Abstract
From a risk assessment perspective, DNA-reactive agents are conventionally assumed to have genotoxic risks at all exposure levels, thus applying a linear extrapolation for low-dose responses. New approaches discussed here, including more diverse and sensitive methods for assessing DNA damage and DNA repair, strongly support the existence of measurable regions where genotoxic responses with increasing doses are insignificant relative to control. Model monofunctional alkylating agents have in vitro and in vivo datasets amenable to determination of points of departure (PoDs) for genotoxic effects. A session at the 2013 Society of Toxicology meeting provided an opportunity to survey the progress in understanding the biological basis of empirically-observed PoDs for DNA alkylating agents. Together with the literature published since, this review discusses cellular pathways activated by endogenous and exogenous alkylation DNA damage. Cells have evolved conserved processes that monitor and counteract a spontaneous steady-state level of DNA damage. The ubiquitous network of DNA repair pathways serves as the first line of defense for clearing of the DNA damage and preventing mutation. Other biological pathways discussed here that are activated by genotoxic stress include post-translational activation of cell cycle networks and transcriptional networks for apoptosis/cell death. The interactions of various DNA repair and DNA damage response pathways provide biological bases for the observed PoD behaviors seen with genotoxic compounds. Thus, after formation of DNA adducts, the activation of cellular pathways can lead to the avoidance a mutagenic outcome. The understanding of the cellular mechanisms acting within the low-dose region will serve to better characterize risks from exposures to DNA-reactive agents at environmentally-relevant concentrations.
Keywords: DNA damage, DNA repair, biological pathways, low-dose, dose-response, points of departure
1 Introduction
Under current human health risk assessment practices, DNA-reactive agents are generally considered by regulatory agencies to have no thresholds for biological outcomes such as mutation and cancer [1]. The debate surrounding the linearity of low-dose effects related to genotoxicity and cancer has been on-going for decades. New understanding in biological mechanism and mode-of-action (MOA), along with new high-content and high-throughput approaches, and increasingly sensitive analytical methods, bring new evidence into this debate. New in vivo and in vitro data have demonstrated the existence of non-linear/bilinear dose-responses for genotoxic effects (i.e. a dose–response curve with a slope not significantly different from zero gradient below the estimated threshold or Break Point Dose (BPD)), where there is no significant difference in mutant frequency between the spontaneous background of control and the low-dose exposure region of DNA-reactive agents [2-6]. In recent years, new statistical approaches have also been developed and applied to analyze low-dose results to establish whether the dose-response is linear or non-linear/bilinear, derive a point of departure (PoD), and determine what impact the spontaneous background genotoxicity should have on risk assessment. These compelling, empirical dose-response data do not address the biological underpinnings of mutation at low-dose exposures per se and require focused investigations of the MOA behind these non-linear/bilinear dose-responses. For an expressed mutation, several key events must occur from the initial DNA adduct formation, including insufficient adduct repair, DNA replication and cell division. Moreover, endogenous DNA adducts are now recognized to be ubiquitously present at quantifiable levels in all living tissues. This new perception of the background exposome is shifting perspective on what is normal vs. adaptive vs. adverse [7, 8]. This review discusses the current understanding of biological, mechanistic processes that explain these PoDs, specifically DNA repair and DNA damage response, and complex interactions between these pathways. The detailed discussion presented here was initiated during a Society of Toxicology 2013 workshop entitled the Biology of the Low-Dose Response for DNA-Reactive Chemicals. A clear focus on molecular and biological approaches to defining and understanding consequences of DNA damage at the cellular level fits well with the 2007 NRC report, “Toxicity Testing in the 21st Century: A Vision and A Strategy” that envisions a future in which all routine testing will use cell-based in vitro assays of toxicity pathways [9, 10].
1.1 Sources of Spontaneous DNA Damage and Cellular DNA Repair Pathways
The genome continuously undergoes damage due to numerous stressors and to the limited DNA chemical stability. Even in the absence of any significant exogenous exposure, mammalian cells sustain thousands of pro-mutagenic DNA lesions every day. Normal metabolic processes are associated with hydrolysis, deamination, alkylation, and oxidation, resulting in base damage, single strand breaks (SSB), double strand breaks (DSB), and interstrand cross-links [20-23]. Under normal conditions, the steady state level of endogenous DNA damage was recently estimated at ≥50,000 lesions per cell; the non-instructional and pro-mutagenic abasic sites are the most common DNA lesions, present daily at ~30,000 nucleoside sites in DNA per cell [18, 22, 24]. DNA repair influences the outcome and dose-response of mutation and chromosome damage following exposure to DNA damaging agents at all exposure levels [11-15]. DNA repair is usually error-free, but there may be rare events where the mis-repair will result in genotoxic outcomes. Under certain conditions or at high exposure doses, DNA repair itself can increase mutation [89-92]. Thousands of times per day, in every cell, DNA lesions are repaired by an integrated defense network that includes five major DNA repair arms: base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), non-homologous end-joining of double strand breaks (NHEJ) and homology-directed repair of DSB, cross-links and broken replication forks (homologous repair; HR). Among these repair activities, BER is considered to be one of the most active pathways, handling thousands of DNA lesions every day such as the major alkylation adducts, oxidized bases, deaminated bases, abasic sites, and SSB and nicks.
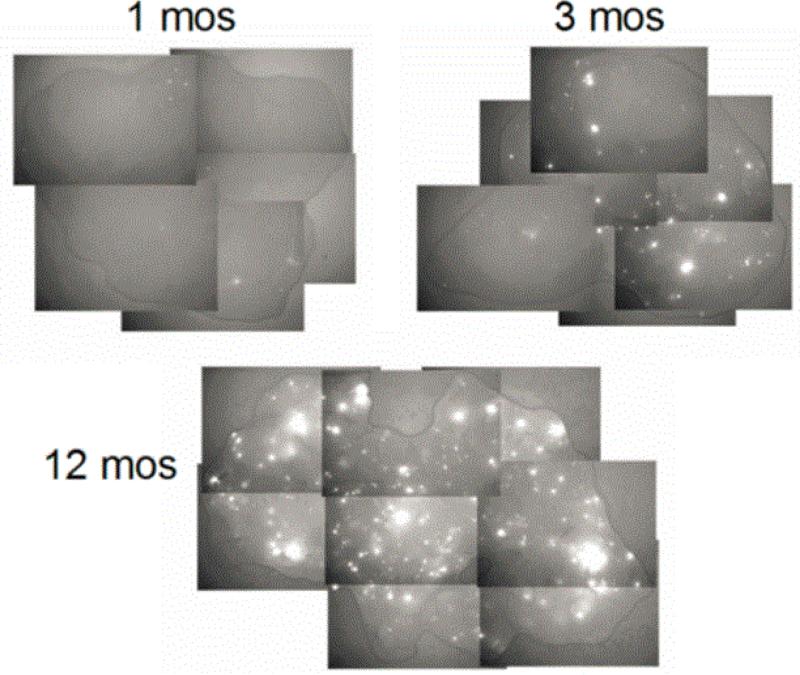
This significant and ubiquitous background of pro-mutagenic DNA damage is likely causative for the normal range of background mutations [5, 18]. Recent work has begun addressing the potential role of endogenous/background DNA damage in background mutagenesis [5, 6, 8, 16-19]. The Engelward laboratory developed a sensitive mouse model in which HR events at an integrated Fluorescent Yellow Direct Repeat (FYDR) transgene give rise to a fluorescent signal. This model provided a clearer understanding of HR background activity, effects due to aging, and HR response after exposure to exogenous agents. This model demonstrated that background rearrangement events in mice accumulate with age at individual rates in different cells and within different tissues (Figure 1) [17, 19].
Figure 1. Mutant cells accumulate with age in tissues under normal conditions.
Wiktor-Brown et al. [19] investigated the effects of aging on the frequency of HR events in the FYDR mice. In pancreas, 23-fold increase in recombinant cell frequency with age was noted in vivo.
2 New Methods to Investigate Responses at Low-Dose Exposures
New understanding of, and new techniques for measuring, the many ways in which normal cells handle DNA damage have led to consideration of the relationship between low-dose DNA damage and DNA repair, in an effort to understand how these processes contribute to cellular homeostasis. This revived interest has resulted in significant efforts to collect low-dose data on dose-response for genotoxic effects and to develop interpretive biological models for those observed dose-response behaviors for DNA damage and mutational consequences. Some of the new methods are discussed below.
2.1 Analytical Approaches for Measuring DNA Adducts
Swenberg et al. [18] developed sensitive analytical approaches to differentiate exogenously- and endogenously-induced DNA adducts. This work demonstrated the ubiquitous presence of a multitude of endogenous DNA lesions, including pro-mutagenic ones that are otherwise identical to many of those induced by DNA-reactive chemicals. The demonstrated steady-state of these adducts, especially of pro-mutagenic ones, has changed perspective on the assumption of mutagenic outcome and, in particular, on the biological plausibility of the one-hit theory for carcinogenesis. Clearly, the presence of an adduct does not equal a mutation, and the availability of DNA repair activity will play a key role in the ultimate fate of a DNA lesion. The recently published data on formaldehyde, demonstrating that low doses of exogenously inhaled formaldehyde induced an increase in exogenous adducts present at levels below the structurally identical, quantifiable endogenous levels, is leading to a paradigm shift concerning the role of such adducts (endogenous and exogenous) in mutation and cancer induction [6, 25].
2.2 High-throughput Investigations of DNA Repair Capacities
High-throughput, high-sensitivity methods have been developed to investigate overall DNA repair activity in response to different types of DNA damage [26]. The “CometChip” technique exploits the traditional Comet assay in which damaged DNA is evaluated by its ability to migrate away from the nucleus during electrophoresis. The assay simultaneously follows DNA damage load over time in a number of cell lines. This method quantifies kinetics and repair capacity within different backgrounds at the level of an individual cell. Initial studies demonstrated differential sensitivity and kinetics of DNA repair of various human cells in response to DNA damaging agents [26].
2.3 Genotoxic Dose-Response Determinations for DNA-Reactive Agents
For quantitative analysis of dose-response relationships for genotoxicity, approaches have been developed to derive various PoD metrics [27-29]. The visual shape of the plotted dose-response data can under certain circumstances be potentially misleading and uninformative [136]. For all dose-response relationships, a region (i.e., low-dose treatment level) exists where the change in adverse response with increasing dose will not be significantly different relative to the control (background). The approaches start with obtaining data from assays containing several doses in lower end of the dose-response curve where no apparent increase over the background is expected in addition to doses in the effect zone. These experimental measurements are then analyzed with appropriate mathematical models and statistical methods to obtain PoD dose-response predictors, and to define exposure levels associated with the acceptable risk [28, 29].
The collection and analysis of datasets specifically designed to address the low-dose dose-response for genotoxic chemicals have now accumulated into an impressive body of empirical evidence that allows derivation of the no-observed-genotoxic-effect-level (NOGEL, i.e., highest dose with no statistically significant response) and other useful PoDs for genotoxic effects [27-30]. Indeed several authoritative bodies have accepted non-linear/bilinear dose-response for certain DNA-reactive chemicals based on the extensive empirical evidence and subsequently have applied a margin-of-exposure approach to their assessment [31-35]. One common element across these cases is generation of data to support a hypothesized or demonstrated MOA, a framework with less detailed description than that required for establishing mechanism, based on identification of key events responsible for the observed effects [36]. There are many well-understood biological processes that could underpin a non-linear/bilinear dose-response for genotoxic effects that include DNA repair processes, redundancy in the genetic code, non-coding regions of DNA, processes associated with DNA replication and damage tolerance, DNA damage response networks, and apoptosis/cell death [11, 13, 30]. For DNA alkylating agents, the involvement of certain DNA repair systems, such as BER, HR, Direct Repair, and MMR, shifts the dose-response curve for genotoxic endpoints. These DNA repair systems are well understood and will be reviewed here.
