Abstract
Objective
To identify glucocorticoid receptor (GR)-associated chromatin sequences and target genes in primary human abdominal subcutaneous fat.
Methods
GR chromatin immunoprecipitation (ChIP)-sequencing methodology in subcutaneous human adipocytes treated ex-vivo with dexamethasone (dex) was optimized to identify genome-wide dex-dependent GR binding regions (GBRs). Gene expression analyses were performed in parallel ± dex treatment.
Results
Fat was obtained from four non-obese female surgical patients with a median age of 50.5 years. ChIP-seq analysis revealed 219 dex-associated GBRs. Of these, 136 GBRs were located within 100 kb of the transcriptional start site and associated with 123 genes. Combining these data with dex-induced gene expression, 70 of the 123 putative direct target genes were significantly up- or downregulated following four hours of dex treatment. Gene expression analysis demonstrated that the top 10 pathways reflected regulation of cellular metabolism and inflammation. DEPTOR, an inhibitor of mTOR, was identified as a potential direct GR target gene.
Conclusions
To our knowledge, this is the first report of genome-wide GR ChIP-seq and gene expression analysis in human fat. The results implicate regulation of key GR target genes that are involved in dampening inflammation and promoting cellular metabolism.
Keywords: Glucocorticoid receptor, adipose tissue, ChIP-sequencing, gene expression array, inflammation, DEPTOR
Introduction
Nuclear hormone receptors play key roles in adipocyte biology, including promoting adipogenesis, secretion of adipokines1, and the development of insulin resistance2,3. The ligand-bound glucocorticoid receptor (GR) regulates genes with important roles in endocrine physiology including adipocyte differentiation4, lipolysis2,3 and, as seen in Cushing’s disease, adipose tissue distribution5. Furthermore, the potent anti-inflammatory effects of synthetic glucocorticoids result in their extensive use for treating a wide variety of diseases. However, systemic glucocorticoid use is complicated by increased risk of infection, impaired wound healing, and inflammatory changes in the skin6. The effects of GR-activation are tissue-specific and fat depot-dependent, likely due to variable GR expression and depot-specific GR-mediated gene expression1. For example, more GR is expressed in visceral fat than abdominal subcutaneous (sc) or femoral sc fat, the latter having the lowest GR expression7. Prolonged exposure to synthetic or endogenous glucocorticoids results in adipose tissue accumulation and insulin resistance in visceral fat that contributes to the development of metabolic syndrome1.
Few studies examining the molecular effects of GR activation in human adipose tissue have been performed and the conclusions are somewhat contradictory. Previous work in human sc adipocytes (isolated following collagenase fat digestion) did not show an effect of glucocorticoid treatment on either insulin-mediated Akt phosphorylation or glucose uptake8. Ex vivo experiments using differentiated human sc preadipocytes found that dexamethasone (dex) treatment did not significantly alter insulin-driven lipogenesis9 but did increase insulin-mediated glucose uptake10. Most recently, primary sc adipocytes pre-treated for 24 hours with cortisol and then stimulated with insulin had relatively increased Akt phosphorylation compared to insulin stimulation alone suggesting that GR activation did not induce insulin resistance in this depot11.
Despite the clinical importance of glucocorticoids and their obvious effects on adipocyte biology, no studies have identified direct GR target genes in human fat. To our knowledge, the only adipocyte GR ChIP-seq results have been published by Yu et al. who performed high-throughput GR ChIP-seq in the murine adipocyte 3T3-L1 cell line. Several GR-binding regions (GBRs) near dex-regulated genes involved in triglyceride synthesis, lipolysis, lipid transport and storage were identified12. To identify GR target genes in sc fat, we developed a GR ChIP-seq protocol that effectively immunoprecipitated GR and cross-linked DNA from isolated human abdominal sc adipocytes. We then identified dex-dependent GBRs and determined their individual locations relative to the closest transcriptional start site (TSS). We also used gene expression array data from dex-treated primary sc adipose tissue and identified significant gene expression changes and compared those genes with genes identified from the proximal TSSs near dex-dependent GBRs. We found that the resulting putative direct GR target genes encoded proteins mainly involved in inflammation and cellular metabolic processes rather than fat synthesis, transport and storage. These findings may help explain the differential effects of glucocorticoids in sc versus other fat depots.
Methods
Adipose Tissue Culture
Otherwise-discarded, de-identified abdominal sc adipose tissue was collected from nine female patients undergoing autologous breast reconstruction with an IRB-approved protocol. Adipose tissue collected from one patient was used for ChIP-seq analysis, three patients for gene expression array analysis and five patients for quantification of mRNA expression of DEPTOR. Patients were not known to be insulin-resistant and were not taking any class of anti-diabetic drug, insulin or glucocorticoids. Patient information that was collected from the reconstructive surgeon included age, self-reported race, body-mass index (BMI), co-morbidities, medications, and post-operative fasting serum glucose. Following surgical excision, sc fat was immediately processed as follows: any grossly visible skin, connective or vascular tissue was removed and the remaining fat was cut into portions with a sharp butcher knife and placed into 50 mL conical tubes containing 20 mL of 1× PBS. Total volumes (PBS plus fat) in each conical tube did not exceed 40 mL. The fat was then hand-minced into small pieces (< 1 mm3) with surgical scissors. Additional stromal/vascular cells were removed by repeated gentle washing in volumes of PBS sufficient to fill a 50-mL conical tube followed by gentle centrifugation at 100 × g. This step was repeated until no additional stromal/vascular cells were observed at the bottom of the tube following centrifugation (generally 3 – 4 times, with some variation between individual subject specimens). Fat was then cultured overnight in 300-cm2 flasks containing 10 mL of low-glucose DMEM fat culture medium (1.0 g/L glucose, 10,000 U/mL gentamicin, and 1% penicillin/streptomycin) for each packed mL of fat.
