Abstract
The purpose of this study was to ascertain the mechanisms by which advanced prostate cancer cells resist bortezomib therapy. Several independent studies have shown that cells are protected from proteasome inhibition by increased autophagic activity. We investigated whether C/EBPβ, a transcription factor involved in the control of autophagic gene expression, regulates resistance to proteasome inhibition. In PC3 cells over-expressing C/EBPβ, turnover of autophagic substrates and expression of core autophagy genes were increased. Conversely, C/EBPβ knockdown suppressed autophagosome–lysosome fusion. We also found that C/EBPβ knockdown suppressed REDD1 expression to delay early autophagy, an effect rescued by exogenous REDD1. Cells with suppressed C/EBPβ levels showed delayed autophagy activation upon bortezomib treatment. Knockdown of C/EBPβ sensitized PC3 cells to bortezomib, and blockade of autophagy by chloroquine did not further increase cell death in cells expressing shRNA targeting C/EBPβ. Lastly, we observed a decreased growth of PC3 cells and xenografts with C/EBPβ knockdown and such xenografts were sensitized to bortezomib treatment. Our results demonstrate that C/EBPβ is a critical effector of autophagy via regulation of autolysosome formation and promotes resistance to proteasome inhibitor treatment by increasing autophagy.
Keywords: Autophagy, C/EBPβ, REDD1, Prostate cancer, Proteasome inhibitor
Introduction
Metastatic prostate cancer (PCa) is an incurable disease and a leading cause of cancer death in Europe and North America. When patients acquire resistance to hormonal therapy, second-line chemotherapy is administered, which can increase patient life span by several years. There has been interest in the use of bortezomib, a proteasome inhibitor, for the treatment of PCa, but clinical studies have shown only modest or no significant improvement in patient benefit when administered in combination with hormonal therapy or docetaxel [1–3]. However, a recent study evaluating microarray data from younger patients (<65 years of age) with localized prostate cancer and treated by radical prostatectomy revealed that gene sets representing proteasome subunits and protein catabolism are the strongest predictors of metastatic progression [4]. Thus, proteasome inhibition may still be of clinical value for certain subsets of patients with PCa.
Autophagy is a catabolic process which serves to turn over long-lived proteins and organelles by engulfing them in double-membrane bound vacuoles, autophagosomes. These structures eventually fuse with lysosomes to degrade their contents and recycle lipids, amino acids, and carbohydrates [5]. Autophagy is initiated by activation of a multi-protein type III PI3K complex, composed of Beclin 1, VPS34 and ATG14, which enriches a region of the endoplasmic reticulum (ER) in phosphotidylinositol-3-phosphate to serve as a platform for the recruitment of other proteins to generate an isolation membrane [6,7]. A group of evolutionary conserved proteins are responsible for elongation and closure of the isolation membrane to form a double-membrane autophagosome by proteolytic cleavage of LC3 and GABARAP family proteins, covalently linking them to phosphatidylethanolamine (PE) and incorporating them into the burgeoning autophagosome [8,9]. Autophagy is not only regulated at the initiating steps, as LC3-II can be delipidated by ATG4b to LC3-I to promote LC3 recycling [10]. Conversely, LC3-II delipidation by ATG4b can be suppressed by REDD1, thereby promoting autophagy [11]. Proteins and organelles destined for degradation are trafficked to autophagosomes by the ubiquitin-binding proteins p62 and NBR1 [12–14].
Autophagy has been recognized as a central process that cancer cells utilize to adapt to stress brought about by hypoxia, anoikis, radiation, or chemotherapy. Recently, it has been suggested that autophagy plays a critical role in the resistance of PCa cells to hormonal therapy or chemotherapy and that inhibition of autophagy synergizes with docetaxel or androgen deprivation [15,16]. Combined treatment of several different cancer cell lines with autophagy suppressive agents and bortezomib increases cell death and improves clinical response in patients with relapsed multiple myeloma [17–19]. Further, proteasome inhibitor-resistant PCa cells are thought to utilize autophagy to resist this class of drugs [20].
Recently, several groups have shown that autophagy is regulated at the transcriptional level [21,22]. C/EBPβ, a basic region-leucine zipper (bZIP) transcription factor, was identified as a regulator of circadian autophagy in the liver by inducing a broad array of autophagy genes [23]. C/EBPβ protein is translated from an intronless transcript from three in-frame start codons as higher molecular weight, liver-enriched activating proteins (LAP and LAP*) or the truncated liver-enriched inhibitory protein (LIP). The dominant-inhibitory LIP isoform lacks all three N-terminal activation domain modules and can inhibit transcription by heterodimerizing with LAPs. Another group also showed that C/EBPβ promotes differentiation of 3T3-L1 preadipocytes by activation of ATG4b gene expression and autophagy [24]. Intriguingly, treatment of Burkitt's lymphoma cells with bortezomib dramatically increases the expression of C/EBPβ [25]. Other proteasome inhibitors, such as lactacystin or MG-132, also increase nuclear levels of C/EBPβ and increase its DNA binding [26,27]. We therefore investigated the role of C/EBPβ in PCa cell autophagy and sensitivity to bortezomib. Our results suggest that autophagy is activated early after bortezomib treatment, that C/EBPβ promotes autophagy in PCa cells via induction of autophagosome–lysosome fusion and via induction of REDD1, and that reducing C/EBPβ expression increases prostate cancer cell sensitivity to bortezomib.
