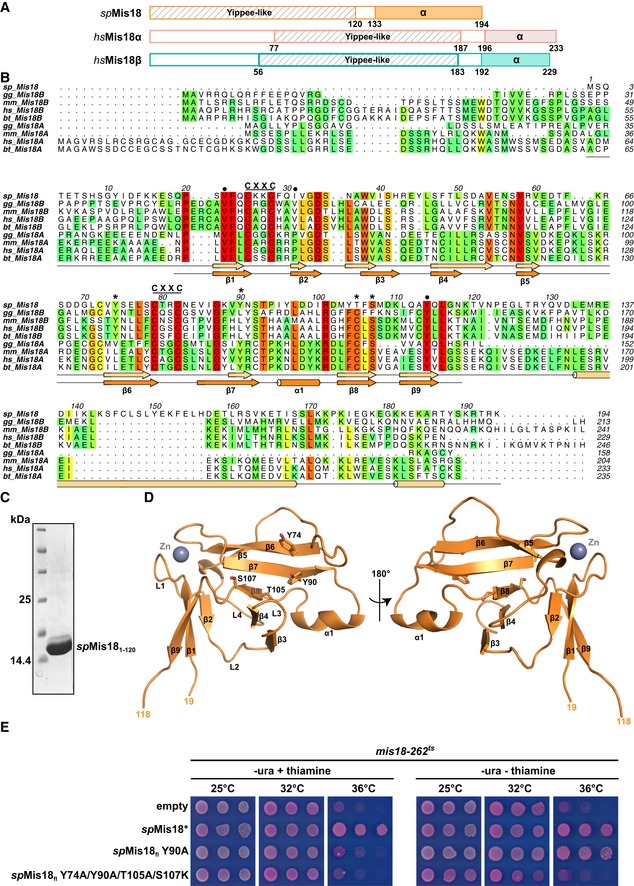
- Schematic representation of domain organization of Mis18 proteins from Schizosaccharomyces pombe and human (as suggested by the Conserved Domain Database (CDD) and secondary structure predictions).
- Amino acid conservation of Mis18 proteins among eukaryotes. The alignment includes orthologs from S. pombe (sp), G. gallus (gg), M. musculus (mm), H. sapiens (hs), and B. taurus (bt). Predicted (light orange; using PsiPred, http://bioinf.cs.ucl.ac.uk/psipred) and observed (bright orange; from the crystal structure shown in D) secondary structure elements are shown below the aligned sequences. Amino acid residues mutated in this study are highlighted with circles (dimer interface residues) and asterisks (putative substrate‐binding pocket residues).
- SDS–PAGE showing a representative fraction of the purified spMis18 N‐terminal Yippee‐like globular domain (amino acid residues 1–120; spMis181–120).
- Cartoon representation of the crystal structure of spMis181–120 in two different orientations, highlighting key residues within the putative substrate‐binding pocket.
- Substrate‐binding pocket mutations Y90A and Y74A/Y90A/T105A/S107K affect the ability of ectopically expressed spMis18fl to rescue the temperature sensitivity of mis18‐262 cells to varying degrees. Fivefold serial dilutions of mis18‐262 cells transformed with plasmids harboring the indicated spMis18fl constructs, spotted on PMG − uracil + phloxine B media supplemented with (repressed) or without (expressed) thiamine, and incubated at the indicated temperatures; dead cells stain dark pink.
Table 1.
Data collection, phasing, and refinement statistics
| Dataset I | Dataset II | |
|---|---|---|
| Data collection | ||
| Space group | P3112 | P3112 |
| Cell dimensions | ||
| a, b, c (Å) | 122.18, 122.18, 73.48 | 121.07, 121.07, 73.21 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Peak | ||
| Wavelength | 1.2825 | 0.97625 |
| Resolution (Å) | 47.0–3.9 (4.0–3.9) | 104.9–2.6 (2.8–2.6) |
| R merge | 9.5 (98.3) | 6.5 (57.3) |
| I/σI | 15.3 (3.7) | 9.7 (1.8) |
| Completeness (%) | 97.2 (93.9) | 99.9 (99.9) |
| Redundancy | 9.9 (10.3) | 4.1 (4.1) |
| Refinement | ||
| Resolution (Å) | 39.6–2.6 | |
| No. reflections | 17,128 | |
| R work/R free | 22.0/26.0 | |
| No. atoms | ||
| Protein | 2,374 | |
| B‐factors | ||
| Protein | 98.0 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.75 | |
| Ramachandran values | ||
| Favored (%) | 93 | |
| Disallowed (%) | 1 | |
Values in parentheses are for highest‐resolution shell.
Figure EV1. Structural characterization of the N‐terminal Yippee‐like globular domain of spMis18.
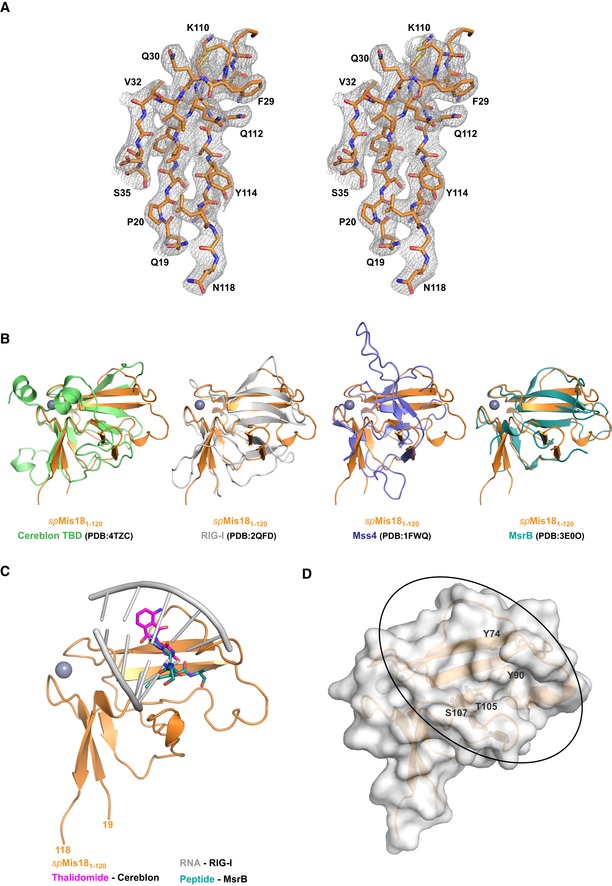
- Stereo image of electron density map (2F o–F c, contoured at 1σ) corresponding to β‐sheet I.
- Structural superposition of spMis181–120 onto its closest structural homologs, Cereblon TBD (PDB: 4TZC), RIG‐I (PDB: 2QFD), Mss4(PDD: 1FWQ), and MsrB (PDB: 3E0O). Structural superpositions were carried out using PDBefold web server (http://www.ebi.ac.uk/msd-srv/ssm).
- Structural superposition of spMis181–120 with its structural homologs bound to their substrates. For clarity, structural homologs of spMis181–120 are not shown.
- Surface representation of spMis181–120 where the cradle‐shaped pocket and amino acid residues mutated in the complementation assay (Fig 1E) are highlighted.