Chromatin Immunoprecipitation (ChIP)
We tested a variety of conditions for GR ChIP and found the following methodology to be optimal. Following overnight culture in fat culture medium, sc fat from one subject was treated for 20 – 30 minutes with fat culture medium containing 1 mg/mL of collagenase (Type 1, Worthington Biochemical Corp) at 37.5 °C in a shaking incubator at a low intensity setting until tissue appeared completely dissociated. Of note, overnight culture allowed the fat to “recover” prior to performing the collagenase digestion in order to minimize adipocyte lysis/cell death and to increase the quantity and quality of DNA and immunoprecipitated GR per volume of adipocytes. Following collagenase digestion, adipocytes were isolated from stromal/vascular cells by filtration through a 350 µm nylon mesh (Small Parts, #: CMN-0350-D), then washing in 10-fold excess 1× PBS, and finally by centrifugation. Isolated adipocytes were then cultured overnight in a 1:10 v/v of fat culture medium.
The following day, the isolated adipocytes were treated for 1 hour with dex (1 µM) or vehicle (EtOH; 0.1% v/v) at 37°C. Crosslinking of proteins and DNA was performed in 1% formaldehyde for 10 minutes at room temperature followed by a 5-minute treatment with 1× glycine. Adipocytes were lysed by sonication in ChIP lysis buffer (5 mM PIPES, 85 mM KCl, and 0.5% Igepal) containing protease inhibitors. DNA sonication (Misonix, Inc. Sonicator model S-4000) was performed in 30-second cycles with 1-minute rest intervals (50 cycles, 90 amps). After centrifugation for 15 minutes at 4000 × g, the supernatant was discarded and the pellet was resuspended in nuclear lysis buffer (50 mM Tris-Cl, 10 mM EDTA, 1% SDS) with cOmplete Protease Inhibitor Cocktail tablets (Roche). ChIP was performed using the Millipore ChIP Assay Kit (17–295). Non-specific reactions were blocked using salmon sperm DNA/protein A (Millipore) and GR with bound, fragmented chromatin was immunoprecipitated on agarose beads (Millipore) using a polyclonal anti-GR IgG antibody (Santa Cruz GR Antibody E-20). DNA was eluted with ChIP elution buffer (10% 1M NaHCO3, 10% SDS, and 80% H2O by volume), reverse cross-linked in 5M NaCl, and purified using Qiagen’s PCR purification kit per kit instructions. Anti-GR ChIP efficiency was confirmed by using qRT-PCR to measure enrichment of a GBR in the SGK1 promoter as described previously13,14. Primers used for amplifying the known GR-bound SGK1 promoter region were: 5′-CCCCTCCCTTCGCTTGTT-3′ (sense), 5′-GGAAGAAGTACAATCTGCATTTCACT-3′ (antisense)13.
Gene Expression Analysis
Following overnight culture, adipose tissue was transferred to fresh fat culture medium and treated for four hours with dex (1 µM) or vehicle (EtOH; 0.1% v/v). Total RNA was harvested using the E.Z.N.A. Total RNA Kit II (Omega Bio-Tek) per kit instructions. Preparation of biotinylated cRNA (Affymetrix IVT Labeling Kit) and hybridization to oligonucleotide arrays (Affymetrix HG U133 Plus 2.0) was performed per manufacturer’s instructions by the University of Chicago Genomics Core Facility. Signal intensity data were normalized using the Robust Multiarray Average (RMA) method15 in the Bioconductor Affy Package (version 2.11)16 for each subject’s sample. Upregulated and downregulated genes were identified as those with greater than 1.3 fold increase or 0.77 decrease between dex- and vehicle-treated array intensity values. Using these genes, significant pathways, with unadjusted P-values < 0.05, were determined using Ingenuity Pathway Analysis® software (IPA®, QIAGEN Redwood City, v23814503)17.
Sequencing and Bioinformatics Analysis
Samples were sequenced using the Illumina Solexa Genome Analyzer II platform at Argonne National Laboratory: dex – input, dex - immunoprecipitated (IP), vehicle – input, vehicle – immunoprecipitated. This experimental design allowed for two levels of controls: 1) input samples acted as controls for IP samples and 2) vehicle acted as a negative control for dex.
A total of 97,195,147 (36 bp) single-end reads were produced and assessed for quality using FastQC v0.10.118. High quality was observed across all samples, where mean quality scores ranged from 35.20 ± 1.81 to 37.50 ± 1.20. Reads were aligned to the human genome (hg18) using BWA aln (v0.7.5a) software (aln parameters: -I -q 15; samse parameters: -n 10; samtools view parameters: -F 4)19. Alignments were filtered to include only uniquely-mapped reads using custom scripts.