Materials and methods
Cell lines, reagents and mice
PC3 and LNCaP cells were maintained in RPMI media with 10% heat inactivated fetal bovine serum (FBS) (Hyclone, Logan, UT) supplemented with penicillin/ streptomycin. Cells were grown in a humidified incubator maintained at 37 °C with 5% CO2. Cell lines transduced with short hairpin RNA (shRNA) or PiggyBac (PB) Teton vectors were grown in media supplemented with 10% tetracycline-screened FBS (Hyclone). For experiments involving inducible expression of shRNA or ectopic C/EBPβ, 0.5 μg/ml doxycycline was added to the media and replaced every 48 hrs. The pCMS-eGFP-RTP801 plasmid was purchased from Addgene (Cambridge, MA; plasmid #65057).
Non-obese diabetic/severe-combined immuno-deficient (NOD/SCID); IL2Rγ−/− (NSG) mice were obtained from a breeding colony established in the Johns Hopkins University division of animal resources. The animals were subcutaneously en-grafted with 2E06 PC3 cells in PBS in a 1:1 mixture with Matrigel. The animals were monitored daily after transplantation. When tumor volumes reached 100 mm3, the mice were placed on doxycycline-laced feed. When tumors reached a volume between 100 and 300 mm3, the animals were randomly assigned to receive either DMSO vehicle or bortezomib, by intraperitoneal (IP) injection (1 mg/kg of body weight) on days 1, 4 and 8. The mice were sacrificed 11 days after engraftment. Tumor volumes were determined by caliper measurement using the ellipsoidal formula: length × width × height × 0.5236 [28].
All animal studies were conducted under protocols approved by the Johns Hopkins Institutional Animal Care and Use Committee in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Western blotting
Protein samples from whole cell lysates or nuclear extracts were prepared and subjected to Western blotting as described [29]. Each experiment was repeated at least twice. Anti-β-actin (AC15) and anti-vinculin (V9131) antibodies were from Sigma-Aldrich (St. Louis, MO). Anti-C/EBPβ (C-19), anti-C/EBPβ (H-7), and anti-SQSTM1/p62 (H-290) antibodies were from Santa Cruz Biotechnologies (Dallas, TX). Anti-LC3B (2775), anti-ATG3 (3415), anti-ATG5 (8540), and anti-ATG-7 (8558) antibodies were from Cell Signaling Technologies (Danvers, MA). Anti-REDD1 antibody (10638-1-AP) was from Proteintech (Rosemont, IL).
Quantitative real-time PCR
RNA extraction, cDNA synthesis, and PCR reactions were performed as previously described [29]. Oligonucleotides were designed using PrimerBlast and custom ordered from Sigma-Aldrich. Their sequences are presented in Table 1.
Table 1.
PCR primers.
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| ACTB | GACCTGGCTGGCCGGGACCT | GGCCATCTCTTGCTCGAAGT |
| ATG4B | ATGAGCATCGCGGAGCTTG | CTAGGGACAGGTTCAGGACG |
| ATG5 | GAAGCTGTTTCGTCCTGTGG | GATGTTCACTCAGCCACTGC |
| ATG7 | ACAGCTTGTTCTTCCAAAGTTCT | TCTCAGATGGTCTCATCATCGC |
| CTSL | CCTCAGCATGAGGAACACGA | AAAAGGTGCTGGTGGAGGTT |
| BNIP3 | CCTCAGCATGAGGAACACGA | AAAAGGTGCTGGTGGAGGTT |
| BNIP3L | TGGAGCCATGAAGAAAGGGG | ACTTCACAGGTCACACGCAT |
| GABARAP | GGCTCCCAAAGCTCGGATAG | CTGGTACAGCTGACCCATTGT |
| REDD1 | CGAACTCCCACCCCAGATCG | AACGACACCCCATCCAGGTA |
| p62 | GATTCGCCGCTTCAGCTTCTG | GTCACTGGAAAAGGCAACCAA |
| ULK1 | CGCCATCTCCTCAAGTTGGA | TGCAAGTCAGACAGGTTGGG |
Ectopic C/EBPβ expression and shRNA
LNCaP cells stably transfected with the TET-on, transposon based PiggyBac vector PB-TRE, or PB-CEBPB expressing murine C/EBPβ, were previously described [29]. PC3 cells were transfected with these same vectors, and cells with stable integration were positively selected with puromycin (2 mg/ml). Lentiviral transduction of PC3 cells with pTRIPZ lentiviral vectors targeting CEBPB was also performed as previously described for LNCaP cells [29].