The overall fold of spMis181–120 is formed by antiparallel β‐sheets: a three‐stranded (β1‐β2‐β9: β‐sheet I) and a six‐stranded (β3‐β4‐β8‐β7‐β6‐β5: β‐sheet II) sheet, arranged approximately perpendicular to each other (Fig 1D). The two β‐sheets are held together by a Zn2+ ion coordinated via the C‐X‐X‐C motifs from loops L1 and L5 (Fig 1D). Structural comparison of spMis181–120 with the available structures in the protein data bank (PDB) identified the thalidomide‐binding domain of Cereblon (PDB: 4tzc; Q‐score 20: 0.47; RMSD: 2.12 Å), a component of an E3 ubiquitin ligase complex implicated in DNA repair, replication, and transcription 21, as the closest structural homolog (Fig EV1B). Other proteins that share a similar fold include RIG‐I (PDB: 2qfd; Q‐score: 0.38; RMSD: 2.19 Å), a nucleic acid‐binding protein involved in innate anti‐viral immunity 22; Mss4 (PDB: 1fwq; Q‐score: 0.34; RMSD: 2.03 Å), a guanine nucleotide exchange factor 23; and MsrB (methionine sulfoxide reductase‐B, PDB: 3e0o; Q‐score: 0.32; RMSD: 1.81 Å), an oxidative reductase implicated in aging 24 (Fig EV1B). This is in agreement with a recent bioinformatics study suggesting an evolutionary relationship between Cereblon, Yippee, and Mis18 proteins 25. Although these proteins recognize substrates as diverse as nucleic acids to proteins, they do so via a common cradle‐shaped binding pocket formed by β‐sheet II (Fig EV1C and D). This observation suggested that the putative substrate‐binding site of spMis181–120 might play an important role in Mis18 function. To test whether the putative substrate‐binding pocket was required for spMis18 function in vivo, we tested the ability of additional spMis18 expressed from a plasmid to complement the growth phenotype of mis18‐262 (G117D) cells, which exhibit loss of function for spMis18 at the restrictive temperature (36°C) 10. While expression of wild‐type spMis18 restored growth at 36°C, expressing the pocket mutant (Y74A/Y90A/T105A/S107K, Figs 1D and EV1D) failed to complement the loss of spMis18 function, demonstrating the requirement of this pocket for Mis18 function (Fig 1E).
spMis181–120 forms a homodimer
The asymmetric unit of the spMis181–120 crystals contained three copies of spMis181–120 assembled in a linear arrangement via two different interfaces (interface I and interface II), resulting in two potential dimeric structures (dimer I and dimer II) (Fig 2A). While the interface stabilizing dimer I is formed by the stacking of loops L5 and L7 of one monomer over their dimeric counterpart, the dimer II interface is formed by the stacking of β‐sheet I (Fig 2A). The oligomeric assembly observed in the crystals prompted us to characterize the oligomeric structure of spMis181–120 in solution. The molecular weight of spMis181–120, as measured using SEC‐MALS (size exclusion combined with multi‐angle light scattering), was 33,338 Da. Given that the calculated molecular weight of an spMis181–120 monomer is 16,160 Da whereas a dimer would be 32,320 Da, this analysis independently confirms that spMis181–120 forms a dimer in solution (Figs 2B and EV2A).
Figure 2. Yippee‐like globular domains of Mis18 proteins possess an intrinsic preference to form homo/heterodimers.
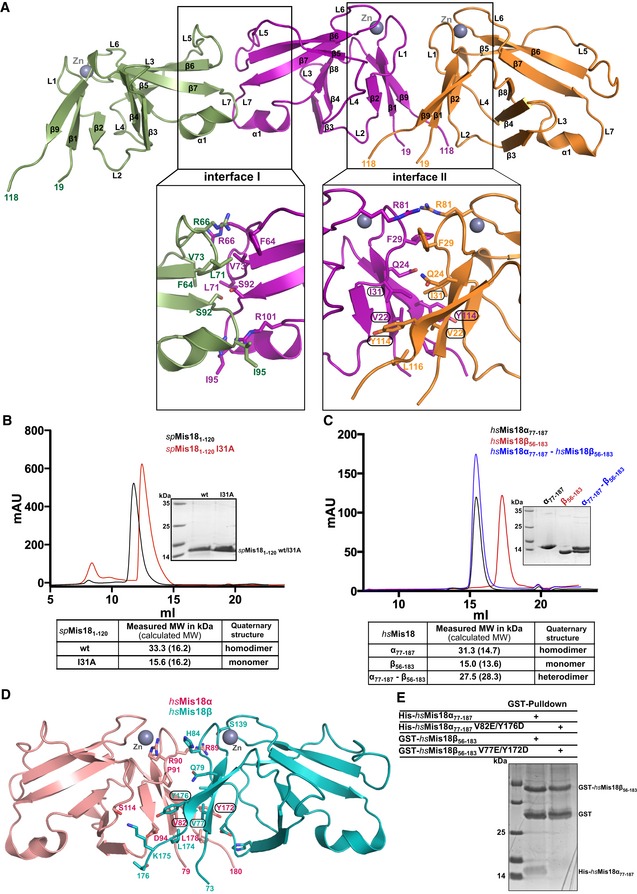
- Cartoon representation of the assembly of spMis18 Yippee‐like domains (spMis181–120) as observed in the crystal asymmetric unit. Close‐up views highlight the amino acid composition and interactions stabilizing the dimeric arrangements. Interface I is composed of conserved residues and an extensive binding surface as compared to interface I (see text for details), suggesting dimer II to be a physiologically relevant dimer. Residues mutated in this study are highlighted with circles.
- Size‐exclusion chromatography (SEC) and SEC‐MALS (SEC combined with multi‐angle light scattering) analyses of the recombinant wild‐type (wt) and dimer‐disrupting mutant (I31A) of spMis181–120. Measured molecular weights from SEC‐MALS confirm that spMis181–120 wt is a dimer in solution and spMis181–120I31A is a monomer.
- Evaluation of the ability of Yippee‐like domains of human Mis18 proteins (hsMis18α77–187 and hsMis18β56–183) to form a heterodimer. SEC and SEC‐MALS analyses demonstrate that while hsMis18α77–187 elutes at 15.5 ml as a dimer (31.3 kDa), hsMis18β56–183 elutes at 17.3 ml as a monomer (15.0 kDa). Purified hsMis18α77–187 and hsMis18β56–183 when mixed together elute at 15.4 ml as a heterodimer (27.5 kDa).
- Cartoon representation of the structure of the homology‐modeled human Mis18α77–187–Mis18β56–183 heterodimer using the crystal structure of spMis181–120 reported here as a template. Residues mutated in this study are highlighted with circles. Modeling was carried out using Phyre2 web server (www.sbg.bio.ic.ac.uk/phyre2/).
- SDS–PAGE analysis of the GST pull‐down assay where wt and dimer‐disrupting mutants of hsMis18α77–187 and hsMis18β56–183 were co‐expressed as His‐ and GST‐tagged proteins, respectively, in E. coli. While wt GST‐hsMis18β56–183 showed interaction with wt His‐hsMis18α77–187, hsMis18 proteins harboring dimer‐disrupting mutations, GST‐hsMis18β56–183V77E/Y172D, and His‐hsMis18α77–187V82E/Y176D did not show noticeable interaction. The corresponding Ni‐NTA pull‐downs showing the expression of His‐tagged proteins are shown in Fig EV2D.