Peaks were analyzed using two methods. In Method 1, peaks were detected by comparing IP samples to input samples (MACS2 parameters, v2.1.0.20140616: -B – SPMR –bw 500 -p 0.05 –to- large -m 5, 50 -p 0.05)20. Then, the enriched peaks were compared between dex and vehicle samples using MAnorm21. For Method 2, the vehicle IP sample was directly compared to the dex IP sample (using the same MACS2 parameters as Method 1). We note that the standard quality statistics for ChIP-seq experiments22 suggested that the experimental data was of relatively low quality (dex ip: cross-correlation = 0.147, phantom peak location = 35, NSC = 1.02, RSC = 0.30; vehicle ip: cross-correlation = 0.20, phantom peak location = 35, NSC = 1.02, RSC = 0.30). This may be due to the purity of chromatin immunoprecipitated from lipid-laden fat. Fortunately, the use of multiple methods to detect high quality peaks and knowing a priori candidate genes (e.g. SGK1) produced robust, biologically-relevant results. Due to the parameters used for peak detection, a great number of peaks were detected in both Methods 1 and 2; after filtering and detecting peaks that were concordant between the two methods, 201 peaks remained (92% of Method 1 peaks), suggesting high concordance between methods. Thus, we focused only on the 219-filtered Method 1 peaks for the remainder of the analyses. The peaks were annotated by Homer v4.623 with default parameters and filtered for peaks located within 100 kb of a nearest TSS using custom scripts.
The lists of differentially expressed genes (within each subject; n=3) from the gene expression analyses were then further characterized: 1) ”up” for upregulated genes, 2) ”down” for downregulated genes, and 3) ”both” for genes with multiple probes exhibiting different regulation patterns.
qRT-PCR
qRT-PCR was used to confirm differential expression of DEPTOR, determined to be a putative direct target gene based on multiple proximal (< 10 kb from the TSS) GBRs. Following surgical excision, sc fat from five additional subjects was immediately processed as follows: any grossly visible skin, connective or vascular tissue was removed and the remaining fat was cut into portions with a sharp butcher knife and washed in 1× PBS to remove any additional stromal/vascular tissue. Fat was then cultured overnight in 300-cm2 flasks containing 10 mL of fat culture medium for each packed mL of fat. Following overnight culture, adipose tissue was transferred to fresh fat culture medium and treated ex vivo for four hours with dex (1 µM) or vehicle (EtOH; 0.1% v/v). Total RNA was then extracted. cDNA was generated using qScript cDNA SuperMix (Quanta) and PCR performed using PerfeCTa SYBR Green FastMix (Quanta). The following primers were used for amplification: DEPTOR 5′-GCATGAGTTTAATTGCCGTCTC-3′ (sense), 5′-CGTCATCATCTCAAGACCTACC-3′ (antisense). GILZ induction was used as a positive control for glucocorticoid effect; primers used were: 5'-ACAGGCCATGGATCTGGTGA-3’ (sense), 5'-CAGCTCTCGGATCTGCTCCTT-3’ (antisense). β-Actin was used for normalization and amplified with the following primers: 5'-CAGCGGAACCGCTCATTGCCAATGG-3’ (sense), 5'-TCACCCCCTGTGCCCATCTACGA-3’ (antisense). A mixed-effects analysis of variance (ANOVA) model was fitted with Ct (cycle threshold) as the response variable; treatment (dex vs. vehicle), gene type (DEPTOR vs. β-Actin), and treatment × gene interaction as the fixed effects; and patient as the random effect24. A linear contrast was constructed to estimate ΔΔCt and its confidence interval, and the results were exponentiated to obtain the estimate of 2−ΔΔCt. Statistical analysis was performed using SAS software (version 9.4, SAS Institute Inc, Cary, NC).
Results
Subject characteristics
Otherwise-discarded abdominal sc fat was collected from four female patients with a median age of 50.5 (range 39 – 80). Three subjects self-reported Caucasian and one reported African-American ancestry (Table 1). Median BMI was 26.5 kg/m2 (range 22.3 – 29.0 kg/m2), i.e. non-obese. Subjects were without metabolic co-morbidities and were not taking glucocorticoids or any class of anti-diabetic drug. Post-operative serum glucose levels while subjects were NPO (nothing by mouth) and only on intravenous fluids indicated that subjects were normoglycemic and likely not diabetic.
TABLE 1.
Subject characteristics
| Subject | Age | Self-reported race | BMI (kg/m2) |
Relevant Co-morbidities |
Relevant Medications |
Analysis of Tissue |
|---|---|---|---|---|---|---|
| A | 54 | Caucasian | 22.3 | None | None | ChIP-seq |
| 1 | 39 | Caucasian | 24.6 | None | None | Expression microarray |
| 2 | 80 | Caucasian | 28.3 | None | None | Expression microarray |
| 3 | 47 | African-American | 29.0 | None | None | Expression microarray |
Abdominal subcutaneous adipose tissue was collected from female subjects undergoing autologous breast reconstruction and then cultured overnight prior to analysis. BMI, body-mass index.