TALEN construction and CEBPB gene editing
TALEN DNA constructs targeting the human CEBPB open reading frame (ORF) were constructed as previously described [29]. CEBPB homology arm 1 (HA1) was amplified from 293T DNA with primers TGCtctagaCTGGTGGGAACAATGCCACC and ACTtgatcaTGGTGGCATTGTTCCCACCAG; restriction enzyme sites are in lower case. The resulting fragment was digested with XbaI and SpeI and ligated into the pSEPT donor plasmid [30]. CEBPB homology arm 2 (HA2) was amplified using GCatcgatGAACTTGTTCAAGCAGCTGCC and AGTcagctgAGGCTCCGGAATCTCTTCTC primers. The resulting fragment was digested with ClaI and SalI and ligated into the pSEPT plasmid containing CEBPB HA1. PC3 cells were co-transfected with TALEN expression vectors targeting CEBPB and the pSEPT donor plasmid at a 1:1 ratio and after 48 hours were seeded into a 96-well plate at 1 cell/well with G418. Individual clones were screened for C/EBPβ expression by Western blotting.
Chromatin immunoprecipitation (ChIP)
1E06 PC3 cells were used in each ChIP reaction, using anti-rabbit C/EBPβ antiserum or normal rabbit IgG (Santa Cruz Biotechnology) as previously described [31]. DNA fragments corresponding to the promoter of REDD1 were detected by qPCR using the primers presented in Table 2.
Table 2.
ChIP primers.
| Region | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| REDD1 [–198 : –17] | GGGTTCGACTGCTGCGAGCTTTC | AGCAGCCTATAAGGACTAGCG |
| REDD1 [–720 : –534] | CTCTCCTTGGGAGCAGTTGG | TGCAAAGGCTCGGGATATGG |
Cell viability and proliferation assays
To assess cell proliferation, 1E05 PC3 cells were seeded in growth media containing 0.5 μg/ml doxycycline. On days 2 and 4, cells were trypsinized and stained with Trypan blue dye, and viable cells were enumerated using a hemocytometer. Experiments were repeated three times. For clonogenic assays, PC3 cells were trypsinized and seeded into 6-well plates at 200 cells per well. 12 days after plating, cells were fixed and stained with 4% formaldehyde and 0.05% crystal violet. After washing with tap water, colonies were manually counted under a bright field microscope. Clusters of cells containing more than 50 cells were counted as a colony.
Autophagosome–lysosome fusion assay
PC3 cells were plated on 60 mm dishes to obtain 80% confluence on the next day. Cells were transfected with tandem mRFP-GFP fluorescent-tagged LC3 (ptfLC3) (Addgene #21074) [32]. On day two post-transfection, cells were trypsinized and plated on sterile glass bottom dishes (Mattek, Ashland, MA) at a 40–50% confluency. Cells were allowed to adhere to glass and spread for 24 hrs. Cells were then imaged using a live cell Zeiss LSM780-FCS Single-point, laser scanning confocal microscope at the JHU Core Imaging facility. Images were processed using ImageJ software. Cells with predominantly yellow RFP/GFP (autophagosome) or red RFP (autolysosome) punctae were counted and analyzed as described [32].
Q74-EGFP degradation assay
PC3 cells seeded in 24-well plates were transfected with 500 ng of Q74-EGFP (Addgene #40262) [33] or pMax-GFP (Lonza, Basel, Switzerland). Forty-eight hours after transfection, fluorescent micrographs of GFP-positive cells were taken using a Nikon Ti-E inverted microscope at 400× magnification from at least five non-overlapping regions of interest. Cells were manually counted using ImageJ software and the percentage of GFP-positive cells displaying punctuate staining was determined.
Statistical analysis
Statistical comparison of the two groups was conducted using Student's t-test. Comparisons of multiple groups were performed using analysis of variance followed by multiple comparisons with Student's t-test and the Holm–Bonferonni correction. P-values were ordered from highest to lowest and α was adjusted to α/(m+1-k) for p(k), where m = number of comparisons and corresponds to the highest p-value. k corresponds to the p-value rank. α was set to 0.05 for comparisons. For analysis of tumor growth, linear regression analysis was used to determine statistical significance in growth rates between groups of mice using Graph Pad Prism statistical software.