Figure EV2. Evaluation of the dimerization ability of Yippee‐like domains of Mis18 proteins.
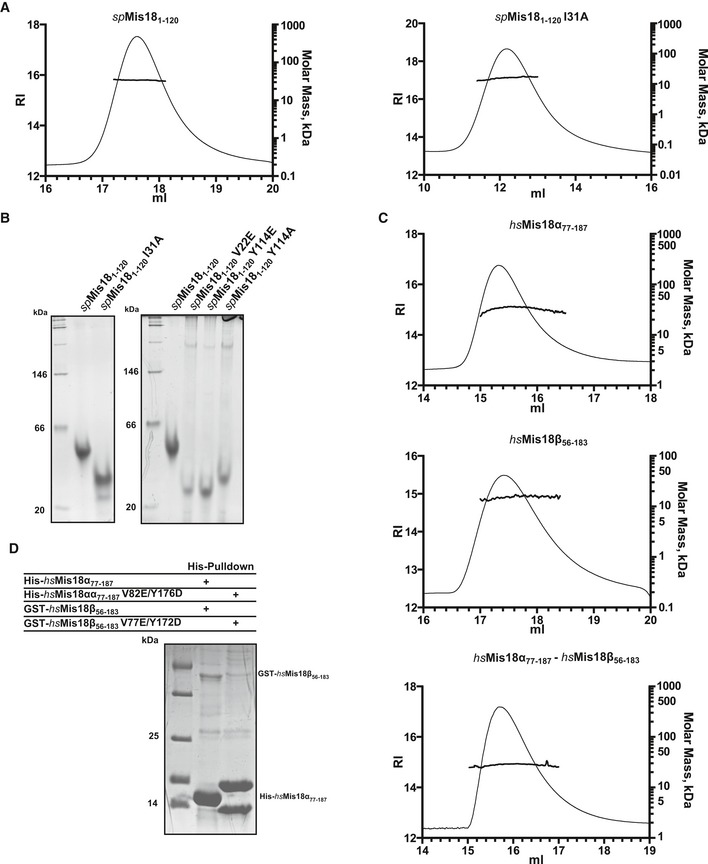
- SEC‐MALS profiles of His‐spMis181–120 (left panel) and His‐spMis181–120I31A (right panel). Refractive index (RI, left y‐axis) and molar mass (MW, right y‐axis) profiles show that His‐spMis181–120 (predicted MW of a monomer: 16.2 kDa) is a dimer (measured MW: 33.3 kDa) and His‐spMis181–120I31A (predicted MW of monomer: 16.2 kDa) is a monomer (measured MW: 15.6 kDa).
- Native PAGE analysis of dimer interface II mutants. Interface mutants I31A, V22E, Y114E, and Y114A migrated faster than wt spMis181–120, confirming the effect these mutations have on the oligomeric structure of spMis181–120.
- SEC‐MALS profiles of His‐hsMis18α77–187 (top panel), His‐hsMis18β56–183 (middle panel), and His‐hsMis18α77–187–His‐hsMis18β56–183 (bottom panel). While His‐hsMis18α77–187 (predicted MW of a monomer: 14.7 kDa) formed a homodimer (measured MW: 31.3 kDa), His‐hsMis18β56–183 (predicted MW of a monomer: 13.6 kDa) remained as a monomer (measured MW: 15.0 kDa). His‐hsMis18α77–187–His‐hsMis18β56–183 (predicted MW of a monomer: 28.3 kDa) formed a heterodimer (measured MW: 27.5 kDa).
- SDS–PAGE analysis of Ni‐NTA pull‐down experiment, where His‐hsMis18α77–187 and GST‐hsMis18β56–183 were co‐expressed in E. coli as wt or dimer‐disrupting mutant proteins. While the GST pull‐down assay shown in Fig 2E confirmed the inability of GST‐hsMis18β56–183V77E/Y172D to interact with His‐hsMis18α77–187V82E/Y176D, the corresponding Ni‐NTA pull‐down shown here confirms the abundant expression of His‐hsMis18α77–187V82E/Y176D in the input. We note that His‐hsMis18α77–187V82E/Y176D migrates as a doublet. Western blot analysis (data not shown) using anti‐His antibody confirmed the presence of His‐tag on both bands suggesting the unstable nature of His‐hsMis18α77–187V82E/Y176D mutant (possibly due to its inability to homo‐ or heterodimerize).
To identify the physiologically relevant dimer, we compared the extent of amino acid conservation of the interface residues and the buried surface area within the dimer I and dimer II interfaces. Dimer I is mainly stabilized by poorly conserved amino acid residues (F64, R66, L71, V73, S92, and I95 with the exception of R101) and involves 1,077 Å2 buried surface area. In contrast, dimer II is stabilized by highly conserved residues (V22, I31, Y114, and L116) and has an extensive binding interface (as compared to dimer I) with a 1,431 Å2 buried surface area. Based on this observation, we reasoned that dimer II is likely the physiologically relevant dimer. To test the basis of dimer formation, we generated several single point mutations at the dimer II interface (I31A, Y114A, Y114E, V22E) and determined whether they perturb the ability of spMis181–120 to dimerize. In size‐exclusion chromatography (Fig 2B), recombinant spMis181–120I31A mutant protein eluted later (Ve = 12.4 ml) than wild‐type spMis181–120 (Ve = 11.8 ml), suggesting that this mutant protein forms a smaller entity than wild‐type. Further analysis of spMis181–120I31A using SEC‐MALS confirmed that it is monomeric with a measured molecular weight of 15,955 Da (calculated molecular weight of a monomer is 16,160 Da) (Figs 2B and EV2A). Consistent with this, the other dimer II interface mutants Y114A, Y114E, and V22E also behaved as smaller entities compared to the wild‐type protein when analyzed by native PAGE (Fig EV2B), thus demonstrating that the dimerization of spMis181–120 is mediated via the dimer II interface (Fig 2A).
Yippee‐like globular domains of Mis18 proteins possess an intrinsic ability to form dimers
To test whether Yippee‐like globular domains of other Mis18 orthologs also display an intrinsic ability to form dimers, we expressed and purified recombinant fragments of human Mis18α (residues 77–187; hsMis18α77–187) and Mis18β (56–183; hsMis18β56–183) containing their Yippee‐like globular domains. SEC and SEC‐MALS analyses were conducted to assess their ability to form oligomers. While hsMis18α77–187 eluted as a dimer at 15.5 ml in SEC with a measured molecular weight of 31,254 Da in SEC‐MALS (calculated MW of a dimer is 29,324 Da), the corresponding values for hsMis18β56‐183 were 17.3 ml and 14,993 Da (calculated MW of a monomer is 13,591 Da), respectively, indicating that hsMis18β56–183 exists as a monomer in solution (Fig 2C). We next tested whether hsMis18α77–187 and hsMis18β56–183 could interact to form a heterodimer, by mixing equimolar quantities of recombinant hsMis18α77–187 and hsMis18β56–183. In SEC, hsMis18β56–183 co‐elutes with hsMis18α77–187 at 15.4 ml suggesting that they form a heterodimer (Fig 2C). The molecular weight of this entity as measured by SEC‐MALS was 27,476 Da, confirming that hsMis18α and hsMis18β can heterodimerize through their respective Yippee‐like domains (Figs 2C and EV2C).