3 Alkylating Agents
3.1 Endogenous and Exogenous Sources
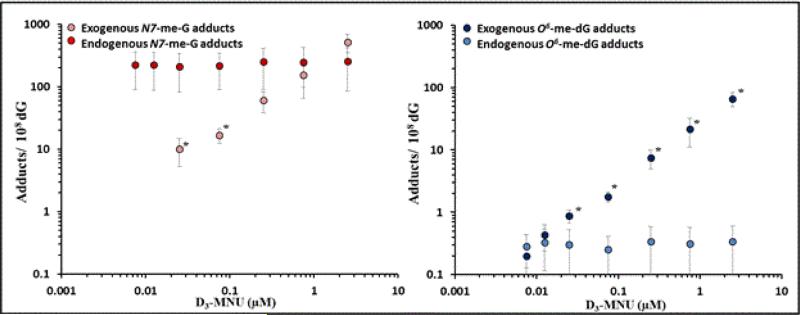
Alkylating agents are ubiquitous in the environment and within living cells. Endogenous DNA alkylation adducts are considered to be the major contributor to the total background levels of all DNA adducts present at steady-state levels in cells [5, 8, 18, 37]. Endogenous alkylating DNA adducts can arise from several different sources, for example from metabolic activity of gut bacteria, or as byproducts of lipid peroxidation, or reacting with cellular methyl donors such as S-adenosylmethionine, a common cofactor in cellular methylation reactions [38, 39]. Major exogenous sources of alkylating agents come from natural and anthropogenic constituents of air, water, and food, as well as from tobacco smoke and fuel combustion products [40-42]. Therefore, it will be difficult to distinguish a small risk at low-dose exposures within the normal distribution of the background range of mutation. Certain alkylating agents are also used as chemotherapeutic drugs, with the goal of killing cancerous cells and treating cancer [43]. For example, chemotherapeutic agent Temozolomide produces a distribution of DNA adducts similar to that of N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) and methylnitrosourea (MNU) [44]. Recently, work has begun addressing the issue of spontaneous/background adducts and exogenous ones (Figure 2).
Figure 2. Dose-response for endogenous and exogenous methylating adducts.
Endogenous (unlabeled) and exogenous (D3-labelled) N7-MeG and O6-MeG were measured after treatment with D3-MNU in AHH-1 cells (from Sharma et al. [5]). Exogenous N7-MeG adducts did not significantly contribute to the total N7-MeG adduct load under low-dose treatment conditions.
3.2 Alkylation DNA Base Adducts
The most abundant adduct produced by alkylating agents is at the N-7 position of guanine, a site that has the highest negative electrostatic potential in DNA (Figure 3) [45]. Adduct profile data show that 67% and 82% of adducts induced by methylmethane sulfonate (MMS) and MNNG, respectively, are N-7-methylguanine (N7-MeG) adducts [46]. N-7 guanine adducts are not cytotoxic or mutagenic as they do not block DNA replication and are not miscoding [47-50].
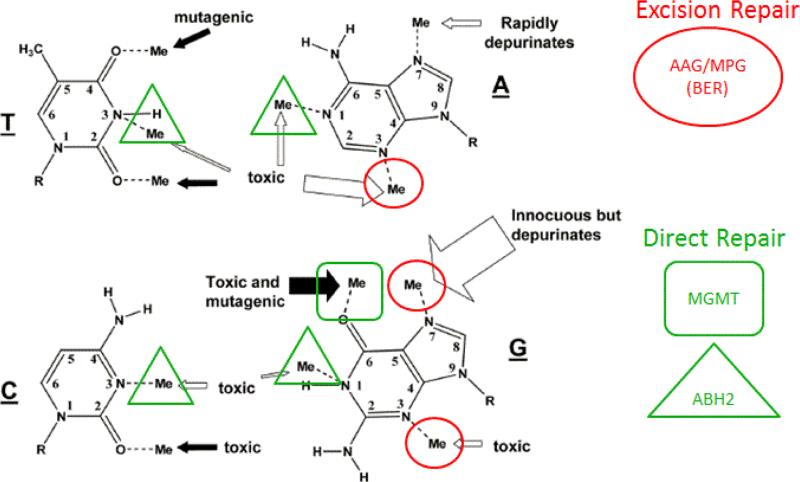
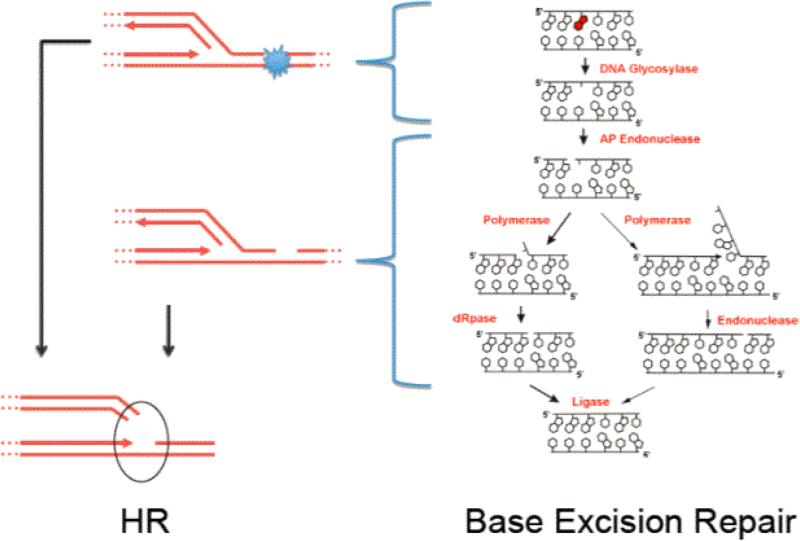
Figure 3. Overlapping DNA repair systems are involved in removal of alkylation DNA adducts.
BER and MGMT substrates are also handled by NER. Unrepaired damage or repair intermediates can be funneled to tolerance mechanisms HR, NHEJ or TLS. Black arrows indicate adducts induced in significant proportion by SN1 alkylating agents only. White arrows indicate adducts induced by both SN1 and SN2 alkylating agents. Arrow thickness correlates with the frequency of induced adducts. Adapted from Wyatt and Pittmann [69].
The next most abundant adduct (<15%) produced by methylating agents is the N-3-methyladenine (N3-MeA; Figure 3), as the N-3 position of adenine has the second highest negative electrostatic potential [45]. MMS is known to produce 11% N3-MeA whereas MNNG produces 12% N3-MeA [46]. Unlike the N7-MeG, unrepaired N3-MeA is highly cytotoxic, inhibiting DNA synthesis and preventing the formation of mutation [51, 52]. Alternatively, N3-MeA was shown to cause low levels of A-to-T transversions [53]. In test systems deficient in alkyladenine DNA glycosylase (Aag), Aag−/− cells with a high proportion of N3-MeA progress through S-phase more slowly due to the DNA replication-blocking capability of these adducts [51].
In double stranded DNA, adducts on the O6 guanine position are also prevalent, and are produced by the SN1 alkylating agents that demonstrate greater reactivity towards base oxygen atoms (Figure 3). MNNG produces 7% of O6-methylguanine (O6-MeG), whereas MMS produces only 0.3% of O6-MeG [46]. O6-MeG is the major pro-mutagenic adduct that induces G:C to A:T mutations as it readily mispairs with thymine [54]; however, it is also highly cytotoxic [55]. As will be discussed later, the mutagenic vs. cytotoxic response to O6-MeG lesion depends on the status of MMR.
The remaining alkylating lesions occur at 10 to 100-fold lower levels than the previously described adducts. Base sites in double stranded DNA with lesser electrostatic potential are at the N-3 position of guanine, O2 position of cytosine, N-7 position of adenine, and O4 and O2 positions of thymine [45]. Alkylation adducts are produced by MMS or MNNG at N-1 and N-7 positions of adenine, N-1 and N-3 positions of guanine, N-3 and O2 positions of cytosine, as well as N-3, O2 and O4 positions of thymine all of these comprise <5% of the total adducts in double stranded DNA [46]. The alkylation adducts can cause mutations or block essential biological processes such as DNA replication leading to cell death (Figure 3). It was also demonstrated that specific alkylation lesions (N1-A, N3-A and N3-C) can be both mutagenic and cytotoxic [137]. Overall, as the spectrum of alkylation DNA base adducts and biological responses to these adducts are well understood, it allows for the generation of MOA data and estimation of PoD values for this class of chemicals.
3.3 Endogenous vs. Exogenous DNA Base Adducts
Methylating agents provide well-studied examples of the contribution of endogenous and exogenous DNA base adducts to background and induced mutation. Work in D3-MNU-treated AHH-1 cells demonstrated linear dose-responses for both exogenous D3-N7-MeG and exogenous D3-O6-MeG adducts at low-dose exposures, and a steady-state of endogenous/background formation (Figure 2; [5]). As the endogenous load of N7-MeG is significant, exogenous adducts did not contribute significantly to the total load until the highest concentrations utilized in the study. On the other hand, endogenous O6-MeG adducts were much less abundant. When compared with the mutation dose-response curves, these exogenous O6-MeG adducts are likely to drive the mutation dose-response above the background/spontaneous mutation frequency [5].
3.4 Genotoxic Dose-Response Datasets
Several datasets exist that offer a detailed view of dose-response for induction of genotoxic effects. For alkylating agents, representative studies include three in vitro [56-58] and three in vivo [2, 3, 138] low-dose dose-response genotoxic datasets (an example is presented in Figure 4). The genotoxic chemicals used in these studies included direct-acting, monofunctional ethylmethanesulfonate (EMS), MMS, ethylnitrosourea (ENU), and MNU, although not all of the studies used all four chemicals. All of these studies were designed with an extended number of low-dose treatments, and with higher numbers of replicates than is typical for genotoxicity studies. They were conducted with different experimental systems, including gene mutations in mammalian cells in vitro (human-derived TK6 and AHH-1 lymphoblastoid cell lines; L5178Y mouse lymphoma cells) and in lymphocytes from in vivo exposure of transgenic (lacZ; gpt-delta) or normal (Pig-a) mice. Micronuclei (MN) were assessed both in vitro (TK6 and AHH-1 cells) and in vivo (mouse bone marrow). All these datasets were statistically evaluated with a methodology that directly compared goodness-of-fit of a linear dose-response model to that of a non-linear/bilinear dose-response model [27, 29, 59]. These studies demonstrated non-linear/bilinear dose-response curves for genotoxic effects with the SN2 alkylating agents EMS and MMS [3, 56-58] and with the SN1 alkylating nitrosoureas, ENU and MNU [3, 56-58, 60, 61].