Results
C/EBPβ LAP : LIP isoform ratio regulates autophagy in prostate cancer cells
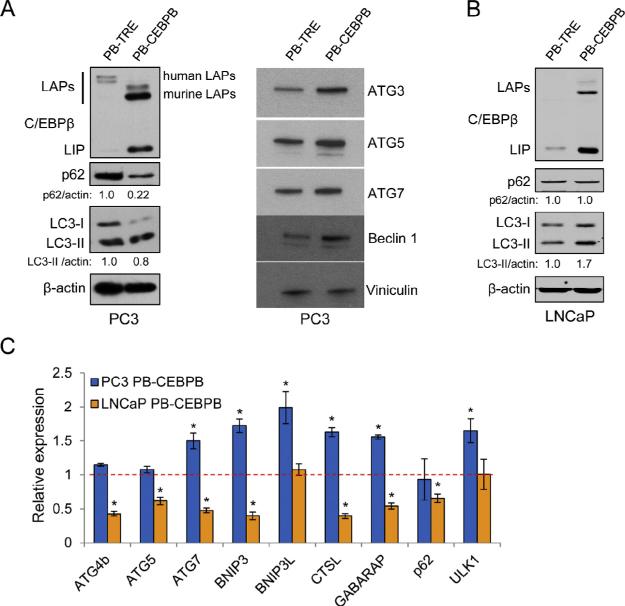
To understand the role of C/EBPβ in prostate cancer cell autophagy, we first ectopically expressed murine C/EBPβ, which shares 98.8% similarity to the human C/EBPβbZIP domain and 80–100% similarity to the human trans-activation and regulatory domains, in the PC3 PCa cell lines using the PB-TRE Tet-on PiggyBac inducible expression vector we had used previously to obtain LNCaP cells harboring doxycycline-inducible C/EBPβ [29]. Induction of PiggyBac-CEBPB (PB-CEBPB) in PC3 cells led to a decline in the total levels of LC3-I and LC3-II and p62 compared to cells expressing the empty PB-TRE vector (Fig. 1A, left). Decreases in p62 levels typically indicate increased autophagic degradation, while changes in LC3-II can imply blockade or activation of autophagosome formation [34]. We further evaluated other proteins involved in autophagosome nucleation and elongation and found increases in ATG3, ATG5, ATG7 and Beclin-1, suggesting that autophagy is increased upon induction of C/EBPβ (Fig. 1A, right).
Fig. 1.
C/EBPβ isoforms LAP and LIP differentially regulate autophagy in PCa cells. (A) PC3 cells stably transduced with PB-TRE or PB-CEBPB were treated for 72 hrs with 0.5 μg/ml doxycycline and cell lysates were analyzed by Western blotting for the indicated proteins. Numbers below the blots indicate the relative band density normalized to β-actin. (B) LNCaP cells stably transduced with PB-TRE or PB-CEBPB were analyzed similarly for C/EBPβ, p62, LC3, and β-actin. (C) PC3 or LNCaP cells transduced with PB-TRE or PB-CEBPB were cultured similarly followed by qPCR analysis for the indicated genes. The dashed line represents gene expression levels in control cell lines expressing PB-TRE. Bar graphs represent the average of three experiments, error bars represent standard error of the mean (SEM) (*p < 0.05).
In contrast with these results in PC3 cells, LNCaP cells showed increased total levels of LC3-I and LC3-II, and no change in p62 in response to C/EBPβ induction (Fig. 1B). Of note, ectopic C/EBPβ is predominantly expressed as the shorter, dominant inhibitory LIP isoform in LNCaP cells as is endogenous C/EBPβ. Thus induction of CEBPB in LNCaP cells may reduce C/EBPβ trans-activation activity to impair autophagy.
As C/EBPβ was shown to directly regulate autophagic genes at the transcriptional level [23,24,35], we also evaluated the expression of several such genes by quantitative real-time PCR (qPCR). We found a modest increase in CTSL, GABARAP, ATG7, BNIP3, BNIP3L and ULK1 mRNAs in PC3 cells and nearly all of these genes were suppressed upon ectopic C/EBPβ expression in LNCaP cells (Fig. 1C).