To test whether the mode of dimerization (dimer II mediated) is conserved from fission yeast to humans, we generated a homology model of the human hsMis18α77–187–hsMis18β56–183 heterodimer using the spMis181–120 crystal structure described above as a template (Phyre2 server: http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) (Fig 2D). Our analysis of the modeled dimer interface (containing Mis18α residues Val 82, Arg 89, Pro 91, Asp 94, Tyr 176, and Leu 178, and Mis18β residues Val 77, His 84, Val 86, His 92, Tyr 172, Leu 174, Lys 175, and Thr 176) did not show any steric clashes and involved 1,387 Å2 buried surface area, similar to the spMis181–120 dimer II interface. Moreover, mutations at the dimer interface (Mis18α V82E/Y176D and Mis18β V77E/Y172D) were sufficient to perturb the ability of hsMis18α77–187 and hsMis18β56–183 to form a heterodimer (Figs 2E and EV2D). This confirms that the Yippee‐like domains within Mis18 proteins employ a conserved mode of dimerization.
Structural and biochemical analyses of Yippee‐like domains from other proteins (Cereblon, RIG‐I and MsrB) have so far yielded no direct evidence that they possess an innate tendency to oligomerize in vitro. We therefore refer to the Yippee‐like domain of Mis18 orthologs as Mis eighteen Dimerization in Yippee (MeDiY) domain hereafter.
The C‐terminal α‐helical domain induces tetramerization of S. pombe Mis18
In addition to the N‐terminal MeDiY domain, Mis18 orthologs possess a C‐terminal α‐helical domain spMis18C‐term‐α (aa residues 121–end). To test whether spMis18C‐term‐α can influence the overall oligomeric state of the protein, we purified recombinant full‐length spMis18 (spMis18fl). Obtaining intact samples of spMis18fl proved difficult either with or without His/His‐GFP tag, as it was sensitive to degradation from the C‐terminus (Fig EV3A). The stable partially degraded His‐GFP‐spMis18fl when analyzed by SEC‐MALS appeared to form a tetramer (Fig 3A). Mass spectrometric peptide sequence coverage analysis of this sample revealed the loss of approximately 20 amino acids at the C‐terminus. Close examination of the amino acid sequence revealed the presence of a low‐complexity region at the extreme C‐terminus (amino acid residues 171–end, 15 out of 20 amino acid residues being Lys/Arg) that is unique to spMis18 (Fig EV3B). We therefore expressed and purified recombinant spMis18 with a C‐terminal truncation (spMis18ΔC; amino acids 1–168; Fig EV3C). SEC‐MALS analysis of spMis18ΔC revealed that it predominantly formed a tetramer (Fig 3B) and demonstrated a role for the α‐helical region downstream of the MeDiY domain (residues 121–168; Fig 1B) in spMis18 tetramerization. Since this α‐helical region is structurally conserved (based on secondary structure prediction; Fig 1B) between Mis18 proteins from evolutionarily distant eukaryotes, we propose that Mis18 oligomerization (either homo or hetero) mediated through the C‐terminus is also likely to be highly conserved.
Figure EV3. Characterization of the overall oligomeric state of spMis18fl .
-
ASEC profile and respective SDS–PAGE analysis of fractions (bottom panel) for His‐GFP‐spMis18fl. Superose 6 10/300 column was used for His‐GFP‐spMis18fl.
-
BMultiple sequence alignment of spMis18 with its orthologs highlights the presence of a low‐complexity region (Lys/Arg‐rich region) at the extreme C‐terminus unique to spMis18.
-
C, DSEC profiles (top panels) and respective SDS–PAGE analyses of their fractions (bottom panels) for His‐spMis18ΔC and His‐GFP‐spMis18C‐term‐α, respectively. Superdex 200 increase 10/300 column was used for His‐spMis18ΔC and His‐GFP‐spMis18C‐term‐α.
Figure 3. spMis18fl forms a tetramer in vitro .
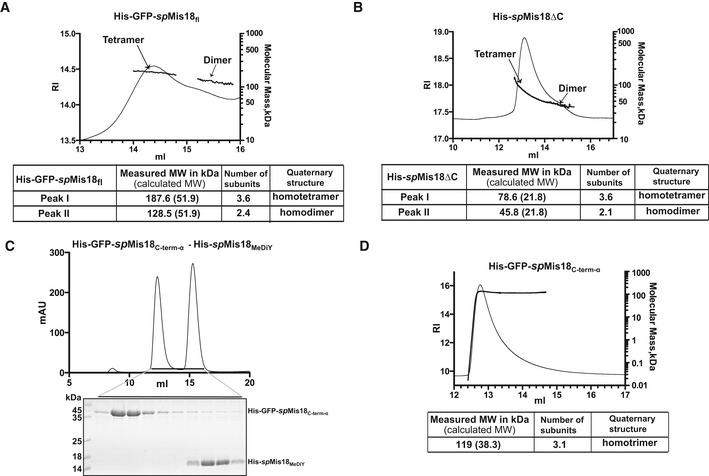
-
A, BSEC‐MALS analysis of His‐GFP‐spMis18fl and His‐spMis18ΔC, respectively. Refractive index (RI, left y‐axis) and molecular mass (kDa, right y‐axis) profiles show that both His‐GFP‐spMis18fl (predicted MW of a monomer is 51.9 kDa) and His‐spMis18ΔC (predicted MW of a monomer 21.8 kDa) predominantly exist as tetramers (187.6 and 78.6 kDa, respectively) with a small population of dimer (128.5 and 45.8 kDa, respectively).
-
CSEC profile of a sample containing an equimolar mixture of purified His‐GFP‐spMis18C‐term‐α and His‐spMis18MeDiY (upper panel) and SDS–PAGE analysis of SEC fractions (bottom panel). His‐GFP‐spMis18C‐term‐α and His‐spMis18MeDiY eluted separately at 12.3 and 15.3 ml, respectively, demonstrating the inability of these domains to interact with each other.
-
DSEC‐MALS analysis of His‐GFP‐spMis18C‐term‐α shows that it exists as a homotrimer with a measured molecular weight of 119.8 kDa (theoretically calculated MW of a monomer is 38.3 kDa).
MeDiY and the C‐terminal α‐helical domain are independent structural modules
To obtain further insights into the overall architecture of the spMis18 oligomer, we tested whether spMis18MeDiY and spMis18C‐term‐α could interact with each other, or whether they existed as structurally independent modules. His‐spMis18MeDiY and His‐GFP‐spMis18C‐term‐α were purified individually and analyzed for complex formation using size‐exclusion chromatography. His‐GFP‐spMis18C‐term‐α and His‐spMis18MeDiY eluted separately at distinct elution volumes (12.3 and 15.3 ml, respectively). This demonstrates that the spMis18MeDiY domain alone is unable to associate with spMis18C‐term‐α (Figs 3C and EV3D).
We next tested whether spMis18C‐term‐α can self‐associate to form oligomers. SEC‐MALS analysis of His‐GFP‐spMis18C‐term‐α (untagged spMis18C‐term‐α (8,775 Da) is too small for accurate mass detection) revealed that it is a homotrimer with a measured molecular weight of 119,886 Da (theoretical molecular weight of a monomer is 38,337 Da while that of a homotrimer would be 115,011 Da) (Fig 3D). We therefore conclude that spMis18 has two structurally independent domains, and they each possess the ability to homo‐oligomerize.