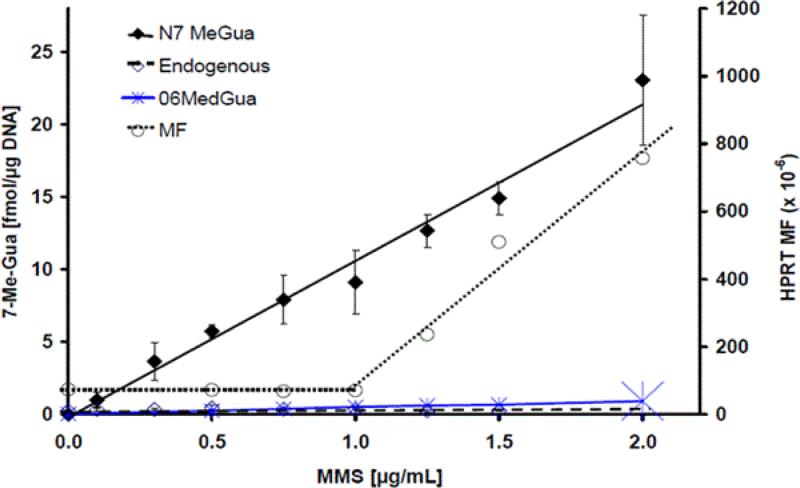
Figure 4. Dose-response curves for mutation and DNA adducts.
Exogenous N7-MeG and O6-MeG adducts were measured following 24 hour treatment with 13C-labelled MMS [62]. Adducts demonstrated linear dose response. Mutation induction at HPRT locus, demonstrating bilinear dose-response, with similarly treated AHH-1 cells was taken from Doak et al. [56].
The EMS dataset for induction of MN in vivo was a particularly compelling example demonstrating a non-linear/bilinear dose-response for genotoxic effects with the biomarker hemoglobin adducts being quantified as a measure of internal (systemic) dose in the blood cells [3]. The data clearly demonstrated an increasing level of hydroxyethylvaline hemoglobin adducts corresponding with increasing administered dose of EMS, but with no increase in corresponding MN induced until the administered dose exceeded the 80 mg/kg bw/day dose in mouse. Mutational in vivo dose-response with EMS was also recently investigated in adult gpt-delta transgenic mice within different tissues at the gpt transgene and at the Pig-a endogenous gene [2]. In the study, measured mutation NOGELs were identified at below 13 mg/kg/day and the lowest calculated PoD value, BMDL10, of 0.038 mg/kg/day was found in the lung of gpt-delta mice.
Furthermore, the mutation induction at the HRPT locus was compared with formation of DNA base adducts (Figure 4). The major (N7-MeG) and the key pro-mutagenic (O6-MeG) adducts were quantified in AHH-1 cells following exposure to 13C-MMS [62]. 13C-labelled adducts increased linearly with the treatment dose. These analytical results were compared with the non-linear mutational frequency data for HPRT locus obtained under the same low-dose MMS treatment [56]. Clear differences in the shape of dose-response curves were seen for exogenous adduct formation and mutation induction at low doses, even for pro-mutagenic DNA adducts.
However, these compelling empirical dose-response data do not address the biological underpinnings of mutation at low-dose exposures per se, and thus investigation of the MOA behind these non-linear/bilinear dose-responses represents a necessary next step. One proposed MOA for mutation describes formation of a pro-mutagenic DNA adduct as an initial/early key event [63-65]. However, as adducts do not equal mutations, there are several additional necessary key events proposed that require functioning cellular responses such DNA replication and cell division, prior to causing a mutation. Thus, formation of adducts, even pro-mutagenic ones, will not necessarily guarantee a mutagenic outcome [66, 67].
3.5 DNA Repair
The biological responses to alkylating agents are quite complex due to the variety of alkylation adducts produced in DNA, and the variability this imparts to their biological significance [4, 63, 65, 68]. Under certain conditions such as imbalanced DNA excision or abnormally high levels of activity, DNA repair itself can be a cause of mutation [89-92]. Not every adduct is considered to be pro-mutagenic whereas adducts that are pro-mutagenic can be processed by a number of biological pathways, preventing the formation of mutation. The major repair pathways involved in the removal of alkylation adducts have different targets that result in different outcomes (Figure 3).
BER is a key DNA repair pathway that removes thousands of DNA lesions every day, including alkylated and oxidized bases, SSB, and abasic sites (Figure 5). The BER pathway repairs the N-alkyl lesions including N3-MeA and N7-MeG. BER is initiated by DNA glycosylases that remove the N-alkyl adducted bases in all species examined [69]. DNA glycosylases of BER decrease the half-lives of N7-MeG and N3-MeA in DNA to minutes and ensure that abasic sites are processed by BER [70, 71]. Both adducts have rapid rates of spontaneous depurination as alkylation of purines destabilizes the N-glycosidic bond and renders these bases more susceptible to hydrolysis with half-lives of approximately 30 h and 70 h, respectively, for N3-MeA and N7-MeG at 39°C [72]. In fact, spontaneous depurination is the major fate of N7-alkyl adducts [48, 73, 74]. The resulting abasic/apurinic sites can be processed by a number of other repair pathways, including HR, NER, or translesion DNA synthesis (TLS), a DNA tolerance pathway [75-77]. Indeed abasic sites have been shown to represent the predominant endogenous lesion present in DNA at a steady-state, with as many as ~30,000 abasic sites being present per cell [18, 22, 24].
Figure 5. Repair of alkylated DNA bases by the BER repair pathway and channeling of DNA repair intermediates into the HR pathway during DNA replication.
Unrepaired DNA adducts, abasic sites, gaps and DNA nicks in S-phase are handled by the HR pathway.
N3-MeA and N7-MeG adducts also appear to be substrates for the NER pathway [78-80]. Direct reversal of DNA damage and TLS are important response pathways for alkylation damage. Direct reversal pathways remove adducts from the DNA restoring the original base directly in an error-free mechanism. In mammals, two direct repair pathways handle alkylation DNA damage, the O6 methylguanine-DNA methyltransferase (MGMT) that restores guanine from O6-MeG and the AlkB homologue (ABH) family of Fe(II)/α-ketoglutarate dioxygenases that directly repair alkylation damage in DNA and RNA at base-pairing sites [81]. In contrast, TLS continues DNA replication across the adducted base blocks encountered by replicative DNA polymerases, either in an error-free or an error-prone manner.
Manipulation of DNA repair activities in cells with molecular biology approaches resulted in a shift in the PoD values following treatment with alkylating agents, thus demonstrating a key role for DNA repair in PoDs for mutational/genotoxic effects [61, 82].
3.6 DNA Damage Response Pathways
DNA stress response pathways, both p53-dependent and p53-independent, are also activated in an effort to counteract DNA damage induced by alkylating agents. These can remove cells harboring DNA damage from the population. ATM signaling kinase and p53 tumor suppressor are key players in DNA damage signaling pathways, activated in response to DNA lesions. Apoptosis induced by the key mutagenic and cytotoxic adduct, O6-MeG, can proceed independently of ATM and p53 [83-85]. N-alkyl lesions can also trigger apoptosis via p53-independent pathways [86]. Thus, as cells continually deal with endogenous/background DNA alkylation, the existence of mutational thresholds at low exposures/doses is not surprising and can be explained based on the adaptive and homeostatic responses known to operate in biological systems in response to these stressors.
4 DNA Repair and Break Point Doses
Manipulations of DNA repair activity have been known to influence genotoxicity and cancer predisposition. Two prototypical types of DNA-reactive agents are discussed below.
4.1 Alkylation DNA Damage
Methods to investigate the influence of DNA repair on low-dose genotoxic outcomes were developed in Johnson's laboratory, such as DNA glycosylase knockdown [61]. Gene expression analysis has been used in cell lines with inhibited DNA repair activity to investigate the potential links between specific DNA repair pathways and shifts in PoDs, such as NOGELs, for gene mutation and chromosome damage endpoints. This integrated methodology has linked methylpurine glycosylase (MPG)/AAG DNA glycosylase to a shift in the NOGEL for MN induction in the human-derived lymphoblastoid AHH-1 cell line following exposure to EMS [82]; a causal role has not been documented for any specific EMS-induced DNA adduct in MN induction. An increase in MPG/AAG glycosylase gene expression occurred above and below NOGEL for chromosome damage as measured through MN induction at certain time points, but not for the HPRT gene mutation dose-response [82]. This small decrease in gene mutation frequency slope in treated cells is in line with evidence that suggests decreased repair of certain alkylation lesions could be protective against mutagenic and cytotoxic effects of abasic site and SSB BER intermediates [86-88].
For the pro-mutagenic O6-MeG adduct derived from MNU treatment, a clear link between the levels of MGMT repair protein (discussed below) and the HPRT gene mutation PoD/NOGEL metric was seen in human-derived AHH-1 cells [61]. This effect was manifested as a shift in the PoD/NOGEL to a lower MNU dose when MGMT was pre-depleted with O6-benzylguanine. Thus, decreased MGMT repair activity reduced the PoD value following MNU treatment compared to PoD obtained with MNU treatment under normal repair conditions. Sequencing of the HPRT mutants showed the expected increase in O6-MeG-induced G-to-A transitions. Even though the mechanisms of action differ for chromosome breaks compared to gene mutation, these differences are likely due to specific DNA lesions and repair by their respective DNA repair pathways. Nonetheless, the underlying MOAs behind the demonstrated shifts in NOGELs and PoDs for both endpoints appear to rely upon changes in DNA repair capacity.
4.2. Double Strand DNA Damage
DSBs are induced by variety of agents, including ionizing radiation, radiomimetic drugs and alkylating agents. HR is an essential pathway for resolution of broken replication forks and DSB in the S/G2 phases of the cell cycle whether they result from endogenous or exogenous processes. HR can be stimulated by DSB, DNA nicks and increased levels of abasic sites (Figure 5). Rare errors in HR repair may result in sequence rearrangements and loss of heterozygosity, two prominent features of tumor cells. Since HR and BER pathways are active in response to spontaneous DNA damage, they would respond to low-dose radiation and genotoxic chemicals. Both pathways are usually error-free, but there may be rare events where there are misalignments during HR and misinsertions during BER that result in genotoxic outcomes. The level of DNA repair activity is tightly controlled at the cellular level as too much or too little repair activity of either of these pathways can lead to increases in genotoxic outcomes [89-92].
Biological consequence of low-dose γ-radiation, the prototypical agent that induces HR, was explored by Olipitz et al. [93]. This work evaluated several endpoints such as accumulation of DNA damage, sequence rearrangements, and gene expression, following whole body exposure to low doses of radiation. The innovative exposure apparatus made it possible for mice to live in the continued presence of radiation at exposures that were approximately 400-fold higher than background for an extended period of time. The exposure to 10.5 cGy is expected to induce additional ~400 base lesions per cell [93]. Using these sensitive techniques, non-linear/bilinear dose-response relationships were observed for several biomarkers following these low-level radiation exposures. After exposure to 400-fold background radiation for five weeks, there was no evidence of any increase in key DNA base lesions in mouse tissues, no increase in DSB, or in HR events (Figure 6). Importantly, when the same total amount of radiation was given in one acute dose, rather than over a 5-week timeframe, DSB and DNA stress responses were in fact detected. Clearly, dose-rate is a critical factor when considering the adverse consequences of low-dose exogenous exposures, presumably because DNA repair keeps up with and repairs DNA damage at low doses, whereas at high exposure doses, available DNA repair capacity is overwhelmed and can lead to adverse outcomes including mutation. Importantly, the original paradigm for the one-hit theory in fact came from very early studies with γ-radiation, the same DNA damage inducer as that used by Olipitz et al. [93] to show non-linear/bilinear dose-responses [93, 94].