C/EBPβ promotes autophagosome–lysosome fusion in PC3 cells
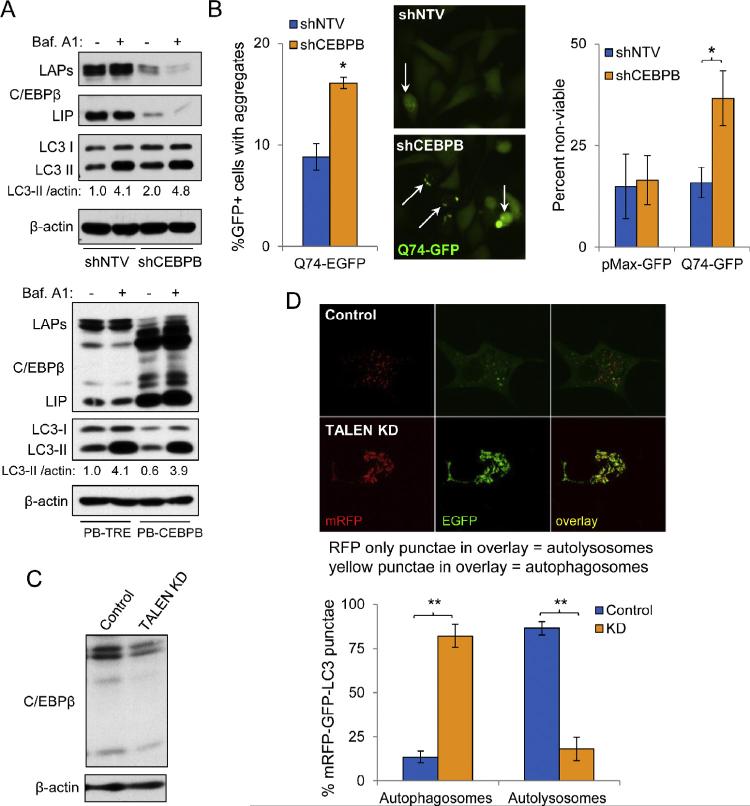
Bafilomycin A1, a vacuolar-ATPase inhibitor, increases lysosomal pH and alters Ca2+ gradients of the lysosome, subsequently inhibiting fusion with autophagosomes, blocking autophagic flux and leading to accumulation of LC3-I and LC3-II [36,37]. We treated cells for four hours with bafilomycin A1 and then evaluated lysates by Western blot analysis. Although the total levels of LC3-I and LC3-II were increased in PC3 cells expressing shCEBPB, the ultimate level of LC3-II achieved in response to bafilomycin A1 was not different from the control cells transduced with shNTV (Fig. 2A, top). In addition, ectopic expression of C/EBPβ did not enhance autophagic flux in bafilomycin A1-treated cells, as again the peak levels of LC3-II, 4.1 and 3.9, were nearly identical (Fig. 2A, bottom). These data suggest that C/EBPβ does not play a dominant role in autophagosome formation, as otherwise CEBPB shRNA might have been predicted to reduce and ectopic C/EBPβ to increase the change in LC3-II in the presence of bafilomycin A1.
Fig. 2.
C/EBPβ promotes autophagosome–lysosome fusion in PC3 cells. (A) Western blot analyses in PC3 cells expressing NTV or CEBPB shRNAs (top panels) or ectopically expressing mouse C/EBPβ from a PiggyBac vector (PB-CEBPB) or the PB-TRE control vector (bottom panels). In both sets of experiments, PC3 cells were treated for 72 hrs with 0.5 μg/ml doxycycline and then treated with either vehicle (DMSO) or 100 nM bafilomycin A1 for 4 hrs. Western blotting for C/EBPβ, LC3, and β-actin was then conducted. (B) Quantification of EGFP-Q74 aggregates in GFP+, transfected PC3 cells expressing shNTV or shCEBPB (left). Sample fluorescence micrograph (center); white arrows indicate cells with aggregates. Cell viability was quantified by Trypan blue dye exclusion in cultures transfected with control pMax-GFP or EGFP-Q74 (right). (C) Western blot analysis of cell lysates from PC3 control and TALEN KD subclones. (D) Confocal micrographs of PC3 TALEN control and CEBPB KD cells transfected with the tandem fluorescent-tagged mRFP-GFP-LC3 (tfLC3) construct (top panels). Quantification of percent of cells with LC3 present in autophagosomes or autolysosomes is shown below (bottom). In the red/green overlay images, yellow foci represent autophagosomes and red foci autolysosomes. Bar graphs throughout represent the average of three experiments, error bars represent SEM (*p < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We next tested whether C/EBPβ regulates turnover of autophagy substrates. We transiently expressed mutant huntingtin-EGFP (Q74-EGFP) fusion protein in PC3 cells together with either shNTV or shCEBPB [33]. Expression of Q74-EGFP results in protein aggregation, which can be observed as punctuate GFP-positive staining. Forty-eight hours after transfection, we quantified the number of GFP-positive cells displaying aggregates and found that there were significantly more cells with GFP aggregates in PC3 cells expressing shRNA targeting CEBPB (Fig. 2B, left and center). These data indicate that turnover of autophagosomes is augmented by C/EBPβ. In addition, there were significantly more non-viable, Trypan blue dye positive cells in cultures lacking C/EBPβ (Fig. 2B, right).
The first step in autophagosome turnover is fusion with lysosomes. We therefore next evaluated autophagosome–lysosome fusion in PC3 cells using plasmid ptfLC3, expressing a mRFP-GFP-LC3 fusion protein [32]. As GFP fluorescence is quenched by the acidic environment of the lysosome, yellow punctae visible under fluorescence microscopy, representing signal from both RFP and GFP, indicate autophagosomes, whereas red punctae represent autolysosomes. Doxycycline induction of shRNA from pTRIPZ also induces RFP and so is not amenable to this assay. We therefore utilized TALEN targeting of genomic CEBPB as an alternative means of knockdown (KD). We utilized homologous recombination to insert a neomycin resistance cassette and generate a deletion in the CEBPB locus with a pSEPT donor plasmid containing homology arms of CEBPB flanking the TALEN cut site (Supplementary Figure S1) [30]. We generated a neomycin-resistant subclone which expressed decreased levels of C/EBPβ relative to the control subclone (Fig. 2C). When TALEN KD cells were transfected with the mRFP-GFP-LC3 reporter, we found that there was a remarkable, 4-fold increase in the percentage of cells with RFP/GFP-positive autophagosomes relative to control cells and a similar decrease in the percent of cells with RFP-positive autolysosomes, implying that fusion with lysosomes is impaired (Fig. 2D). Collectively, these data demonstrate that C/EBPβ is critical for basal autophagy in PC3 cells by regulating autophagosome– lysosome fusion.