GR ChIP-seq analysis of subcutaneous fat
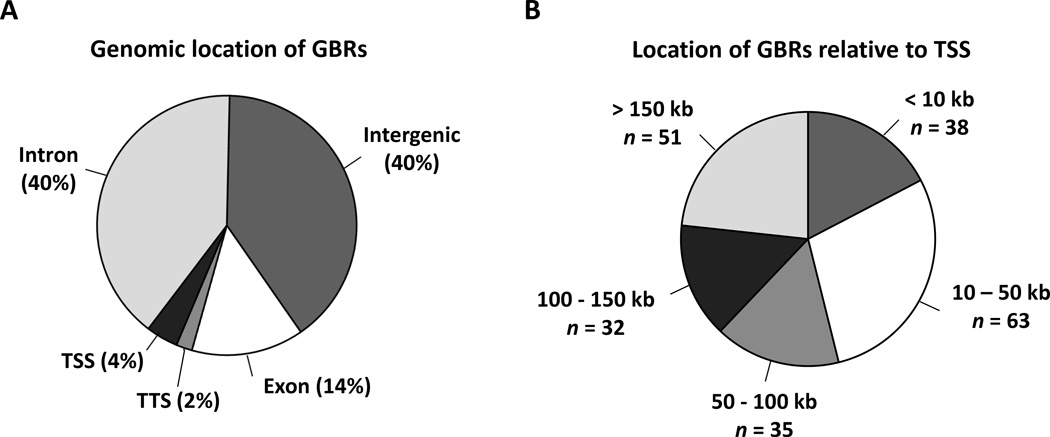
To identify putative direct GR target genes in isolated abdominal sc adipocytes, we performed anti-GR ChIP-seq and determined the location of GBRs with a cutoff of –log2 P ≥ 4 using the MACS2 software. A total of 219 dex-specific GBRs were identified. As shown in Figure 1A, the most GBRs were located in either intergenic (40%) or intronic regions (40%), with a small percentage located in exons (14%), TSSs (4%) or transcriptional termination sites (TTSs; 2%). We were particularly interested in the GBRs within 100 kb of the nearest TSS (Tables 2 and 3). Of these GBRs, 28% (n = 38) were within 10 kb, 46% (n = 63) were 10 – 50 kb and 26% (n = 35) were 50 – 100 kb from the TSS (Figure 1B). These 136 dex-specific GBRs corresponded with 123 genes. The majority of these genes were associated with a single GBR (n=119) while four genes (DEPTOR, COL14A1, NR3C1, POTEA) had multiple GBRs.
Figure 1.
(A) Location of GBRs across the human genome. Dex-specific GBRs were mapped to the human genome (v hg18) and the relative percentages of GBRs found in different regions are indicated. The vast majority (80%) of GBRs were found within intergenic and intronic regions. TTS, transcriptional termination site. (B) Distance of distinct dex-specific GBRs, identified by ChIP-seq analysis, from the transcriptional start site (TSS). Primary human subcutaneous adipose tissue was cultured overnight and adipocytes were isolated by collagenase digestion, washed and treated with dexamethasone (dex) (1 µM) or vehicle for one hour prior to ChIP-seq analysis. A total of 219 dex-specific GR-binding regions (GBRs) were identified, -log2 P ≥ 4. The figure shows the number, n, of GBRs and the distance (in kb) from the TSS.
TABLE 2.
Genes with a TSS < 10 kb from a GBR
| AHSP | EEF1A1 | HIST4H4 | NR3C2 |
| BIVM-ERCC5 | GH1 | ING3 | PABPC1 |
| C16orf3 | HBB | KIF9-AS1 | PAIP2 |
| CCKBR | HIST1H2BC | LRRCC1 | SGK1 |
| CNKSR2 | HIST1H4B | MIR4271 | SLC18B1 |
| DDX11L5 | HIST1H4C | MOV10 | USP14 |
| DEPTOR (2) | HIST1H4H | NAF1 | VPS26A |
| DSTYK | HIST1H4I | NR2F1-AS1 | ZNF219 |
| EDC3 | HIST1H4L | NR3C1 (2) | ZNF385B |
A total of 36 genes were identified within a TSS within 10kb (either direction) of a dex-dependent GBR. A gene listed with “(2)” denotes two distinct GBRs within 10 kb of the same TSS. Additional significant (−log2 P ≥ 4) dex-dependent GBRs [n = 181, (Figure 1B)] located greater than 10 kb from the nearest TSS of a gene are listed in supplemental Table 1.
TABLE 3.