Oligomerization of spMis18 via the MeDiY domain is required for its function
We next tested whether spMis18 dimerization is functionally important in vivo. Co‐immunoprecipitation assays were performed on S. pombe cells expressing endogenous spMis18fl with a C‐terminal 8xMyc tag, and spMis18fl or spMis18flI31A expressed ectopically as a C‐terminally GFP‐tagged fusion protein. Endogenous wild‐type spMis18fl‐Myc associated efficiently with ectopic wild‐type spMis18fl‐GFP, confirming the interaction of spMis18fl with itself in vivo (Fig 4A). Such self‐association of wild‐type spMis18 has also been demonstrated previously through yeast two‐hybrid assays 13, 15. In contrast, the spMis18flI31A dimerization mutant showed significantly reduced association with endogenous spMis18fl‐Myc. Additionally, spMis18MeDiY alone was sufficient to associate with spMis18fl‐Myc (Fig 4A). This highlights the role of spMis18MeDiY in the overall oligomeric state of spMis18 in vivo.
Figure 4. Dimerization mediated by the MeDiY domain promotes spMis18 function and centromere association.
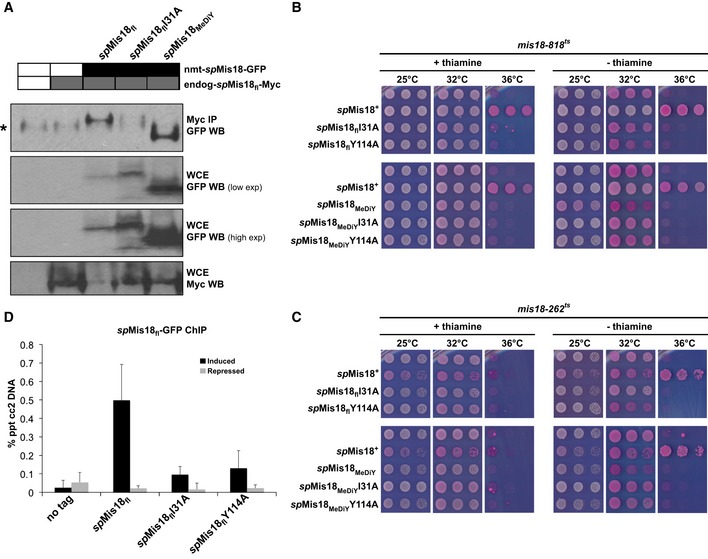
-
AspMis18fl‐Myc co‐immunoprecipitates with both GFP‐tagged spMis18fl and spMis18MeDiY, but shows reduced association when MeDiY‐mediated dimerization is disrupted in spMis18flI31A. The asterisk (*) in the top panel denotes the IgG heavy chain.
-
B, CDimer II interface mutations I31A and Y114A affect the ability of ectopically expressed spMis18fl to rescue the temperature sensitivity of mis18‐818 and mis18‐262 cells, while expression of spMis18MeDiY alone confers a dominant‐negative effect on growth in a MeDiY dimerization‐dependent manner. Fivefold serial dilutions of cells expressing the indicated spMis18 constructs integrated at the leu1 locus in the genome, spotted on complete PMG + phloxine B media supplemented with (repressed) or without (expressed) thiamine, and incubated at the indicated temperatures; dead cells stain dark pink.
-
DMutations that disrupt MeDiY dimerization lead to reduced levels of spMis18fl association with centromeres. qChIP analyses of spMis18fl‐GFP association with centromere 2 (cc2) in the indicated strains when grown in complete PMG media supplemented with (repressed) or without (expressed) thiamine. Error bars represent standard deviation between at least three biological replicates.
To test whether spMis18MeDiY‐mediated oligomerization is essential for spMis18 function, we performed genetic complementation assays in which we evaluated the ability of dimerization mutants I31A and Y114A (in the context of both spMis18fl and spMis18MeDiY), to rescue the temperature‐sensitive growth phenotype of mis18‐818 (T49A) and mis18‐262 (G117D) cells in vivo (Fig 4B and C) 10. Dimer‐disrupting mutants spMis18flI31A and spMis18flY114A failed to rescue the viability defect at restrictive temperature in both mis18‐262 and mis18‐818 cells. Expression of spMis18MeDiY alone, while failing to complement the loss of spMis18 function at 36°C, reproducibly conferred a dominant‐negative effect on growth in both mis18‐262 and mis18‐818 cells at 32°C. This inhibitory effect on growth of mis18‐262 and mis18‐818 cells depended on the ability of spMis18MeDiY to dimerize, as the spMis18MeDiYI31A and Y114A mutations caused no such negative influence on growth (Fig 4B and C). We conclude that dimerization of spMis18 via the MeDiY domain is crucial for spMis18 function in vivo.
Centromeric localization of spMis18 depends on MeDiY‐mediated dimerization
spMis18 is known to associate with centromeres, where it is required for the incorporation of CENP‐ACnp1 10. Chromatin immunoprecipitation analyses were performed to determine whether dimerization mediated by the MeDiY domain is required for the association of spMis18fl with centromeres. Disruption of spMis18 MeDiY‐mediated dimerization through spMis18flI31A or spMis18flY114A mutations resulted in reduced association of ectopically expressed spMis18fl‐GFP with centromeres in wild‐type cells (Fig 4D), although no significant defects in subcellular localization of spMis18fl‐GFP were observed (Fig EV4A). Additionally, cell growth and CENP‐ACnp1 association with centromeres were essentially unaffected in these cells (Fig EV4B and C). Thus, MeDiY domain‐mediated dimerization ensures optimal spMis18 association with centromeres.
Figure EV4. Ectopically expressed spMis18fl proteins localize to the nucleus and do not alter cell growth.
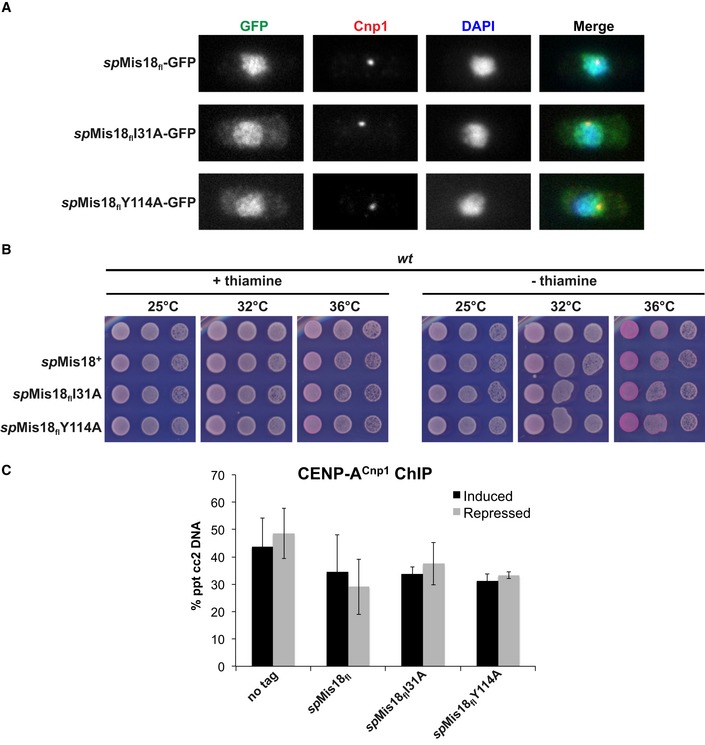
- Ectopically expressed spMis18fl‐GFP localizes throughout the nucleus in a MeDiY dimerization‐independent manner. Immunofluorescence of wild‐type S. pombe cells ectopically expressing GFP‐tagged spMis18fl (wt or mutants), stained with antibodies to GFP (green) and CENP‐AC np1 (red), and DAPI (blue).