C/EBPβ LAP promotes autophagy in PC3 cells by augmenting REDD1 expression
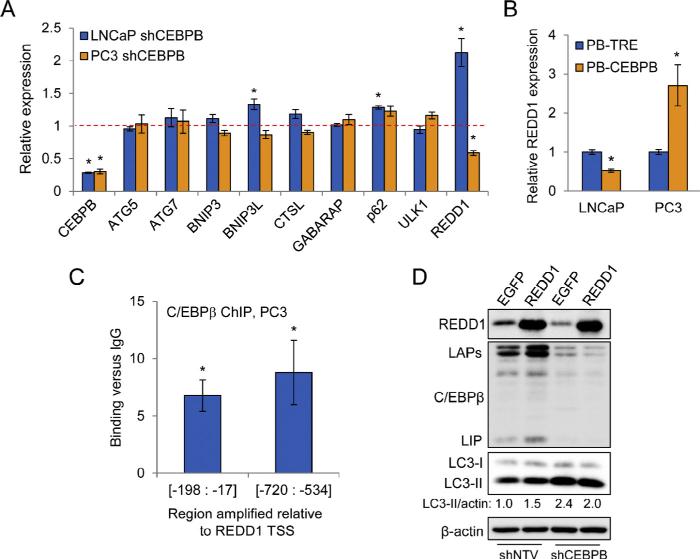
Although C/EBPβ is reported to broadly regulate the expression of autophagy genes in the liver [23], we did not observe differential expression of all genes analyzed in PC3 cells over-expressing C/EBPβ. Recently, it was shown that REDD1 could promote autophagy by suppressing ATG4b-mediated LC3-II delipidation to LC3-I, an effect most critical during periods of metabolic stress [11]. This function of REDD1 prevents recycling of LC3-II and leads to greater turnover of autophagosomes. REDD1 can also inhibit mTORC1, which suppresses autophagy by phosphorylating ULK1 and ATG13, upstream autophagy regulators [38]. C/EBPβ was previously shown to cooperate with ATF4 to promote REDD1 transcription by binding to a non-consensus site centered 1004 bp upstream from the REDD1 transcription start site (TSS), but only in cells under oxidative stress [39,40]. We evaluated REDD1 gene expression changes in PCa cells expressing shRNA targeting CEBPB by qPCR. In contrast to cells over-expressing C/EBPβ, we found that none of the genes that we had analyzed in Fig. 1 were affected by C/EBPβ KD in LNCaP or PC3 cells (Fig. 3A). However, in PC3 cells with CEBPB deficiency, we found a significant 2-fold down-regulation of REDD1 transcript levels (Fig. 3A). We also found that LNCaP cells showed increased REDD1 upon shCEBPB induction, likely due to decreased expression of the dominant-inhibitory LIP isoform (Fig. 3A). Notably, REDD1 mRNA showed stronger up-regulation upon C/EBPβ over-expression in PC3 cells than other autophagy genes analyzed (Fig. 3B). Conversely, over-expression of C/EBPβ in LNCaP cells suppressed REDD1 transcript levels (Fig. 3B).
Fig. 3.
C/EBPβ promotes autophagy in PC3 cells by regulation of REDD1 expression. (A) qPCR analysis of autophagic genes in PC3 and LNCaP cells expressing shRNA targeting CEBPB. The dashed line represents gene expression levels in control cell lines expressing shNTV. (B) REDD1 gene expression in cells expressing C/EBPβ from the PBCEBPB vector or harboring the control PB-TRE vector after 72 hr culture with doxycycline. (C) qPCR analysis, using two primer pairs, of genomic DNA extracted from PC3 cell chromatin following immunoprecipitation with anti-C/EBPβ antibody or IgG control. Fold-increase in C/EBPβ binding versus IgG is shown. (D) PC3 cells expressing shNTV or shCEBPB were transiently transfected with pMax-GFP or pCMS-EGFP-REDD1, and 48 hours later whole cell lysates were subject to Western blot analysis for the indicated proteins. Data presented are representative of three independent experiments. Bar graphs represent the average of three experiments, error bars represent SEM (*p < 0.05).