Genes with a TSS > 10 kb from a GBR, n = 181
| > 150 kb | |||
| ACTL7B | DPYD-AS1 | MKRN9P | RP11−799P8.1 |
| ACTRT1 (2) | DST | NFIA | SNX16 |
| APCDD1 | DYM | NR3C2 (7) | SPIN4 |
| APOBEC4 | FLJ41200 | OPHN1 | ST6GAL2 (2) |
| BTBD9 | FNDC3B | PCDH17 | ST6GALNAC5 |
| C4orf32 | LOC100289230 | PJA2 | TEX41 |
| CCDC102B | LOC101927697 | PRG4 | TOM1L1 |
| CDKN2B | LOC401497 | PRR20C | TSN |
| CEP128 | MEI4 | RAG2 | UBE2E2 |
| CSNK1G3 | MIR4799 | RBFOX1 | ZFHX3 |
| DMRT2 | MIR548AU | RP11-265P11.2 | |
| 100 – 150 kb | |||
| AHR | FAM76B | NAMPT | RP11-575F12.1 |
| COL14A1 (8) | FBXL17 | NR3C1 (8) | RP11−65M17.1 |
| CWH43 | KYNU | POT1 | SLC35F4 |
| CYB561 | LINC01080 | RP11-460N16.1 | TTC29 |
| EBF1 | MIR548AN | ||
| 50 – 100 kb | |||
| AC079776.3 | EIF3M | PDE8A | SEMA3A |
| ACTRT1 | ESYT3 | POTEA (3) | SMAD6 |
| ACVR1C | FERMT2 | ROCK1P1 | TSHZ2 |
| AKAP6 | KAT6A | RP11−81H3.2 | ZMYM2 |
| BARX2 | KDM6A | RRAGB | ZNF217 |
| COL14A1 (2) | MAT2B | SCAF8 | ZNF730 |
| DEPTOR (6) | NR3C1 (2) | ||
| 10 – 50 kb | |||
| AIMP1 | KDELR3 | MXD3 | RTN4 |
| AKAP7 | KIAA1147 | MYO16 | SELPLG |
| BRAP | KIAA1614 | NRG2 | SENCR |
| C1orf140 | KLHL31 | NTPCR | SLC13A1 |
| CA8 | LAMA3 | OR4K5 | SLC23A2 |
| CBR4 | LCORL | PALLD | SLC7A14 |
| CCNE2 | LOC284009 | PAWR | SNX1 |
| DYNC1I2 | LRRIQ3 | POM121L4P | SPANXN2 |
| EDEM3 | LRRN3 | POPDC3 | STK24 |
| GABPB1-AS1 | LTN1 | PRKAR1A | TINAG |
| GPC4 | MAGEB18 | PRKG1 | TRPM3 |
| GPR128 | MIR3975 | RALA | UBE2E3 |
| HLF | MIR591 | RECQL | VPS13D |
| IL12RB2 | MIR618 | RNF2 | YIPF5 |
| INSM2 | MIR7844 | RP5−1043L13.1 | ZBTB2 |
| KCTD20 | MSRB3 | RP5−899E9.1 |
A gene listed with (n) denotes the number (n) of distinct GBRs located in that region in either direction of the TSS.
GR ChIP-seq and glucocorticoid-regulated gene expression identify GR target genes and associated pathways
Human abdominal sc fat from three subjects was treated for four hours with vehicle or dex and RNA was harvested to determine glucocorticoid-mediated gene expression. A total of 9766 genes were differentially regulated (> 1.3– or < 0.77-fold change) with dex treatment (Table 4). Subject 1 had 2395 genes differentially regulated of which 1042 were upregulated, 1312 were downregulated and 41 were both up- and downregulated (different probes for a single gene demonstrated differential regulation patterns). Subject 2 had 3154 genes differentially regulated, 1169 were upregulated, 1937 were downregulated and 48 were both. Subject 3 had a larger number of genes differentially regulated; of 7891 genes, 3599 were upregulated, 3925 were downregulated and 367 were both. Gene expression data were then compared to GBRs within 100 kb of a TSS to determine putative direct GR target genes. We found GBRs within 100 kb of a TSS for 17/123 (14%) genes that were upregulated, 38/123 (31%) genes that were downregulated and 15/123 (12%) genes that were both up- and downregulated; the remaining 53 genes (43%) had no change in regulation with dex treatment in any of the three subjects (Table 5). This suggests that in primary fat, activated GR binds to many chromatin sites throughout the genome without affecting four-hour gene expression; among those GR-associated genes that are subject to dex-mediated regulation, most are downregulated.
TABLE 4.
Glucocorticoid-regulated gene expression
| Subject | Total number of genes differentially regulated |
Upregulated (≥ 1.3-fold) |
Downregulated (≤ 0.77-fold) |
Both |
|---|---|---|---|---|
| 1 | 2395 | 1042 | 1312 | 41 |
| 2 | 3154 | 1169 | 1937 | 48 |
| 3 | 7891 | 3599 | 3925 | 367 |
“Both” signifies genes that were upregulated and downregulated (i.e. probes for a single gene demonstrated differential expression patterns).
TABLE 5.
Glucocorticoid-regulated genes associated with GBRs < 100 kb from their TSS (N=3 subjects)
| Direction of regulation | Number of genes (N=123) |
|---|---|
| Upregulated (≥ 1.3-fold) | 17 |
| Downregulated (≤ 0.77-fold) | 38 |
| Both | 15 |
| No change | 53 |
Minced human abdominal subcutaneous adipose tissue was treated ex vivo with dexamethasone (1 µM) or vehicle (EtOH) for 4 hours prior to RNA harvest. “Both” signifies genes that were upregulated in at least one subject and downregulated in another subject.
We next used the dex-induced gene expression data (independently of ChIP-seq analysis) to determine associated biological pathways. The top 10 pathways and their P-values are shown for each subject in Table 6. Pathways related to immune, cancer (cell metabolism) and GR function were identified for all subjects. “Molecular Mechanisms of Cancer” was the first or second pathway for all subjects, suggesting that adipocyte cell metabolism is modified by GR activation. Other significant pathways included immune pathways such as “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis” and “B Cell Receptor Signaling.” “Glucocorticoid Receptor Signaling” was a top 10 pathway for two subjects. Thus, although the quantity of GR-regulated genes in human fat varied among the three subjects, the pathways identified had significant functional overlap and suggested the direct regulation of cellular metabolism and inflammation.
TABLE 6.
Top 10 glucocorticoid-dependent gene expression pathways in primary human subcutaneous abdominal adipose tissue
| Subject 1 (P-value) | Subject 2 (P-value) | Subject 3 (P-value) |
|---|---|---|
|
|
|
Pathways that are in boldface appear in more than one patient.
= pathway present in all three patients.