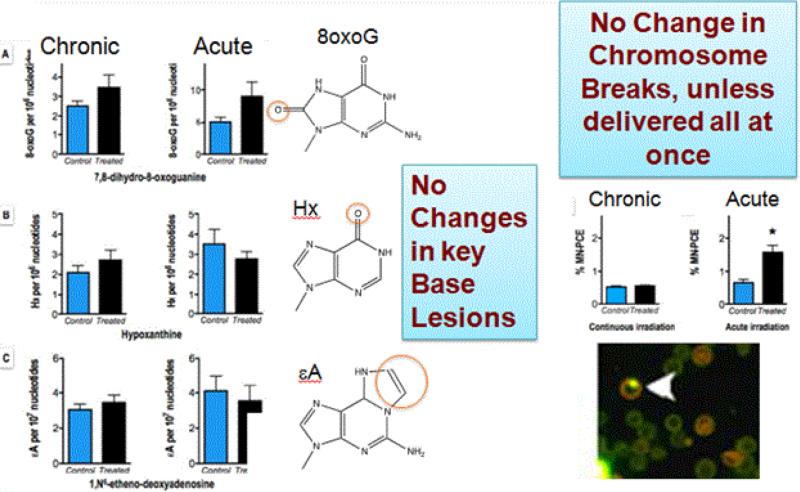
Figure 6. No detected changes in DNA damage or responses after repeated exposure to low level radiation.
After continuous exposure over 5 weeks to 0.0002 cGy/min radiation (400-fold over background radiation), the exposed mice did not demonstrate increased levels of DNA nucleobase damage (hypoxanthine, 8oxoG, 1,N6-ethenoadenine, or 3,N4-ethenocytosine) or DNA fragmentation (MNT assay and double strand break–induced HR) above background levels. In addition, low dose-rate radiation did not induce Cdkn1a, Gadd45a, Mdm2, Atm, or Dbd2 gene expression before and after irradiation (from Olipitz et al. [93]). The same total dose delivered acutely induced both MN and transcriptional responses.
5 Interactions between DNA Repair and DDR Pathways
The realization that complex interactions exist between different DNA repair pathways is an important development. These complex biological relationships can combine to manifest as the non-linear dose-response for genotoxic effects.
5.1 BER and HR
The Engelward laboratory has focused on the ways in which one DNA repair pathway affects another. For instance, excessive activity in one pathway creates repair intermediates that stimulate a different pathway. This paradigm is evident in the interactions between BER that promote HR events (Figure 5). The Engelward laboratory has shown that conditions inhibiting the initiation of BER actually suppress HR in vivo, presumably because the BER intermediates (including SSB or abasic sites) are more recombinogenic than some of the original adducted base substrates of BER [95]. These results show that changes in the balance of activity and protein products in one pathway (BER) can affect processing via another pathway (HR) and may put cells at increased risk of mutation.
5.2 MGMT and MMR
Another well-studied network of repair pathways handles the pro-mutagenic O6-MeG adducts. Exposure to SN1 MNNG or MNU nitrosoureas results in a variety of DNA base lesions. The most toxic of these is O6-MeG, produced at ≤10% of all nitrosourea-induced alkylated bases [46]. Repair of this adduct involves the DNA MGMT protein, which directly transfers the methyl group from O6-MeG to its active site's cysteine, followed by ubiquitin-mediated degradation of the now-methylated MGMT protein [96, 97]. When levels of MGMT are not adequate or depleted, the MMR pathway serves as a backup repair pathway by eventually eliminating cells with O6-MeG bases and preventing mutation (Figure 7). If the cells enter S-phase prior to O6-MeG repair, the DNA polymerase commonly mispairs O6-MeG with thymine resulting in a O6-MeG:T mismatch and fixation of a G-to-A transition mutation in the second round of replication. This new mismatch, however, is readily recognized by the heterodimer MSH2-MSH6 of the MMR pathway [98]. SCEs, chromosomal aberrations, and HR events can also be induced by O6-MeG adduct via an MMR-dependent pathway [99-104]. Under normal circumstances, MMR repairs spontaneous single-base mispairs and small insertion or deletion loops created by the DNA polymerase during replication; loss of MMR function results in a nearly 1000-fold increase in spontaneous mutation rate [105].
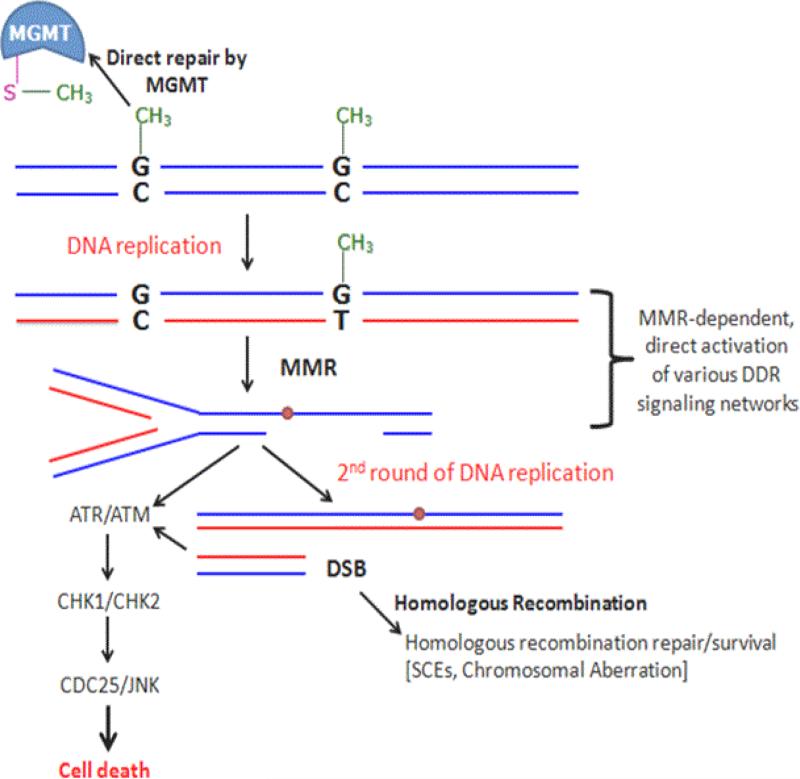
Figure 7. Cellular processing and repair of O6-MeG adducts in DNA.
MGMT directly repairs O6-MeG adducts. If unrepaired, O6-MeG preferably mispairs with T during DNA replication leading to G:C-to-A:T transitions. Alternatively, O6-MeG lesion induces apoptosis via an MMR-dependent pathway. O6-MeG/MMR-dependent DNA damage response includes multi-pathway, multi-time scale signaling network activation led by early ATM, H2AX, ATR-CHK1, and p53 phosphorylation, then followed by late phosphorylation of ATM-CHK2 and JNK kinase, as well as dramatic increases in p53 levels and p53 transcriptional targets [85]. Sister chromatid exchanges (SCE) and chromosomal aberrations are induced by O6-MeG lesions via an MMR-dependent pathway in the second cell cycle [102]. The gaps and nicks present during this phase can form DSB that are handled by HR. With loss of MMR, cells become “methylation-tolerant” accumulating mutations and escaping cell death in the presence of unrepaired O6-MeG.
5.3 MMR and DDR
In addition to its role in post-replication repair, MMR also responds to certain forms of DNA damage including alkylation damage, where MMR brings about the induction of cell cycle checkpoints and apoptosis by one of two proposed models; either by futile cycling [106-108] or direct signaling. The direct signaling model is supported by observed interactions between MSH2 or MLH1 with signaling kinases such as ATM, ATR, CHK1, and CHK2 [109-112].
MMR proteins MSH2 and MLH1 were shown to be recruited to chromatin containing the O6-MeG:T mismatches in a number of in vitro cell systems during the first S-phase following exposure [113]. The cells typically complete another round of the cell cycle before arresting in the second G2/M phase after damage [108, 114]. MSH2 and its partner MSH6 recognize O6-MeG:T mismatches, following MSH2/MSH6 heterodimer conversion to an ATP-dependent sliding clamp on DNA, and recruit the second MMR heterodimer of MLH1-PMS2. Together, these MMR complexes coordinate excision of the newly synthesized daughter strand and re-synthesis across from the O6-MeG adduct by exonuclease (Exo) I [115-118]. The replication activity also involves the function of the polymerase processivity factor PCNA which coimmunoprecipitates with MSH2 and MLH1 on damaged chromatin in the first S-phase [113]. In the futile cycle model [119, 120], the repeated processing of the O6-MeG:T mismatch by the MMR pathway results in prolonged single strand nicks or unreplicated gaps that remain past S-phase through to the next cell cycle. As the replication fork encounters these sustained nicks and/or gaps in the second S-phase after treatment, they are converted into DSBs and activate DNA damage response signaling cascades, including CHK1 and CHK2 DNA damage response kinases in the second S-phase after damage [85, 108, 113]. Moreover, results from the Heinen laboratory also raise the possibility that the continued presence of MSH2 and MLH1 on chromatin in the second cell cycle may serve as protein blockades that impede replication fork progression during the second S-phase leading to DNA damage response. Interestingly, in pluripotent stem cells (PSCs), MMR-dependent alkylation damage response results in a robust apoptotic response that occurs in the first S-phase after DNA damage [121]. These results indicate either that that PSCs are extremely sensitive to perturbations in DNA replication caused by iterative MMR cycles, or that MMR proteins may be involved in direct signaling of damage immediately upon recognition in the first cell cycle.
Collectively, these results provide evidence that cells utilize multiple DNA repair mechanisms to protect themselves from the threat of endogenous and exogenous mutagenesis caused by alkylating agents. In addition, DNA damage responses in different test systems may differ while still functioning together to prevent formation of mutation.
6 Profiling of Biological Pathways and Genotoxic Dose-Response Relationships
High-throughput and high-content assays can inform on chemical-specific perturbations of toxicity pathways. Cells respond to physical and chemical stressors by activating signal transduction cascades that can lead to various cellular outcomes or even cell death. These biological outcomes can be analyzed together and modeled in order to predict the BPD for mutational outcomes in relation to activation of various biological pathways.
6.1 Computational Approaches
Cellular repair foci arising from protein recruitment to sites of DNA damage are necessary to complete lesion repair and reduce the probability of mutation. The molecular mechanisms involved in formation of these complexes, dependent on phosphorylation status and protein abundance, are expected to be key determinants of the repair and mutational outcome at the low-dose exposures. Increasing knowledge of the interplay of these processes can guide construction of predictive computational systems biology pathway (CSBP) models of mutational outcome that can provide mechanistic understanding of BPD behaviors [122]. The p53-mdm2 stress response pathway is a key cellular stress response whose dynamics should serve as the underpinning for the mechanistic basis of non-linear/bilinear dose-response for mutagenesis [123]. Hamner-Unilever work on DNA damage pathway modeling networks has developed from a case study approach for implementing key recommendations from the 2007 NRC report, Toxicity Testing in the 21st Century: A Vision and A Strategy [9]. The merits of case study approaches for DNA damage assessment were outlined earlier and a more complete discussion of the approach for consumer product applications has recently appeared [124, 125].