As we observed that REDD1 expression correlated with full-length C/EBPβ levels in the absence of oxidative stress, we analyzed the human REDD1 promoter to identify additional binding sites for C/EBPβ. We found a site matching the C/EBPβ consensus at −603 bp and a near-consensus site at −99 bp (Supplementary Figure S2). To test whether C/EBPβ binds to these regions, we subjected nuclear extracts from PC3 cells to chromatin immunoprecipitation (ChIP). Using two different primer pairs, each flanking one of the putative C/EBPβ binding sites, we observed between 6 and 8-fold greater levels of amplified product in extracts precipitated with anti-C/ EBPβ antiserum relative to normal rabbit IgG (Fig. 3C), indicating that endogenous C/EBPβ binds to these regions of the REDD1 promoter.
C/EBPβ regulation of REDD1 suggests that C/EBPβ not only is critical for autophagosome–lysosome fusion, but also might play a role in autophagosome maturation. Residual REDD1 present upon shCEBPB-mediated CEBPB KD must be sufficient to prevent a complete block at this step in autophagy. Knockdown of REDD1 was problematic, as all siRNAs tested also reduced C/EBPβ (not shown), and of note REDD1(−/−) MEFs show greatly reduced autophagosome formation [11]. Therefore, to further test whether C/EBPβ drives early autophagy via REDD1, we transiently over-expressed REDD1 in PC3 cells. We found that expression of REDD1 from the pCMS-EGFP-REDD1 vector [41] increased LC3-II in PC3 cells harboring shNTV, as predicted from its role in early autophagy, but mildly reduced LC3-II in shCEBPB cells, compared to controls expressing the pMax-GFP plasmid (Fig. 3D). These results suggest that C/EBPβ promotes autophagy in part by regulating REDD1 expression.
C/EBPβ promotes autophagy following bortezomib treatment
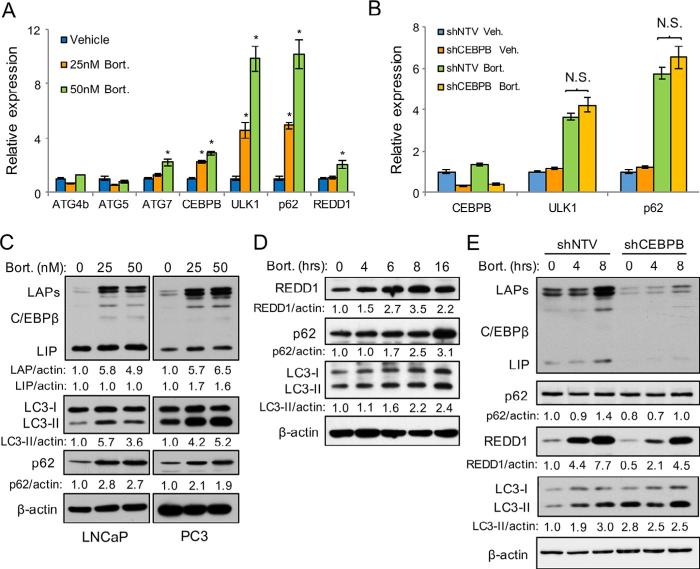
Previous reports suggest that proteasome inhibitors increase expression and activity of C/EBPβ [25,26]. As proteasome inhibitors also activate autophagy [42,43], we tested whether C/EBPβ is required for autophagy following bortezomib treatment. Gene expression analysis of several autophagy genes revealed that ULK1 and p62 were markedly increased 16 hours after bortezomib treatment of PC3 cells (Fig. 4A). ATG5 and ATG4B were unchanged and ATG7 and REDD1 were mildly upregulated by bortezomib. The changes in p62 and ULK1 were not mediated by C/EBPβ because shCEBPB cells showed the same increase in gene expression upon bortezomib treatment (Fig. 4B). Treatment of LNCaP or PC3 cells with bortezomib for 24 hours markedly increased C/EBPβ (LAP), LC3-II, and p62 protein expression (Fig. 4C). To further evaluate the effects of bortezomib on PCa cell autophagy, we analyzed LC3 and p62 by time course analysis. We found that REDD1 increased from 4 to 8 hours following 25 nM bortezomib treatment, but then decreased between 8 and 16 hours (Fig. 4D). LC3-II and p62 protein levels showed the strongest increase 16 hours after treatment. Lastly, we tested whether C/EBPβ was critical for autophagy following bortezomib exposure. We treated PC3 cells expressing shCEBPB with bortezomib and evaluated autophagy markers by time course analysis. C/EBPβ deficiency decreased REDD1 expression and prevented increases in LC3-II, though shCEBPB cells had higher basal levels of LC3-II (Fig. 4E). These data indicate that bortezomib promotes autophagy and that C/EBPβ is critical for activating autophagy early after bortezomib treatment.
Fig. 4.