Verification of a direct GR target gene by qRT-PCR
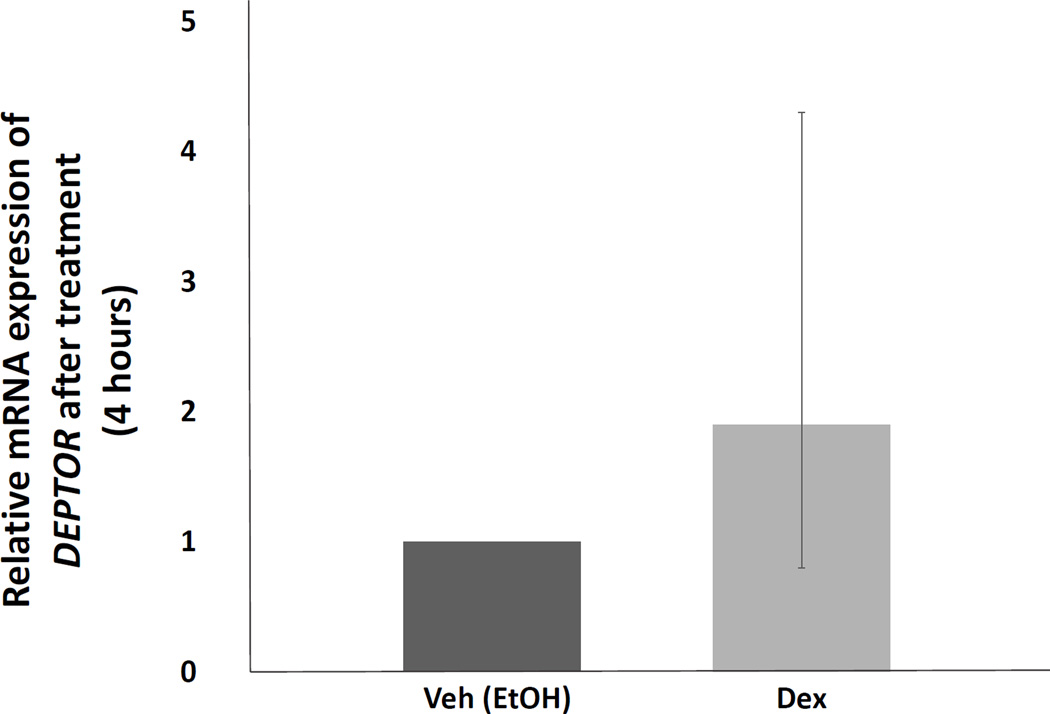
Of the putative direct GR target genes, DEPTOR, an inhibitor of the mechanistic target of rapamycin (mTOR)25, demonstrated both the largest and most numerous peak number (2 peaks < 10 kb and 6 peaks 10 – 100 kb from the TSS). In addition, DEPTOR was upregulated an average of 1.47-fold (n = 3) in the gene expression array analysis (probe 218858_at). We then performed DEPTOR qRT-PCR using mRNA collected from five additional subjects’ abdominal sc fat. DEPTOR mRNA transcript levels increased an average of 1.9-fold (95% CI: 0.81–4.26; P = 0.14) following four-hour dex treatment (Figure 2). We also noted increased expression of DEPTOR protein with 12- and 24- hour dex treatment of human fat compared to vehicle (data not shown).
Figure 2.
Relative mRNA expression of putative GR target gene, DEPTOR, by qRT-PCR. RNA was harvested from primary minced abdominal subcutaneous fat (from additional subjects, N = 5) treated with dex (1 µM) or veh (EtOH) for four hours and qRT-PCR for DEPTOR was performed. There was a trend toward significance with an average 1.9-fold (95% CI: 0.8 – 4.3; P = 0.14) increase in mRNA expression of DEPTOR with dex treatment. Error bars represent 95% CI. Veh = vehicle. Dex = dexamethasone.
Discussion
Although the effects of GR activation in visceral fat have been investigated by multiple groups, the mechanisms by which GR regulates gene expression in sc fat is less well-understood. Therefore, we performed a genomic analysis to investigate GR-mediated gene expression in human sc fat. To our knowledge, this is the first report of GR ChIP-seq in human sc fat and therefore, the first identification of putative direct GR target genes in this depot.
Yu and colleagues performed GR ChIP-seq in the murine adipocyte 3T3-L1 cell line and reported many GBRs near genes involved in triglyceride homeostasis12. In contrast, our data did not identify GBRs associated with triglyceride metabolism genes likely reflecting the differences in GR-regulated functions in human subcutaneous fat versus a cultured cell line derived from a mouse embryonic fibroblast. Lee et al. conducted longer-term gene expression studies (without ChIP-seq) using human omental and abdominal sc fat treated with insulin and dex for seven days in culture. Although some immune function genes were induced, there was significant suppression of immune and inflammatory pathways in both fat depots supporting the potent anti-inflammatory effects of synthetic glucocorticoids26. Interestingly, these results and our work demonstrate acute and chronic regulation of inflammatory pathways by GR activation. This is further supported by the in vivo studies of Pavlatou and colleagues who found that evening cortisol, the ratio of morning/evening cortisol, and 24-hour urinary-free cortisol levels measured in overweight and obese subjects correlated with mRNA expression of cellular metabolism and inflammatory genes in abdominal sc fat27.