- Ectopic expression of dimer II interface mutants does not affect growth of wild‐type S. pombe cells. Fivefold serial dilutions of cells expressing the indicated spMis18fl constructs integrated at the leu1 locus in the genome, spotted on complete PMG + phloxine B media supplemented with (repressed) or without (expressed) thiamine, and incubated at the indicated temperatures; dead cells stain dark pink.
- Mutations that disrupt MeDiY dimerization cause no significant change in CENP‐AC np1 association with centromeres in wild‐type cells. qChIP analyses of CENP‐AC np1 association with centromere 2 (cc2) in the indicated strains when grown in complete PMG media supplemented with (repressed) or without (expressed) thiamine. Error bars represent standard deviation between at least three biological replicates.
Centromeric association of the Mis18 complex is one of the early key steps involved in the establishment of CENP‐A nucleosomes at centromeres 17. Although some of the components of the Mis18 complex show variations among different species (particularly the absence of Mis18BP1/KNL2 in fission yeast), the function of the Mis18 complex in targeting HJURP to centromeres is well conserved 14, 15, 26, 27, 28. The molecular mechanism by which the Mis18 complex is targeted to centromeres, however, remains to be determined, and any structural insights into the molecular architecture of the Mis18 complex and/or its components have the potential to advance our understanding of centromere establishment.
The Mis18 complex is critical for CENP‐A deposition at centromeres; however, Mis18 is poorly characterized in terms of its structure and function. Here, we show that fission yeast Mis18 and its human isoforms, Mis18α and Mis18β, possess two structurally distinct domains: an N‐terminal “Yippee‐like” globular domain and a C‐terminal α‐helical domain. Structural alignments revealed unambiguous structural homology between the N‐terminal “Yippee‐like” globular domain and proteins of diverse function such as Cereblon 21, RIG‐I 22, MSS4 23, and MsrB 24, as well as identified a conserved substrate‐binding pocket in Mis18 required for its function. However, predicting the identity and nature of Mis18 binding partner(s) seems non‐trivial, as the amino acids that make up the binding pocket and their spatial orientation are markedly different among the various homologous proteins.
A key finding of our analyses is the ability of the “Yippee‐like” globular domain (MeDiY) of S. pombe and human Mis18 proteins to dimerize. To date, none of the other related domains are known to form dimers. Interestingly, S. pombe Mis18 and also human Mis18 29, in the presence of Mis18C‐term‐α, forms a tetramer. Notably, ectopic expression of the MeDiY domain alone, while failing to complement the loss of spMis18 function conferred a dominant‐negative effect on the growth of mis18 mutants, implicating both MeDiY and C‐terminal α‐helical domains in ensuring the required oligomeric structure for function. Specifically, perturbing the dimeric interface of the MeDiY domain affects the centromere localization of spMis18. Identifying the binding partners of MeDiY and determining how (and if) Mis18 oligomerization can influence substrate recognition are important questions to be addressed in the future to unravel the molecular mechanisms that mediate centromere localization and function of the Mis18 complex. A previous study has shown that HJURP dimerization is required for stable deposition of CENP‐A nucleosomes at centromeres and implicated a direct role for HJURP dimerization in forming octameric CENP‐A nucleosomes 30. These observations highlight protein oligomerization‐mediated regulation as an emerging regulatory theme for the inherently complex process of centromere establishment and maintenance.
Materials and Methods
Expression and purification of recombinant S. pombe proteins
Schizosaccharomyces pombe Mis18fl, Mis18MeDiY, Mis18ΔC, and Mis18C‐term‐α codon‐optimized sequences (GeneArt) were cloned into pEC‐K‐3C‐His or 9GFP (Addgene) LIC vectors with N‐terminal His or His‐GFP tags cleavable with either a 3C or TEV site, respectively. Mutations were introduced using QuikChange site‐directed mutagenesis protocol (Stratagene). All proteins were expressed using E. coli BL21 Gold; His‐spMis18MeDiY was grown in Super Broth and induced for 6 h at 25°C using 0.3 mM IPTG. His‐spMis18ΔC was grown in Super Broth, while His‐GFP spMis18C‐term‐α and His‐GFP‐spMis18fl were grown in 2× TY before inducing for 16 h at 18°C by adding 0.3 mM IPTG.
All proteins were lysed via sonication in a lysis buffer containing 20 mM Tris pH 8 (or 8.5 for His‐spMis18MeDiY), 35 mM imidazole and 2 mM BME supplemented with 10 μg/ml DNase, 1 mM PMSF, and complete EDTA‐free (Roche). The following NaCl concentrations were used in the lysis buffer: 500 mM for His‐spMis18MeDiY, 50 mM for His‐spMis18ΔC, and 100 mM for His‐GFP‐spMis18C‐term‐α or His‐GFP‐spMis18fl. After clarification, His‐GFP‐spMis18C‐term‐α, His‐GFP‐spMis18fl, and His‐spMis18MeDiY proteins were purified using a 5‐ml HisTrap HP column (GE Healthcare), while His‐spMis18ΔC was purified by batch mode using HisPur Ni‐NTA resin (Thermo Scientific). Resin was washed with lysis buffer, and then, His‐spMis181–120, His‐GFP‐spMis18C‐term‐α or His‐GFP‐spMis18fl received additional washes with 20 mM Tris pH 8/8.5, 35 mM imidazole, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, and 2 mM BME. The following NaCl concentrations were used: 500 mM for His‐spMis18MeDiY or 1 M for His‐GFP‐spMis18C‐term‐α and His‐GFP‐spMis18fl. Proteins were finally washed in lysis buffer and then eluted with 20 mM Tris pH 8/8.5, 500 mM imidazole, and 2 mM BME. The following NaCl concentrations were used in the elution buffer: 500 mM for His‐spMis18MeDiY, His‐spMis18ΔC, and His‐GFP‐spMis18C‐term‐α, and 100 mM NaCl for His‐GFP‐spMis18fl. Eluted His‐spMis181–120 was used directly for crystallization trial, while all other proteins were subjected to size‐exclusion chromatography. His‐spMis18ΔC was loaded onto Superdex 200 Hi‐load 16/600 (GE Healthcare) equilibrated with 20 mM Tris pH 8, 400 mM NaCl, and 1 mM TCEP. Appropriate fractions were pooled and injected onto a Superdex 200 increase 10/300 column (GE Healthcare) equilibrated with 20 mM Tris pH 8, 50 mM NaCl, and 4 mM DTT. His‐GFP‐spMis18C‐term‐α was applied to a Superdex 200 increase 10/300 column equilibrated with 20 mM Tris pH 8, 150 mM NaCl, and 2 mM DTT. His‐GFP‐spMis18fl was applied to a Superose 6 10/300 column (GE Healthcare) equilibrated with 20 mM Tris pH 8, 300 mM NaCl, and 2 mM DTT. Fractions were analyzed on SDS–PAGE and stained with Coomassie blue.