6.2 Transcriptome Responses of Biological Pathways
To develop a robust dataset for understanding the p53-mdm2 pathway mechanistically, Hamner and Unilever have used the HT1080 human-derived fibrosarcoma cell line with wild-type p53 protein, to investigate p53-mdm2 pathway activation. Cells were exposed to a variety of compounds, including SN2 alkylator MMS, topoisomerase inhibitor etoposide (ETP) inducing DSB, β-irradiation mimic neocarzinostatin (NCS) producing DSB, and quercetin (QUE), a polyphenol [126]. The CSBP model construction began using a simple negative feedback stress response pathway model designed to account for either proportionate control or perfect adaptation [127]. In proportionate control, there is some, small increase in response (e.g., mutation rate or MN formation) to an increasing stressor level. Perfect adaptation is the ability of a cell or an organism to maintain a constant net level of damage (i.e., mutation rate) throughout a range of increasing stressor and is capable of producing a threshold response [128].
To fill out details of these DNA-repair and DDR networks, a high coverage approach, measuring whole genome transcriptomics and various proteins/phosphorylated proteins in the pathway, including p21, mdm2, p53, phospho-p53 (ser-15) and γ-H2AX was used. In addition, cell cycle, apoptosis, necrosis, and MN were also quantified over a broad, 18-point dose range spanning a low-dose range for mutational PoD. These data streams and representation of the changes in the signaling pathway were visualized using a composite suite of endpoints shown in increasing color intensities to capture the changes between compounds and across doses [126].
6.3 Genotoxic Responses
The dose-response for MN formation with MMS, under low-dose exposure conditions, showed evidence of a non-linear/bilinear dose-response [59]. Surprisingly, the lowest observed genotoxic effect levels for MN formation were similar to or even lower than those identified for the markers of the p53 pathway, including gene expression, protein modification, and cell-based measures of response (Figure 8). In addition, no significant gene expression changes at dose levels below the benchmark doses (BMDs)/PoDs for MN formation were seen with any of the test compounds employed in the study. With MMS, there were few transcriptional responses in the region of the MN-BMDL (Figure 9). Feedback models of perfect adaptation involving transcriptional regulation of repair genes and p53 pathway components were not at all consistent across these experimental results. In the work with MN, control of mutagenicity following low levels of DNA damage did not arise from cell cycle arrest, apoptosis, or up-regulation of DNA repair genes. All of these cellular pathways had BMDs similar to or greater than those for MN formation. Previous work had already shown that transcriptional regulation in response to DNA-damage was a high dose phenomenon and unlikely to be responsible for bilinear curves [129]. Surprisingly, none of the evaluated markers had BMDs below those for MN formation.
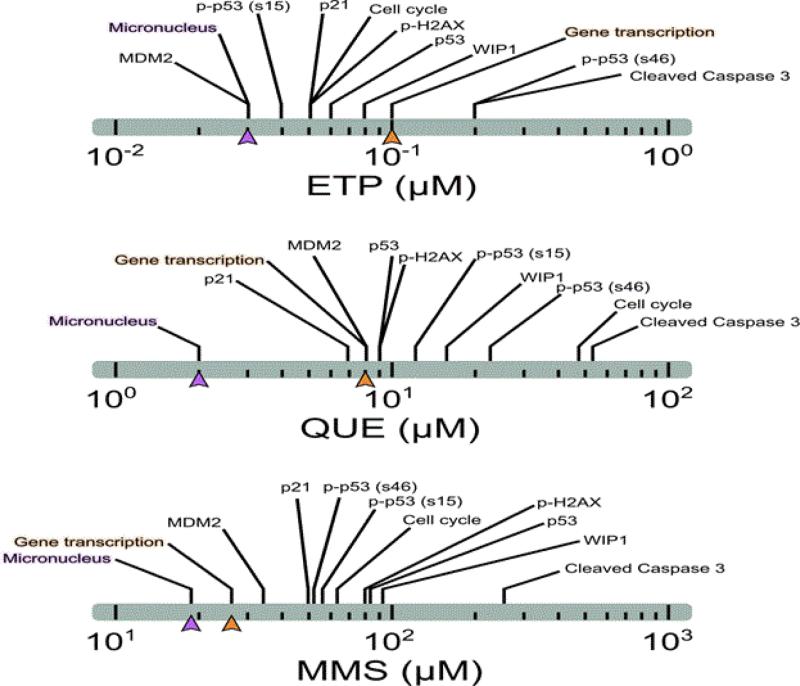
Figure 8. Comparison of responses across endpoints for DNA damage response for three DNA-damaging compounds.
Values shown are for BMDLs (lower 95% confidence limit for BMD). For each chemical, MN induction (purple) occurred at lower doses than gene transcription changes (orange). With MMS, the BMDL for the transcriptional activation was closest to the MN-BMDL for any of the compounds, but was still greater than the MN-BMDL.
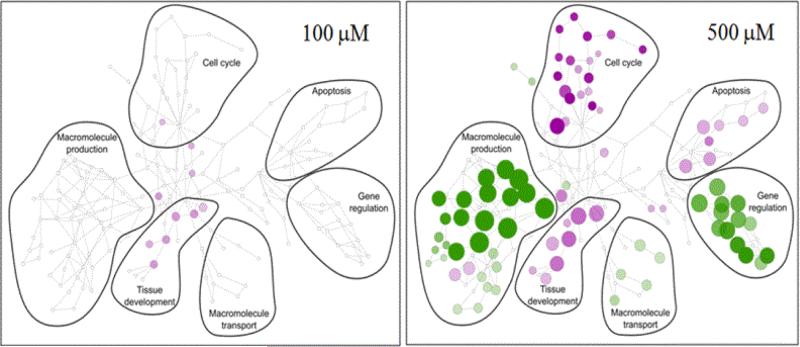
Figure 9. Organization of gene transcriptional changes 24 hr after treating HT-1080 cells with various concentrations of MMS.
Cells were treated with up to 500 μM MMS. Responses at key doses are presented. The union of all significantly changed genes was used to assess all GO ontology categories that were significantly enriched at any treatment. This organization provided the structure of categories shown by the various encircled patterns. The colors, green (up-regulated) and purple (down-regulated), show the groupings that were significantly changed at each treatment and the size of circles represents the numbers of genes changed in particular GO-categories. At 100 μM, the transcriptional changes were minimal even though this concentration was 10-fold above the MN-BMDL. The visualization tools were developed in work with nuclear receptors [133, 134].
6.4 Post-Transcriptional Modification
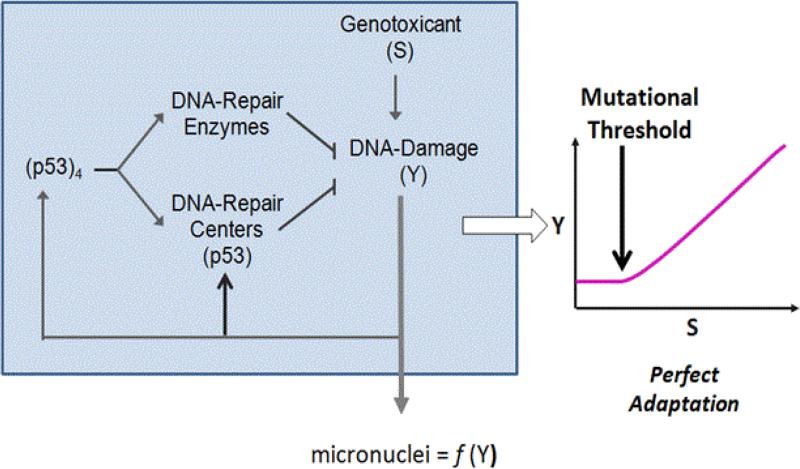
The next steps in the development of DNA-damage pathway dose-response modeling moved toward consideration of processes that contained two regulatory responses. One pathway was the transcriptional activation at higher doses coupled with a more rapid-responding, lower dose pathway involving activation of repair through post-translational modifications of existing repair proteins [130]. A post-translational modification (PTM) for DNA repair may serve as a negative feedback loop to enhance repair rates through increased formation of DNA repair centers (DRCs) [131]. In this two-pathway DNA-repair circuitry, both arms required p53 forming repair centers at low DNA damage levels, or as a tetrameric transcription factor at high levels (Figure 10). Idealized model for perfect adaptation demonstrated thresholds occurred when zero-order degradation of key phosphoproteins involved in the DRC formation/maintenance were included. Progress in examining the biological basis of bilinear responses with MMS will likely require better tools to look at formation, resolution and persistence of the DRCs after low-dose treatments.
Figure 10. Computational Model for Threshold Response.
A working model explaining threshold responses, demonstrated for MN formation with increasing exposures, has two response pathways – a fast acting, post-translational pathway that works to maintain perfect control (and threshold behaviors) and a transcriptional pathway with p53 tetramer that contributes at much higher levels of damage (thus higher doses), after there is an increase in MN formation.
DRCs formation was also examined after treatment with NCS. The dose-response following treatment with NCS was strikingly bilinear. With this compound, there were increases in DRCs at doses below those causing mutations. The resolution of DRCs at sub-threshold doses for MN with NCS was rapid, but became much slower at doses that increased MN formation (Figure 11). Examination of DRC kinetics following treatment with various DNA-damaging agents has to become more common to allow the development of a mechanistic understanding of BPD. This work to date indicated that the threshold behaviors noted with DNA-damaging agents most likely arise due to post-translational activation of DRCs accompanied by processes such as zero-order clearance of components of the repair centers [132].
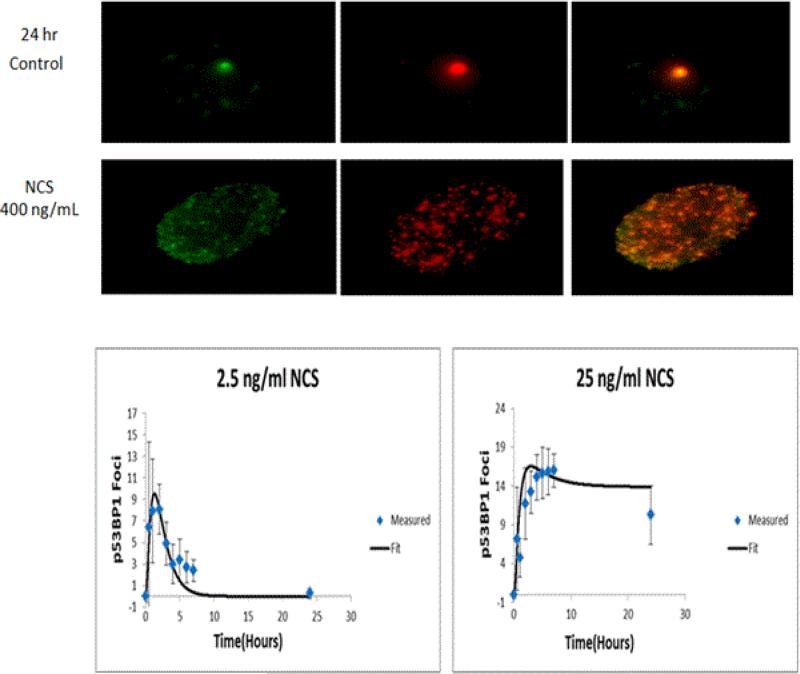
Figure 11. Time-Course Behaviors of DNA-Repair Centers (DRCs) at low and higher doses.