Of the 123 genes associated with at least one GBR (within 100 kb from the nearest TSS), 70 genes were regulated by GR activation four hours following ex-vivo dex treatment. GR is known to associate with chromatin in a dynamic, temporal fashion and thus, may differentially regulate gene transcription at different timepoints28. In addition, differential mRNA half-lives will lead to different time frames of steady-state mRNA induction or repression following GR activation of target genes. Nevertheless, we found several predicted GR target genes as well as unexpected direct target genes. The latter include three histone-encoding genes, HIST1H2BC, HIST1H4C and HIST1H4H, all of which demonstrated a GBR within 10 kb of their TSS and were also significantly regulated (Table 2). Thus, GR activation may regulate histone expression and thereby, chromatin remodeling. NR3C1, which encodes GR itself, and NR3C2, which encodes the mineralocorticoid receptor (MR), both had associated GBRs; significant downregulation of NR3C1 was seen in one subject’s gene expression analysis. The proximity of GBRs to the TSS of both these genes supports a negative feedback mechanism of GR on GR29 and MR transcription in human fat.
There were 53 genes that had an associated GBR but were not significantly regulated by GR after four-hour dex treatment by gene expression analysis. These GR-bound, four-hour unregulated genes may be regulated at another timepoint in human fat. One example was SGK1 which was found to have a proximal GBR but not significantly regulated with four-hour dex treatment. SGK1 is known to be regulated by GR30,31 but also by other nuclear receptors (NRs) such as the progesterone receptor and MR31 suggesting the complex cross-talk between NRs to regulate gene expression. We identified two significant dex-dependent GBRs within 10 kb of the DEPTOR TSS and confirmed dex-dependent induction of DEPTOR mRNA steady-state levels in five additional subjects’ RNA. DEPTOR encodes an endogenous inhibitor of mTOR25 and Laplante et al. reported it to be a glucocorticoid-induced gene in 3T3-L1 cells32. Furthermore, they demonstrated that dex treatment induced DEPTOR protein expression in undifferentiated mouse embryonic fibroblasts, in undifferentiated 3T3-L1 cells and in 3T3-L1 cells and mouse embryonic fibroblasts that were differentiated into mature adipocytes. They also showed that RU-486, an antagonist of GR, inhibited DEPTOR protein expression induced by dex in these cells32. We confirmed increased DEPTOR protein expression in primary enriched human abdominal subcutaneous fat with dex treatment. DEPTOR promotes lipogenesis and adipogenesis and is associated with obesity in mice and humans30. Increased central adiposity is seen in Cushing’s disease and as the result of pharmacological glucocorticoid treatment1. Although this increased central adiposity is primary in the visceral fat depot, there is adipocyte hypertrophy in the abdominal sc depot and increased expansion of the abdominal sc depot compared to the femoral and gluteal sc adipose tissue depots1. Induction of DEPTOR suggests a likely mechanism for the glucocorticoid-induced increase in subcutaneous adiposity; however because of the differential biology of the subcutaneous and visceral fat depots, we do not assume that changes in DEPTOR transcription would be identical in the visceral depot. Identifying GR modulators with anti-inflammatory properties that do not induce DEPTOR expression might be a useful strategy to screen for dissociated GR agonists that are less likely to cause fat accumulation.
One of the limitations of this study is that all experiments were performed in vitro and in the absence of all other endocrine hormones. Thus, it is likely that some GR-regulated genes were not identified in the absence of cross-talk with other nuclear receptors. Additionally, gene expression was only measured four hours after treatment and therefore genes regulated at other time points may have been missed. Another caveat is that although we used enriched adipose tissue for gene expression analyses, some macrophages could be present in the adipocyte fraction and contribute to the inflammation pathways we identified. Finally, the role of GR in sc fat may differ from other fat depots. An interesting area of future study will be to compare GR-mediated gene expression in various adipose tissue depots from the same subject.
In conclusion, we report the first genome-wide GR ChIP-seq and gene expression analysis in human fat. Our results identify direct GR target genes encoding proteins that regulate inflammation and modulate cellular metabolism. The identification of direct GR target genes in human sc fat allows for a better understanding of GR signaling in this fat depot and could help identify novel GR modulators with reduced deleterious effects on human subcutaneous fat and subsequent physiology.
What is already known about this subject?
What does your study add?
Three short bullet-point answers
The glucocorticoid receptor has a significant role in metabolic and inflammatory processes in visceral fat; its role in the regulation of primary human subcutaneous adipose tissue gene expression is less well-defined.
We identified GR target genes in primary human subcutaneous adipose tissue by performing GR ChIP-seq and gene expression analysis.
Pathways identified by GR-regulated gene expression analyses include cellular metabolism and inflammation.
Acknowledgments
We thank the University of Chicago Department of Surgery for salary support for Puneet Singh, MD. We thank the University of Chicago Genomics Core Facility and Pieter Faber, PhD for expertise in gene expression microarray analysis and the Bioinformatics Core Facility including Jorge Andrade, PhD for bioinformatic expertise with ChIP-sequencing. We also thank Amanda Spratt for assistance in the IRB-approval process, Brenda Copley for entering patients on the fat collection protocol, and Husain Sattar, MD as well as the University of Chicago Human Tissue Resource Center (HTRC) for critical help with pathology and procurement of fat. We thank members of the Brady and Conzen laboratories for sharing their expertise in fat biology.
Funding: The University of Chicago Comprehensive Cancer Center NIH P30-CA014599, The Specialized Program of Research Excellence in Breast Cancer P50-CA125183, NIH R01 CA089208 (SDC), HHMI Medical Research Fellows Program (COB), John D. Arnold MD Scientific Research Prize (COB)
Drs. Kocherginsky and Conzen have a patent issued "Methods and compositions related to glucocorticoid receptor (GR) antagonists and breast cancer." This patent is not directly relevant to this work in normal human adipose tissue. The method is to target the GR in breast epithelial cancers. However, it may be considered broadly relevant to the work.