Expression and purification of recombinant human proteins
Human Mis18αfl, Mis18α77–187, Mis18βfl, and Mis18β56–183 codon‐optimized sequences (GeneArt) were cloned into pEC‐K‐3C‐His LIC vector or pGEX‐6P‐1 (GE Healthcare). Human proteins were expressed separately or co‐expressed using pGEX‐6P‐1 and pEC‐K‐3C‐His in BL21 Gold in a similar manner to spMis18ΔC. His‐hsMis18α77–187 was lysed via sonication in 20 mM Tris pH 8, 500 mM NaCl, 35 mM imidazole, and 4 mM BME with 10 μg/ml DNase, 1 mM PMSF, and complete EDTA‐free. Clarified lysate was loaded onto a 5‐ml HisTrap HP column and washed with lysis buffer and then with 20 mM Tris pH 8, 1 M NaCl, 35 mM imidazole, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, and 4 mM BME and re‐equilibrated in lysis buffer before eluting with 20 mM Tris pH 8, 500 mM NaCl, 500 mM imidazole, and 4 mM BME. GST‐hsMis18β56–183 was sonicated in 20 mM Tris pH 8, 500 mM NaCl, and 4 mM DTT with 10 μg/ml DNase, 1 mM PMSF, and complete EDTA‐free. Cleared lysate was loaded on 12 ml glutathione–Sepharose (GE Healthcare) in batch mode and washed in lysis buffer and then with 20 mM Tris pH 8, 1 M NaCl, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, and 4 mM DTT followed by lysis buffer. 3C protease was added to cleave the tag on the beads overnight and untagged protein collected. To obtain the hsMis18α77–187/Mis18β56–183 complex, proteins were co‐expressed and purified as described for GST‐hsMis18β56–183. All proteins were separated on Superose 6 10/300 pre‐equilibrated with 20 mM Tris pH 8, 200 mM NaCl, and 5 mM DTT.
His/GST pull‐down assays
To test for protein–protein interactions, purified His‐tagged spMis18MeDiY and His‐GFP‐tagged spMis18C‐term‐α proteins were mixed and applied to a Superdex 200 increase 10/300 column equilibrated with 20 mM Tris pH 8, 150 mM NaCl, and 2 mM DTT and fractions analyzed via SDS–PAGE. To test for interaction of human hsMis18α and β, 10 ml of culture of co‐expressed proteins was grown and sonicated in 20 mM Tris pH 8, 100 mM NaCl, and 2 mM BME. After clarification, supernatants were split and 35 mM imidazole added to half before incubation for 1 h with either glutathione–Sepharose or HisPur Ni‐NTA resin and then washed with buffer. Purified proteins were run on SDS–PAGE.
Native PAGE
12% native PAGE gels (22 mM Tris pH 8.8, 12% acrylamide with stacking 320 mM Tris pH 8.8, 4% acrylamide) were run in 25 mM Tris and 192 mM glycine at 150 V for 30 mins. Proteins in 1× sample buffer (31.25 mM Tris pH 6.8, 12.5% glycerol, 0.5% bromophenol blue) and Native Marker (Life Technologies) were loaded and then run at 150 V for 3 h.
Crystallization and data collection
Crystallization trials were performed using a nanoliter crystallization robot at the Edinburgh crystallization facility. Crystals were grown by vapor diffusion method. Diffraction quality crystals were obtained using well buffer containing 0.2 M ammonium chloride/formate/acetate/phosphate and 20% PEG 3350 (with the measured pH of the solution in the range of 6.2–8). 15–20 mg/ml protein sample was mixed with the well buffer in a 1:1 ratio. Crystals were briefly transferred to cryoprotectant solution (crystallization solutions were supplemented with glycerol or ethylene glycol to a final concentration 25%) before flash cooling in liquid nitrogen. The crystals diffracted to 2.6 Å resolution at the MX beamlines of the Diamond Light Source (Table 1).
Crystal structure solution and refinement
The structure of spMis18MeDiY was determined by the single anomalous dispersion (SAD) method using the anomalous signal of intrinsically bound Zn2+ ion. Data were processed using XIA2 and scaled with SCALA of CCP4 31. SAD phasing and the calculation of the initial map were performed using phenix.autosol from the PHENIX suite of programs 32. The model was built by iterative rounds of manual building with COOT 33, and refinement was done using Refmac5 of CCP4. Data collection, phasing, and refinement statistics are shown in Table 1.
SEC‐MALS
Size‐exclusion chromatography coupled to multi‐angle light scattering (SEC‐MALS) was performed at the Edinburgh Protein Production Facility at room temperature using a ÄKTA FPLC. 100 μl of protein at 1 mg/ml was loaded onto either Superose 6 10/300, Superdex 75 10/300, or Superdex 200 10/300 columns (GE Healthcare) pre‐equilibrated with 50 mM HEPES pH 8, 150 mM NaCl, and 5 mM DTT for His‐spMis18MeDiY, His‐GFP‐spMis18C‐term‐α, and hsMis18α77–187/hsMis18β56–183, or 50 mM HEPES pH 8, 50 mM NaCl, and 5 mM DTT for His‐spMis18ΔC or 50 mM HEPES pH 8, 300 mM NaCl, and 1 mM TCEP for His‐GFP spMis18fl. A MiniDAWN in‐line detector (Wyatt Technology) was used to measure MALS, while a Viscotek RI Detector (Wyatt Technology) was used to detect refractive index. Data were analyzed using ASTRA™ software (Wyatt Technology).
Plasmids and S. pombe strains
For in vivo assays in S. pombe, spMis18 cDNA (wt, I31A, Y114A, Y90A or Y74A Y90A T105A S107K in the context of Mis18fl or Mis18MeDiY) was PCR‐amplified and cloned into the pDUAL‐GFH41 vector, which allows for expression of C‐terminally GFP‐tagged spMis18 (wt or mutants) under the control of the medium‐strength nmt41 promoter that is induced in the absence of thiamine in culture media 34. spMis18 constructs cloned into the pDUAL‐GFH41 vector were then either transformed as such and selected in the absence of uracil in growth media, or integrated into the genome at the leu1 locus, in wt, mis18‐818, or mis18‐262 cells. Genotypes of S. pombe strains used in this study are listed in Table EV1.
Genetic complementation assays
Fivefold serial dilutions of mis18‐262, mis18‐818, or wt cells expressing GFP‐tagged spMis18fl or spMis18MeDiY (wt or mutants) either from an ectopic pDUAL‐GFH41 plasmid (selected for in media lacking uracil; Fig 1E) or from the leu1 locus (integrated in the genome; Figs 4B and C and EV4B), were spotted onto PMG media containing phloxine B supplemented with or without thiamine and incubated at the indicated temperatures for 3–5 days.
Co‐immunoprecipitation and Western analyses
For co‐immunoprecipitation experiments, cells expressing endogenous spMis18fl‐Myc and ectopic spMis18‐GFP (wt or mutants; from the leu1 locus) were cultured in complete PMG media lacking thiamine for 21 h and processed as previously described 11. Immunoprecipitation was performed using rabbit anti‐myc antibody A14 (Santa Cruz Biotech) and Western analyses using monoclonal anti‐GFP (Roche) or anti‐myc 9B11 (Cell Signaling).