Top: Images of DRC foci in control nuclei. Middle section: images of DRCs following treatment with very high doses of NCS. The foci in individual nuclei show the co-location of two repair proteins – p53 binding protein and γ-H2AX. Lower left: dose- and time-response for DRCs (as foci per nucleus) following treatment with NCS. At lower concentrations (left), foci resolve quickly. At higher concentrations, DRCs persist out beyond 24 hrs. The lower doses are in the sub-threshold region for MN-formation and the higher doses are those with increased MN frequencies. Plots are representative of studies reported in other work from the Hamner-Unilever collaboration [135].
7 Conclusions
There is significant interest in understanding the contribution of biological mechanisms to the non-linear/bilinear dose-response curves for DNA-reactive agents. Model monofunctional alkylating agents have datasets amenable to PoD determination for genotoxic effects in both in vitro and in vivo tests systems; these findings were supported by robust statistical analysis of in vitro and in vivo datasets [27-30]. Such new experimental and computational approaches will help develop mechanistic evidence to support the necessary, biological understanding of the MOA for empirically demonstrated PoDs for DNA-reactive agents. The biological evidence for alkylating agents points to multiple DNA repair systems and DDR pathways acting together to prevent mutation, giving rise to non-linear/bilinear dose-responses for genotoxicity. Complex interactions within a particular DNA repair pathway, as well as interplay between different DNA repair and DDR pathways, counteract the effects of ever-present background DNA damage. Together, these pathways appear sufficient to counteract mutation at low exposures/doses and/or the propagation of cells harboring significant DNA damage. The interplay of these biological processes serves as the basis of non-linear/bilinear dose-responses for genotoxic effects.
Continuing efforts should be increasingly focused on the biological underpinnings of PoDs in order to show the biological pathways/networks involved in repair and homeostatic processes for various types of DNA damage. Hence, the examination of these key cellular, mechanistic responses should be integrated into the designs of the MOA studies to demonstrate the PoDs for genotoxic effects for various types of DNA-reactive agents. These diverse data streams can then be applied to risk assessment for genotoxic chemicals to reduce overall uncertainty in the process, specifically for risk characterization and uncertainties surrounding exposures at environmentally-relevant concentrations.
Acknowledgements
The authors are grateful to Dr. B. B. Gollapudi for his support and contributions during development of the workshop and this manuscript.
This work was supported by National Institutes of Health (NIH) (http://www.nih.gov) grant R01-CA079827; U01-ES016045 with partial support from R21-ES019498 (to B.P.E). The MIT Center for Environmental was funded by NIH grant P30-ES002109 (to B.P.E). This work was also supported by Unilever Safety & Environmental Assurance Centre, UK (to M.E.A, R.A.C, Y.A. and P.L.C) and by ExxonMobil Foundation, USA (to M.E.A). Funding was also provided by State of Connecticut Grant 13-SCB-UCHC-06 and by NIH CA181959 (to C.D.H).
Abbreviations
- AAG
alkyladenine DNA glycosylase
- ATM
Ataxia Telangiectasia Mutated
- BER
base excision repair
- BPD
Break Point Dose
- BMD
benchmark dose
- BMDL
benchmark dose lower confidence limit
- CSBP
computational systems biology pathway
- DSB
double strand breaks
- DDR
DNA damage response
- DRCs
DNA repair centers
- EMS
ethylmethanesulfonate
- ENU
ethylnitrosourea
- FYDR
Fluorescent Yellow Direct Repeat
- HR
homologous repair
- MGMT
methylguanine methyl transferase
- MMS
methylmethane sulfonate
- MNNG
nethylnitronitrosoguanidine
- MNU
methylnitrosourea
- MPG
methylpurine glycosylase
- MN
micronucleus/micronuclei
- MMR
mismatch repair
- MOA
mode-of-action
- N3-MeA
N-3-methyladenine
- N7-MeG
N-7-methylguanine
- NCS
neocarzinostatin
- NRC
National Research Council
- NOGEL
no-observed-genotoxic-effect-level
- NER
nucleotide excision repair
- O6-MeG
O6-methylguanine
- γ-H2AX
Phosphorylated (gamma) histone H2AX
- PSCs
pluripotent stem cells
- PoDs
points of departure
- PTM
post-translational modification
- QUE
quercitin
- SSB
single strand break
- TLS
translesion DNA synthesis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.NRC . National Research Council Committee on Improving Risk Analysis Approaches Used by the U. S. EPA Science and Decisions: Advancing Risk Assessment. National Academy of Sciences; Washington D.C.: 2009. [Google Scholar]
- 2.Cao X, Mittelstaedt RA, Pearce MG, Allen BC, Soeteman-Hernandez LG, Johnson GE, Bigger CA, Heflich RH. Quantitative dose-response analysis of ethyl methanesulfonate genotoxicity in adult gpt-delta transgenic mice. Environmental and molecular mutagenesis. 2014;55:385–399. doi: 10.1002/em.21854. [DOI] [PubMed] [Google Scholar]
- 3.Gocke E, Muller L. In vivo studies in the mouse to define a threshold for the genotoxicity of EMS and ENU. Mutation research. 2009;678:101–107. doi: 10.1016/j.mrgentox.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Marsden DA, Jones DJ, Britton RG, Ognibene T, Ubick E, Johnson GE, Farmer PB, Brown K. Dose-response relationships for N7-(2-hydroxyethyl)guanine induced by low-dose [14C]ethylene oxide: evidence for a novel mechanism of endogenous adduct formation. Cancer research. 2009;69:3052–3059. doi: 10.1158/0008-5472.CAN-08-4233. [DOI] [PubMed] [Google Scholar]
- 5.Sharma V, Collins LB, Clement JM, Zhang Z, Nakamura J, Swenberg JA. Molecular dosimetry of endogenous and exogenous O(6)-methyl-dG and N7-methyl-G adducts following low dose [D3]-methylnitrosourea exposures in cultured human cells. Chemical research in toxicology. 2014;27:480–482. doi: 10.1021/tx5000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu R, Lai Y, Hartwell HJ, Moeller BC, Doyle-Eisele M, Kracko D, Bodnar WM, Starr TB, Swenberg JA. Formation, Accumulation, and Hydrolysis of Endogenous and Exogenous Formaldehyde-Induced DNA Damage. Toxicological sciences. 2015;146:170–182. doi: 10.1093/toxsci/kfv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, Lai Y, Moeller B, Lu K, Swenberg J. The endogenous exposome, DNA repair. 2014;19:3–13. doi: 10.1016/j.dnarep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NRC . Toxicity testing in the 21st century: A vision and a strategy. National Academies Press; Washington DC: 2007. [Google Scholar]
- 10.Krewski D, Acosta D, Jr, Andersen M, Anderson H, Bailar JC, 3rd, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S, Jr., Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L. Toxicity testing in the 21st century: a vision and a strategy, Journal of toxicology and environmental health. Part B. Critical reviews. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerard M, Baum M, Bitsch A, Eisenbrand G, Elhajouji A, Epe B, Habermeyer M, Kaina B, Martus HJ, Pfuhler S, Schmitz C, Sutter A, Thomas AD, Ziemann C, Froetschl R. Assessment of mechanisms driving non-linear dose-response relationships in genotoxicity testing. Mutation research. Reviews in mutation research. 2015;763:181–201. doi: 10.1016/j.mrrev.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Kirsch-Volders M, Aardema M, Elhajouji A. Concepts of threshold in mutagenesis and carcinogenesis. Mutation research. 2000;464:3–11. doi: 10.1016/s1383-5718(99)00161-8. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins GJ, Doak SH, Johnson GE, Quick E, Waters EM, Parry JM. Do dose response thresholds exist for genotoxic alkylating agents? Mutagenesis. 2005;20:389–398. doi: 10.1093/mutage/gei054. [DOI] [PubMed] [Google Scholar]
- 14.Parry JM, Jenkins GJ, Haddad F, Bourner R, Parry EM. In vitro and in vivo extrapolations of genotoxin exposures: consideration of factors which influence dose-response thresholds. Mutation research. 2000;464:53–63. doi: 10.1016/s1383-5718(99)00166-7. [DOI] [PubMed] [Google Scholar]
- 15.Thomas AD, Fahrer J, Johnson GE, Kaina B. Theoretical Considerations for Thresholds in Chemical Carcinogenesis. Mutation Research/Reviews in Mutation Research. 2015 doi: 10.1016/j.mrrev.2015.05.001. doi:10.1016/j.mrrev.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Meira LB, Calvo JA, Shah D, Klapacz J, Moroski-Erkul CA, Bronson RT, Samson LD. Repair of endogenous DNA base lesions modulate lifespan in mice. DNA repair. 2014;21:78–86. doi: 10.1016/j.dnarep.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sukup-Jackson MR, Kiraly O, Kay JE, Na L, Rowland EA, Winther KE, Chow DN, Kimoto T, Matsuguchi T, Jonnalagadda VS, Maklakova VI, Singh VR, Wadduwage DN, Rajapakse J, So PT, Collier LS, Engelward BP. Rosa26-GFP direct repeat (RaDR-GFP) mice reveal tissue- and age-dependence of homologous recombination in mammals in vivo. PLoS genetics. 2014;10:e1004299. doi: 10.1371/journal.pgen.1004299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicological sciences : an official journal of the Society of Toxicology. 2011;120:S130–145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiktor-Brown DM, Olipitz W, Hendricks CA, Rugo RE, Engelward BP. Tissue-specific differences in the accumulation of sequence rearrangements with age. DNA repair. 2008;7:694–703. doi: 10.1016/j.dnarep.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ames BN. Endogenous oxidative DNA damage, aging, and cancer. Free radical research communications. 1989;7:121–128. doi: 10.3109/10715768909087933. [DOI] [PubMed] [Google Scholar]
- 21.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 23.Saul RL, Ames BN. Mechanisms of DNA damage and repair. Springer; US, NY: 1986. Background levels of DNA damage in the population; pp. 529–535. [DOI] [PubMed] [Google Scholar]
- 24.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. The Journal of biological chemistry. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swenberg JA, Moeller BC, Lu K, Rager JE, Fry RC, Starr TB. Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicologic pathology. 2013;41:181–189. doi: 10.1177/0192623312466459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gollapudi BB, Johnson GE, Hernandez LG, Pottenger LH, Dearfield KL, Jeffrey AM, Julien E, Kim JH, Lovell DP, Macgregor JT, Moore MM, van Benthem J, White PA, Zeiger E, Thybaud V. Quantitative approaches for assessing dose-response relationships in genetic toxicology studies. Environmental and molecular mutagenesis. 2013;54:8–18. doi: 10.1002/em.21727. [DOI] [PubMed] [Google Scholar]
- 28.MacGregor JT, Frotschl R, White PA, Crump KS, Eastmond DA, Fukushima S, Guerard M, Hayashi M, Soeteman-Hernandez LG, Kasamatsu T, Levy DD, Morita T, Muller L, Schoeny R, Schuler MJ, Thybaud V, Johnson GE. IWGT report on quantitative approaches to genotoxicity risk assessment I. Methods and metrics for defining exposure-response relationships and points of departure (PoDs) Mutation research. Genetic toxicology and environmental mutagenesis. 2015a;783:55–65. doi: 10.1016/j.mrgentox.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Johnson GE, Soeteman-Hernandez LG, Gollapudi BB, Bodger OG, Dearfield KL, Heflich RH, Hixon JG, Lovell DP, MacGregor JT, Pottenger LH, Thompson CM, Abraham L, Thybaud V, Tanir JY, Zeiger E, van Benthem J, White PA. Derivation of point of departure (PoD) estimates in genetic toxicology studies and their potential applications in risk assessment. Environmental and molecular mutagenesis. 2014;55:609–623. doi: 10.1002/em.21870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacGregor JT, Frotschl R, White PA, Crump KS, Eastmond DA, Fukushima S, Guerard M, Hayashi M, Soeteman-Hernandez LG, Johnson GE, Kasamatsu T, Levy DD, Morita T, Muller L, Schoeny R, Schuler MJ, Thybaud V. IWGT report on quantitative approaches to genotoxicity risk assessment II. Use of point-of-departure (PoD) metrics in defining acceptable exposure limits and assessing human risk. Mutation research. Genetic toxicology and environmental mutagenesis. 2015b;783:66–78. doi: 10.1016/j.mrgentox.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 31.SCOEL Recommendation from the Scientific Committee on Occupational Exposure Limits for Formaldehyde. European Commission The Scientific Committee on Occupational Exposure Limits, SCOEL/SUM/125. 2008 [Google Scholar]