Footnotes
Disclosure: There are no other conflicts of interest to report.
References
- 1.Lee M, Pramyothin P, Karastergiou K, Fried SK. Deconstructing the roles of glucocorticoids in adipose tissue biology and the development of central obesity. Biochim Biophys Acta. 2014;1842:473–481. doi: 10.1016/j.bbadis.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rafacho A, Ortsater H, Nadal A, Quesada I. Glucocorticoid treatment and endocrine pancreas function: implications for glucose homeostasis, insulin resistance and diabetes. J Endocrinol. 2014;223:R49–R62. doi: 10.1530/JOE-14-0373. [DOI] [PubMed] [Google Scholar]
- 3.Patel R, Williams-Dautovich J, Cummins CL. Minireview: new molecular mediators of glucocorticoid receptor activity in metabolic tissues. Mol Endocrinol. 2014;28:999–1011. doi: 10.1210/me.2014-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 5.Lonn L, Kvist H, Ernest I, Sjostrom L. Changes in body composition and adipose tissue distribution after treatment of women with Cushing’s syndrome. Metabolism. 1994;43:1517–1522. doi: 10.1016/0026-0495(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 6.Rhen T and Cidlowski JA. Antiinflammatory action of glucorticoids-New mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 7.Rebuffe-Scrive M, Bronnegard M, Nilsson A, Eldh J, Gustafsson JA, Bjorntorp P. Steroid hormone receptors in human adipose tissues. J Clin Endocrinol Metab. 1990;71:1215–1219. doi: 10.1210/jcem-71-5-1215. [DOI] [PubMed] [Google Scholar]
- 8.Lundgren M, Buren J, Ruge T, Myrnas T, Eriksson JW. Glucocorticoids down-regulate glucose uptake capacity and insulin-signaling proteins in omental but not subcutaneous human adipocytes. J Clin Endocrinol Metab. 2004;89:29890–29897. doi: 10.1210/jc.2003-031157. [DOI] [PubMed] [Google Scholar]
- 9.Gathercole LL, Morgan SA, Bujalska IJ, Hauton D, Stewart PM, Tomlinson JW. Regulation of lipogenesis by glucocorticoids and insulin in human adipose tissue. PLoS One. 2011;6:e26223. doi: 10.1371/journal.pone.0026223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gathercole LL, Bujalska IJ, Stewart PM, Tomlinson JW. Glucocorticoid modulation fo insulin signaling in human subcutaneous adipose tissue. J Clin Endocrinol Metab. 2007;92:4332–4339. doi: 10.1210/jc.2007-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazlehurst JM, Gathercole LL, Nasiri M, Armstrong MJ, Borrows S, Yu J, Wagenmakers AJ, Stewart PM, Tomlinson JW. Glucocorticoids fail to cause insulin resistance in human subcutaneous adipose tissue in vivo. J Clin Endocrinol Metab. 2013;98:1631–1640. doi: 10.1210/jc.2012-3523. [DOI] [PubMed] [Google Scholar]
- 12.Yu C, Mayba O, Lee JV, Tran J, Harris C, Speed TP, Wang J. Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS One. 2010;5:e15188. doi: 10.1371/journal.pone.0015188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebbar PB, Archer TK. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem. 2007;282:8284–8591. doi: 10.1074/jbc.M610554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itani OA, Liu KZ, Cornish KL, Campbell JR, Thomas CP. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5’-flanking region. Am J Physiol Endocrinol Metab. 2002;283:E971–E979. doi: 10.1152/ajpendo.00021.2002. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas S, Bonchev D. A survey of current software for network analysis in molecular biology. Hum Genomics. 2010;4:353–360. doi: 10.1186/1479-7364-4-5-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews S. FastQC: A quality control application for high throughput sequence data. Babraham Institute; http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc. [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nature Biotechnology. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biology. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein PB, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen, et al. ChIP-seq guidelines and practices of ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan JS, Reed Ann, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MJ, Gong DW, Burkey BF, Fried SK. Pathways regulated by glucocorticoids in omental and subcutaneous human adipose tissue: a microarray study. Am J Physiol Endocrinol Metab. 2011;300:E571–E580. doi: 10.1152/ajpendo.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavlatou MG, Vickers KC, Varma S, Malek R, Sampson M, Remaley AT, Gold PW, Skarulis MC, Kino T. Circulating cortisol-associated signature of glucocorticoid-related gene expression in subcutaneous fat of obese subjects. Obesity. 2013;21:960–967. doi: 10.1002/oby.20073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 29.Webster JC, Cidlowski JA. Downregulation of the glucocorticoid receptor. A mechanism for physiological adaptation to hormones. Ann N Y Acad Sci. 1994;746:216–220. doi: 10.1111/j.1749-6632.1994.tb39238.x. [DOI] [PubMed] [Google Scholar]
- 30.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011;71:6360–6370. doi: 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang F, Bohmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev. 2006;86:1151–1178. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- 32.Laplante M, Horvat S, Festuccia WT, Birsoy K, Prevorsek Z, Efeyan A, Sabatini DM. DEPTOR cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Cell Metabolism. 2012;16:202–212. doi: 10.1016/j.cmet.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]