Quantitative chromatin immunoprecipitation
Anti‐GFP ChIP, anti‐CENP‐ACnp1 ChIP, and real‐time PCR analyses were performed as previously described 11, on wild‐type S. pombe cells expressing ectopic GFP‐tagged spMis18fl (wt or mutants; from the leu1 locus) cultured in complete PMG media lacking thiamine for 21 h.
Cytology
Immunolocalization and microscopy were performed as previously described 11 on wild‐type S. pombe cells expressing ectopic GFP‐tagged spMis18fl (wt or mutants; from the leu1 locus) cultured in complete PMG media lacking thiamine for 21 h.
Data deposition
Atomic coordinates of the structure and structure factors are deposited in the RCSB protein data bank (www.rcsb.org) with accession code 5HJ0.
Author contributions
LS, BM‐P, RB, FS, RK‐S, GR, and AAJ performed experiments and analyzed data. LS, BM‐P, RCA, and AAJ designed experiments, analyzed data, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank D. Foltz for sharing data prior to publication. We thank the staff of Diamond Light Source for assistance during data collection and staff of Edinburgh protein production facility, and particularly M. Wear for his help with SEC‐MALS data generation and analysis. We thank C. Ponting and L. Sanchez‐Pulido for the initial bioinformatics analysis of the Mis18 Yippee‐like domain. We also thank L. Mackay for his help with the MS‐based protein sequence coverage analysis of Mis18. We are grateful to M. Yanagida and the National Bioresource Project (YGRC) Japan for S. pombe strains, and P. Heun, and members of Jeyaprakash and Allshire laboratory for discussions. We also thank W. Earnshaw, A. Cook, K. Hardwick, J. Welburn, A. Pidoux, and M. A. Abad Fernandez for the critical reading of the manuscript. The Wellcome Trust generously supported this work through a Wellcome Trust Career Development Grant to AAJ (095822), a Principal Research Fellowship to RCA (095021 & 065061), a Wellcome Trust Centre Core Grant (092076), an instrument grant (091020), and the Wellcome Trust‐University of Edinburgh Institutional Strategic Support Fund. The European Commission supported this work through a career integration grant to AAJ and through an EpiGeneSys Network of Excellence grant to RCA (HEALTH‐F4‐2010‐257082. LS was supported by an EC FP7 Marie Curie International Incoming Fellowship (PIIF‐GA‐2010‐275280) and an EMBO Long Term Fellowship (ALTF 1491‐2010). RB was supported by a Wellcome Trust 4‐year PhD program.
EMBO Reports (2016) 17: 496–507
References
- 1. Cheeseman IM (2014) The kinetochore. Cold Spring Harb Perspect Biol 6: a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fukagawa T, Earnshaw WC (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30: 496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Catania S, Allshire RC (2014) Anarchic centromeres: deciphering order from apparent chaos. Curr Opin Cell Biol 26: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gascoigne KE, Cheeseman IM (2013) Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosome Res 21: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falk SJ, Black BE (2013) Centromeric chromatin and the pathway that drives its propagation. Biochim Biophys Acta 1819: 313–321 [PubMed] [Google Scholar]
- 6. Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Partridge JF, Borgstrom B, Allshire RC (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791 [PMC free article] [PubMed] [Google Scholar]
- 8. Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, McColl J, Klenerman D, Carr AM, Sauer M, Allshire RC et al (2012) Quantitative single‐molecule microscopy reveals that CENP‐A(Cnp1) deposition occurs during G2 in fission yeast. Open Biol 2: 120078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foltz DR, Jansen LE, Bailey AO, Yates JR III, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere‐specific assembly of CENP‐a nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP‐A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- 11. Subramanian L, Toda NR, Rappsilber J, Allshire RC (2014) Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP‐A assembly. Open Biol 4: 140043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirai H, Arai K, Kariyazono R, Yamamoto M, Sato M (2014) The kinetochore protein Kis1/Eic1/Mis19 ensures the integrity of mitotic spindles through maintenance of kinetochore factors Mis6/CENP‐I and CENP‐A. PLoS ONE 9: e111905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayashi T, Ebe M, Nagao K, Kokubu A, Sajiki K, Yanagida M (2014) Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP‐A through interacting with NMD factors and the SWI/SNF complex. Genes Cells 19: 541–554 [DOI] [PubMed] [Google Scholar]
- 14. Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X et al (2009) Fission yeast Scm3: a CENP‐A receptor required for integrity of subkinetochore chromatin. Mol Cell 33: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams JS, Hayashi T, Yanagida M, Russell P (2009) Fission yeast Scm3 mediates stable assembly of Cnp1/CENP‐A into centromeric chromatin. Mol Cell 33: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22: 52–63 [DOI] [PubMed] [Google Scholar]
- 17. Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP‐A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- 18. Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS (2010) A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP‐A. Nat Cell Biol 12: 1186–1193 [DOI] [PubMed] [Google Scholar]
- 19. Roxstrom‐Lindquist K, Faye I (2001) The Drosophila gene Yippee reveals a novel family of putative zinc binding proteins highly conserved among eukaryotes. Insect Mol Biol 10: 77–86 [DOI] [PubMed] [Google Scholar]
- 20. Krissinel E, Henrick K (2004) Secondary‐structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- 21. Chamberlain PP, Lopez‐Girona A, Miller K, Carmel G, Pagarigan B, Chie‐Leon B, Rychak E, Corral LG, Ren YJ, Wang M et al (2014) Structure of the human Cereblon‐DDB1‐lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol 21: 803–809 [DOI] [PubMed] [Google Scholar]
- 22. Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP (2008) The C‐terminal regulatory domain is the RNA 5′‐triphosphate sensor of RIG‐I. Mol Cell 29: 169–179 [DOI] [PubMed] [Google Scholar]
- 23. Yu H, Schreiber SL (1995) Structure of guanine‐nucleotide‐exchange factor human Mss4 and identification of its Rab‐interacting surface. Nature 376: 788–791 [DOI] [PubMed] [Google Scholar]
- 24. Kim YK, Shin YJ, Lee WH, Kim HY, Hwang KY (2009) Structural and kinetic analysis of an MsrA‐MsrB fusion protein from Streptococcus pneumoniae . Mol Microbiol 72: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lupas AN, Zhu H, Korycinski M (2015) The thalidomide‐binding domain of cereblon defines the CULT domain family and is a new member of the beta‐tent fold. PLoS Comput Biol 11: e1004023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR (2011) HJURP is a CENP‐A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moree B, Meyer CB, Fuller CJ, Straight AF (2011) CENP‐C recruits M18BP1 to centromeres to promote CENP‐A chromatin assembly. J Cell Biol 194: 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G (2012) CENP‐C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR (2016) Licensing of centromeric chromatin assembly through the Mis18‐Mis18 heterotetramer. Mol Cell. 61: 774–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zasadzinska E, Barnhart‐Dailey MC, Kuich PH, Foltz DR (2013) Dimerization of the CENP‐A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J 32: 2113–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- 32. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, Yoshida M (2004) pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21: 1289–1305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File