- 32.SCOEL Recommendation from the Scientific Committee on Occupational Exposure Limits for propylene oxide. European Commission The Scientific Committee on Occupational Exposure Limits SCOEL/SUM/161. :2010. [Google Scholar]
- 33.U.K. HSE, The carcinogenicity of formaldehyde A subcommittee report to the Advisory Committee on Toxic Substances, Health & Safety Commission, United Kingdom. EH40/2005 Workplace Exposure Limits (second edition) WATCH (Working Group on Action to Control Chemicals) Committee. United Kingdom Health and Safety Executive. 20052011 [Google Scholar]
- 34.EINECS No. 203-545-4 Summary Risk Assessment Report. Final Report. Germany: 2008. Bundesanstalt für Arbeitsschutz und Arbeitsmedizin (BAuA) Vinyl Acetate CAS No 108-05-4. [Google Scholar]
- 35.M7 ICH Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk. International Conference on Harmonization (ICH) of Technical Requirements for Registration of Pharmaceuticals for Human Use; 2014. [Google Scholar]
- 36.Boobis AR, Cohen SM, Dellarco V, McGregor D, Meek ME, Vickers C, Willcocks D, Farland W. IPCS framework for analyzing the relevance of a cancer mode of action for humans. Critical reviews in toxicology. 2006;36:781–792. doi: 10.1080/10408440600977677. [DOI] [PubMed] [Google Scholar]
- 37.Lutz WK. Endogenous genotoxic agents and processes as a basis of spontaneous carcinogenesis. Mutation research. 1990;238:287–295. doi: 10.1016/0165-1110(90)90020-c. [DOI] [PubMed] [Google Scholar]
- 38.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. The EMBO journal. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. Journal of bacteriology. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballschmiter K. Pattern and sources of naturally produced organohalogens in the marine environment: biogenic formation of organohalogens. Chemosphere. 2003;52:313–324. doi: 10.1016/S0045-6535(03)00211-X. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton JT, McRoberts WC, Keppler F, Kalin RM, Harper DB. Chloride methylation by plant pectin: an efficient environmentally significant process. Science. 2003;301:206–209. doi: 10.1126/science.1085036. [DOI] [PubMed] [Google Scholar]
- 42.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutation research. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 43.Kufe DW, Holland JF, Frei E. American Cancer Society. ancer Medicine 6, BC Decker, Hamilton ON. 2003 [Google Scholar]
- 44.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer treatment reviews. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 45.Pullman A, Pullman B. Molecular electrostatic potential of the nucleic acids. Quarterly reviews of biophysics. 1981;14:289–380. doi: 10.1017/s0033583500002341. [DOI] [PubMed] [Google Scholar]
- 46.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutation research. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 47.Boiteux S, Laval J. Imidazole open ring 7-methylguanine: an inhibitor of DNA synthesis. Biochemical and biophysical research communications. 1983;110:552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- 48.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. The formation and biological significance of N7-guanine adducts. Mutation research. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutation research. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 50.Philippin G, Cadet J, Gasparutto D, Mazon G, Fuchs RP. Ethylene oxide and propylene oxide derived N7-alkylguanine adducts are bypassed accurately in vivo. DNA repair. 2014;22:133–136. doi: 10.1016/j.dnarep.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Engelward BP, Allan JM, Dreslin AJ, Kelly JD, Wu MM, Gold B, Samson LD. A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. The Journal of biological chemistry. 1998;273:5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 52.Johnson RE, Yu SL, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Molecular and cellular biology. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fronza G, Gold B. The biological effects of N3-methyladenine. Journal of cellular biochemistry. 2004;91:250–257. doi: 10.1002/jcb.10698. [DOI] [PubMed] [Google Scholar]
- 54.Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proceedings of the National Academy of Sciences. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldmacher VS, Cuzick R, Thilly WG. Isolation and partial characterization of human cell mutants differing in sensitivity to killing and mutation by methylnitrosourea and N-methyl-N'-nitro-N-nitrosoguanidine. Journal of Biological Chemistry. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 56.Doak SH, Jenkins GJ, Johnson GE, Quick E, Parry EM, Parry JM. Mechanistic influences for mutation induction curves after exposure to DNA-reactive carcinogens. Cancer research. 2007;67:3904–3911. doi: 10.1158/0008-5472.CAN-06-4061. [DOI] [PubMed] [Google Scholar]
- 57.Pottenger LH, Schisler MR, Zhang F, Bartels MJ, Fontaine DD, McFadden LG, Bhaskar Gollapudi B. Dose-response and operational thresholds/NOAELs for in vitro mutagenic effects from DNA-reactive mutagens. MMS and MNU, Mutation research. 2009;678:138–147. doi: 10.1016/j.mrgentox.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Bryce SM, Avlasevich SL, Bemis JC, Phonethepswath S, Dertinger SD. Miniaturized flow cytometric in vitro micronucleus assay represents an efficient tool for comprehensively characterizing genotoxicity dose-response relationships. Mutation research. 2010;703:191–199. doi: 10.1016/j.mrgentox.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lutz WK, Lutz RW. Statistical model to estimate a threshold dose and its confidence limits for the analysis of sublinear dose-response relationships, exemplified for mutagenicity data. Mutation research. 2009;678:118–122. doi: 10.1016/j.mrgentox.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Johnson GE, Doak SH, Griffiths SM, Quick EL, Skibinski DO, Zair ZM, Jenkins GJ. Non-linear dose-response of DNA-reactive genotoxins: recommendations for data analysis. Mutation research. 2009;678:95–100. doi: 10.1016/j.mrgentox.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 61.Thomas AD, Jenkins GJ, Kaina B, Bodger OG, Tomaszowski KH, Lewis PD, Doak SH, Johnson GE. Influence of DNA repair on nonlinear dose-responses for mutation. Toxicological sciences : an official journal of the Society of Toxicology. 2013;132:87–95. doi: 10.1093/toxsci/kfs341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swenberg JA, Fryar-Tita E, Jeong YC, Boysen G, Starr T, Walker VE, Albertini RJ. Biomarkers in toxicology and risk assessment: informing critical dose-response relationships. Chemical research in toxicology. 2008;21:253–265. doi: 10.1021/tx700408t. [DOI] [PubMed] [Google Scholar]
- 63.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Creating context for the use of DNA adduct data in cancer risk assessment: I. Data organization. Critical reviews in toxicology. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 64.Pottenger LH, Gollapudi BB. Genotoxicity testing: moving beyond qualitative “screen and bin” approach towards characterization of dose-response and thresholds. Environmental and molecular mutagenesis. 2010;51:792–799. doi: 10.1002/em.20612. [DOI] [PubMed] [Google Scholar]
- 65.Pottenger LH, Andrews LS, Bachman AN, Boogaard PJ, Cadet J, Embry MR, Farmer PB, Himmelstein MW, Jarabek AM, Martin EA, Mauthe RJ, Persaud R, Preston RJ, Schoeny R, Skare J, Swenberg JA, Williams GM, Zeiger E, Zhang F, Kim JH. An organizational approach for the assessment of DNA adduct data in risk assessment: case studies for aflatoxin B1, tamoxifen and vinyl chloride. Critical reviews in toxicology. 2014;44:348–391. doi: 10.3109/10408444.2013.873768. [DOI] [PubMed] [Google Scholar]
- 66.Johnson NM, Egner PA, Baxter VK, Sporn MB, Wible RS, Sutter TR, Groopman JD, Kensler TW, Roebuck BD. Complete protection against aflatoxin B(1)-induced liver cancer with a triterpenoid: DNA adduct dosimetry, molecular signature, and genotoxicity threshold. Cancer prevention research. 2014;7:658–665. doi: 10.1158/1940-6207.CAPR-13-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olden K, Vulimiri SV. Laboratory to community: chemoprevention is the answer. Cancer Prev Res. 2014;7:648–652. doi: 10.1158/1940-6207.CAPR-14-0124. [DOI] [PubMed] [Google Scholar]
- 68.Albertini RJ, Sweeney LM. Propylene oxide: genotoxicity profile of a rodent nasal carcinogen. Critical reviews in toxicology. 2007;37:489–520. doi: 10.1080/10408440701382959. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chemical research in toxicology. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. The Journal of biological chemistry. 2004a;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 71.O'Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. The Journal of biological chemistry. 2004b;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 72.Osborne MR, Phillips DH. Preparation of a methylated DNA standard, and its stability on storage. Chemical research in toxicology. 2000;13:257–261. doi: 10.1021/tx990182e. [DOI] [PubMed] [Google Scholar]
- 73.Shipova E, Gates KS. A fluorimetric assay for the spontaneous release of an N7-alkylguanine residue from duplex DNA. Bioorg Med Chem Lett. 2005;15:2111–2113. doi: 10.1016/j.bmcl.2005.02.058. [DOI] [PubMed] [Google Scholar]
- 74.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chemical research in toxicology. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 75.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 76.Covo S, Blanco L, Livneh Z. Lesion bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. The Journal of biological chemistry. 2004;279:859–865. doi: 10.1074/jbc.M310447200. [DOI] [PubMed] [Google Scholar]
- 77.Adar S, Izhar L, Hendel A, Geacintov N, Livneh Z. Repair of gaps opposite lesions by homologous recombination in mammalian cells. Nucleic acids research. 2009;37:5737–5748. doi: 10.1093/nar/gkp632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scicchitano DA, Hanawalt PC. Repair of N-methylpurines in specific DNA sequences in Chinese hamster ovary cells: absence of strand specificity in the dihydrofolate reductase gene. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:3050–3054. doi: 10.1073/pnas.86.9.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao W, Chow BL. Synergism between yeast nucleotide and base excision repair pathways in the protection against DNA methylation damage. Current genetics. 1998;33:92–99. doi: 10.1007/s002940050313. [DOI] [PubMed] [Google Scholar]
- 80.Plosky B, Samson L, Engelward BP, Gold B, Schlaen B, Millas T, Magnotti M, Schor J, Scicchitano DA. Base excision repair and nucleotide excision repair contribute to the removal of N-methylpurines from active genes. DNA repair. 2002;1:683–696. doi: 10.1016/s1568-7864(02)00075-7. [DOI] [PubMed] [Google Scholar]
- 81.Nay SL, O`Connor TR. Direct Repair in Mammalian Cells. 2013 [Google Scholar]
- 82.Zair ZM, Jenkins GJ, Doak SH, Singh R, Brown K, Johnson GE. N-methylpurine DNA glycosylase plays a pivotal role in the threshold response of ethyl methanesulfonate-induced chromosome damage. Toxicological sciences. 2011;119:346–358. doi: 10.1093/toxsci/kfq341. [DOI] [PubMed] [Google Scholar]
- 83.Debiak M, Nikolova T, Kaina B. Loss of ATM sensitizes against O6-methylguanine triggered apoptosis, SCEs and chromosomal aberrations. DNA repair. 2004;3:359–368. doi: 10.1016/j.dnarep.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 84.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noonan EM, Shah D, Yaffe MB, Lauffenburger DA, Samson LD. O6-Methylguanine DNA lesions induce an intra-S-phase arrest from which cells exit into apoptosis governed by early and late multi-pathway signaling network activation. Integrative biology: quantitative biosciences from nano to macro. 2012;4:1237–1255. doi: 10.1039/c2ib20091k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. The Journal of biological chemistry. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 87.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, Zurer I, Rotter V, Samson LD, Harris CC. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. The Journal of clinical investigation. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer research. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 89.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klapacz J, Lingaraju GM, Guo HH, Shah D, Moar-Shoshani A, Loeb LA, Samson LD. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Molecular cell. 2010;37:843–853. doi: 10.1016/j.molcel.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer research. 2002;62:219–225. [PubMed] [Google Scholar]
- 92.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutation research. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 93.Olipitz W, Wiktor-Brown D, Shuga J, Pang B, McFaline J, Lonkar P, Thomas A, Mutamba JT, Greenberger JS, Samson LD, Dedon PC, Yanch JC, Engelward BP. Integrated molecular analysis indicates undetectable change in DNA damage in mice after continuous irradiation at ~ 400-fold natural background radiation. Environmental health perspectives. 2012;120:1130–1136. doi: 10.1289/ehp.1104294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muller H. Radiation and genetics. Am Nat. 1930;64:220–251. [Google Scholar]
- 95.Kiraly O, Gong G, Roytman MD, Yamada Y, Samson LD, Engelward BP. DNA glycosylase activity and cell proliferation are key factors in modulating homologous recombination in vivo. Carcinogenesis. 2014;35:2495–2502. doi: 10.1093/carcin/bgu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutation research. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 97.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA repair. 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 98.Duckett DR, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, Modrich P. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Armstrong MJ, Galloway SM. Mismatch repair provokes chromosome aberrations in hamster cells treated with methylating agents or 6-thioguanine, but not with ethylating agents. Mutation research. 1997;373:167–178. doi: 10.1016/s0027-5107(96)00234-5. [DOI] [PubMed] [Google Scholar]
- 100.Galloway SM, Greenwood SK, Hill RB, Bradt CI, Bean CL. A role for mismatch repair in production of chromosome aberrations by methylating agents in human cells. Mutation research. 1995;346:231–245. doi: 10.1016/0165-7992(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 101.Kaina B, Fritz G, Coquerelle T. Contribution of O6-alkylguanine and N-alkylpurines to the formation of sister chromatid exchanges, chromosomal aberrations, and gene mutations: new insights gained from studies of genetically engineered mammalian cell lines. Environmental and molecular mutagenesis. 1993;22:283–292. doi: 10.1002/em.2850220418. [DOI] [PubMed] [Google Scholar]
- 102.Kaina B. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenetic and genome research. 2004;104:77–86. doi: 10.1159/000077469. [DOI] [PubMed] [Google Scholar]
- 103.Zhang H, Tsujimura T, Bhattacharyya NP, Maher VM, McCormick JJ. O6-methylguanine induces intrachromosomal homologous recombination in human cells. Carcinogenesis. 1996;17:2229–2235. doi: 10.1093/carcin/17.10.2229. [DOI] [PubMed] [Google Scholar]
- 104.Zhang H, Marra G, Jiricny J, Maher VM, McCormick JJ. Mismatch repair is required for O(6)-methylguanine-induced homologous recombination in human fibroblasts. Carcinogenesis. 2000;21:1639–1646. doi: 10.1093/carcin/21.9.1639. [DOI] [PubMed] [Google Scholar]
- 105.Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 106.Heinen CD, Schmutte C, Fishel R. DNA repair and tumorigenesis: lessons from hereditary cancer syndromes. Cancer biology & therapy. 2002;1:477–485. doi: 10.4161/cbt.1.5.160. [DOI] [PubMed] [Google Scholar]
- 107.Kaina B. DNA damage-triggered apoptosis: critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochemical pharmacology. 2003;66:1547–1554. doi: 10.1016/s0006-2952(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 108.Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes & development. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adamson A, Beardsley D, Kim W, Gao Y, Baskaran R, Brown K. Methylator-induced, mismatch repair-dependent G2 arrest is activated through Chk1 and Chk2. Mol Biol Cell. 2005;16:1513–1526. doi: 10.1091/mbc.E04-02-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Molecular cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway. The Journal of biological chemistry. 2010;285:5974–5982. doi: 10.1074/jbc.M109.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mastrocola AS, Heinen CD. Nuclear reorganization of DNA mismatch repair proteins in response to DNA damage. DNA repair. 2010;9:120–133. doi: 10.1016/j.dnarep.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quiros S, Roos WP, Kaina B. Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell cycle. 2010;9:168–178. doi: 10.4161/cc.9.1.10363. [DOI] [PubMed] [Google Scholar]
- 115.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Molecular cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 117.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes & development. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klapacz J, Meira LB, Luchetti DG, Calvo JA, Bronson RT, Edelmann W, Samson LD. O6-methylguanine-induced cell death involves exonuclease 1 as well as DNA mismatch recognition in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:576–581. doi: 10.1073/pnas.0811991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cejka P, Stojic L, Mojas N, Russell AM, Heinimann K, Cannavo E, di Pietro M, Marra G, Jiricny J. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. The EMBO journal. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaina B, Ziouta A, Ochs K, Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex-, Mex+ and methylation-tolerant mismatch repair compromised cells: facts and models. Mutation research. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- 121.Lin B, Gupta D, Heinen CD. Human pluripotent stem cells have a novel mismatch repair-dependent damage response. The Journal of biological chemistry. 2014;289:24314–24324. doi: 10.1074/jbc.M114.570937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Q, Bhattacharya S, Andersen ME, Conolly RB. Computational systems biology and dose-response modeling in relation to new directions in toxicity testing, Journal of toxicology and environmental health. Part B. Critical reviews. 2010;13:253–276. doi: 10.1080/10937404.2010.483943. [DOI] [PubMed] [Google Scholar]
- 123.Simmons SO, Fan CY, Ramabhadran R. Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicological sciences. 2009;111:202–225. doi: 10.1093/toxsci/kfp140. [DOI] [PubMed] [Google Scholar]
- 124.Bhattacharya S, Zhang Q, Carmichael PL, Boekelheide K, Andersen ME. Toxicity testing in the 21 century: defining new risk assessment approaches based on perturbation of intracellular toxicity pathways. PloS one. 2011;6:e20887. doi: 10.1371/journal.pone.0020887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Adeleye Y, Andersen M, Clewell R, Davies M, Dent M, Edwards S, Fowler P, Malcomber S, Nicol B, Scott A, Scott S, Sun B, Westmoreland C, White A, Zhang Q, Carmichael PL. Implementing Toxicity Testing in the 21st Century (TT21C): Making safety decisions using toxicity pathways, and progress in a prototype risk assessment. Toxicology. 2015;332:102–111. doi: 10.1016/j.tox.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 126.Clewell RA, Sun B, Adeleye Y, Carmichael P, Efremenko A, McMullen PD, Pendse S, Trask OJ, White A, Andersen ME. Profiling dose-dependent activation of p53-mediated signaling pathways by chemicals with distinct mechanisms of DNA damage. Toxicological sciences. 2014;142:56–73. doi: 10.1093/toxsci/kfu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Q, Andersen ME. Dose response relationship in anti-stress gene regulatory networks. PLoS computational biology. 2007;3:e24. doi: 10.1371/journal.pcbi.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Q, Bhattacharya S, Conolly RB, Clewell HJ, Kaminski NE, Andersen ME. Molecular signaling network motifs provide a mechanistic basis for cellular threshold responses. Environmental health perspectives. 2014;122:1261–1270. doi: 10.1289/ehp.1408244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seidel SD, Sparrow BR, Kan HL, Stott WT, Schisler MR, Linscombe VA, Gollapudi BB. Profiles of gene expression changes in L5178Y mouse lymphoma cells treated with methyl methanesulfonate and sodium chloride. Mutagenesis. 2004;19:195–201. doi: 10.1093/mutage/geh027. [DOI] [PubMed] [Google Scholar]
- 130.Zhang Q, Bhattacharya S, Pi J, Clewell R, Carmichael P, Andersen M. Adaptive Posttranslational Control of Cellular Stress Pathways in relation to Toxicity Testing and Safety Assessment. Toxicolical sciences. 2015;147:302–16. doi: 10.1093/toxsci/kfv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neumaier T, Swenson J, Pham C, Polyzos A, Lo AT, Yang P, Dyball J, Asaithamby A, Chen DJ, Bissell MJ, Thalhammer S, Costes SV. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:443–448. doi: 10.1073/pnas.1117849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clewell R, Sun B, Adeleye Y, Carmichael P, Andersen M. Does DNA-Repair Center Formation at Low Doses Account for Mutagenic Thresholds. Toxicological sciences, in review. 2015a [Google Scholar]
- 133.McMullen PD, Bhattacharya S, Woods CG, Sun B, Yarborough K, Ross SM, Miller ME, McBride MT, LeCluyse EL, Clewell RA, Andersen ME. A map of the PPARalpha transcription regulatory network for primary human hepatocytes. Chemico-biological interactions. 2014;209:14–24. doi: 10.1016/j.cbi.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 134.Pendse S, McMullen PD. An Interactive Visualization Tool to Interpret Transcriptomics Data. International Conference on Bioinformatics & Computational Biology. 2014:215. [Google Scholar]
- 135.Clewell RA, Sun B, Adeleye Y, Carmichael PL, Andersen ME. Does DNA-Repair Center Formation at Low Doses Account for Mutagenic Thresholds. Toxicological sciences. 2015 in review. [Google Scholar]
- 136.Slob W, Setzer RW. Shape and steepness of toxicological dose-response relationships of continuous endpoints. Critical Reviews in Toxicology. 2014;44:270–197. doi: 10.3109/10408444.2013.853726. [DOI] [PubMed] [Google Scholar]
- 137.Fu D, Calvo JA, Samson LD. Genomic instability in cancer Balancing repair and tolerance of DNA damage caused by alkylating agents. Nature reviews in cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ji Z, LeBaron MJ, Schisler MR, Zhang F, Bartels MJ, Gollapudi BB, Pottenger LH. Dose–Response for Multiple Biomarkers of Exposure and Genotoxic Effect Following Repeated Treatment of Rats with the Alkylating Agents. MMS and MNU. Mutagenesis, gev035. 2015 doi: 10.1093/mutage/gev035. [DOI] [PubMed] [Google Scholar]