C/EBPβ promotes autophagy driven by bortezomib challenge. (A) qPCR analysis for indicated mRNAs from PC3 cells that had been treated with bortezomib or vehicle for 16 hrs. (B) qPCR analysis of autophagy genes in PC3 cells expressing shNTV or shCEBPB and that had been treated for 16 hrs with 25 nM bortezomib or vehicle. (C) LNCaP or PC3 parental cells were treated for 24 hrs with the indicated concentrations of bortezomib and cell lysates were then subjected to Western blot analysis. (D) PC3 cells were treated with 25 nM bortezomib for the indicated times and cell lysates were then analyzed by Western blot analysis for indicated proteins. (E) PC3 cells expressing NTV or CEBPB shRNAs were treated with 50 nM bortezomib for the indicated times and cell lysates were again analyzed by Western blot analysis. Numbers below blots indicate relative density after normalizing to β-actin. Bar graphs represent the average of three experiments, error bars represent SEM (*p < 0.05).
Reduced C/EBPβ slows PC3 tumor growth and increases sensitivity to bortezomib
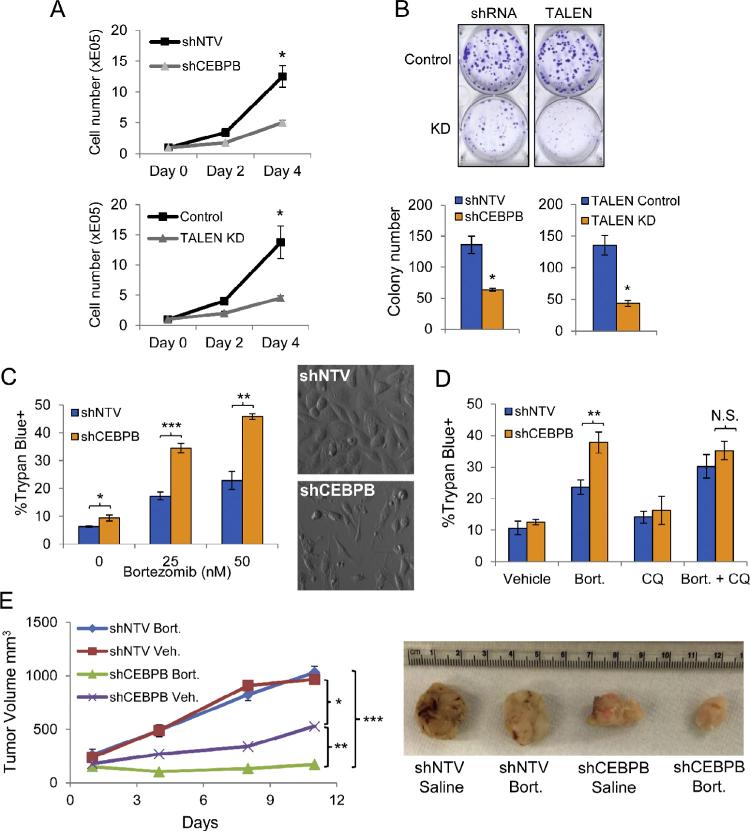
Autophagy positively regulates “cellular fitness” in a variety of cell types [44]. We therefore evaluated the effect of C/EBPβ on PC3 proliferation. We quantified cell growth over a four day time course and found that inhibition of C/EBPβ by shRNA targeting or TALEN reduced cell accumulation by more than 2-fold (Fig. 5A). We also evaluated clonogenic growth and found that cells deficient in C/EBPβ due to TALEN targeting or shRNA expression showed a more than two-fold average decrease in colony number (Fig. 5B). We treated shNTV and shCEBPB PC3 cells with 25 or 50 nM of bortezomib for 24 hours and evaluated viability by Trypan blue dye exclusion. Suppression of C/EBPβ increased cell death relative to shNTV cells by 2-fold with either dose of bortezomib (Fig. 5C, left). Phase contrast images of cells treated with 50 nM bortezomib and shNTV or shCEBPB are also shown (Fig. 5C, right). To evaluate whether autophagy contributed to increased cell death in shCEBPB cells, we treated cells with chloroquine (CQ) in combination with bortezomib. Chloroquine is a weak base that accumulates in the acidic compartments of late endosomes and lysosomes, alkalinizes these structures, and subsequently inhibits autophagosome–lysosome fusion [45,46]. We found that there was no difference in shNTV versus shCEBPB cell death in cultures treated with the combination of CQ and bortezomib, indicating that increased death in shCEBPB cultures was due to a block in autophagy (Fig. 5D). We next evaluated PC3 tumor growth and sensitivity to bortezomib in vivo. NSG mice subcutaneously engrafted with shNTV or shCEBPB PC3 cells were treated with either vehicle or bortezomib (1 mg/kg) on days 1, 4 and 8 (Fig. 5E). Consistent with our in vitro findings, we observed a decreased rate of growth of shCEBPB PC3 tumors (p = 0.026). Further, we observed no difference in the rate of growth in control tumors receiving either vehicle or bortezomib. In contrast, bortezomib treatment and CEBPB KD strikingly synergized to further decrease tumor growth (p = 0.017). These results suggest that C/EBPβ is critical for prostate tumor growth and that decreasing C/EBPβ increases sensitivity to bortezomib in vivo.
Fig. 5.
C/EBPβ deficiency suppresses PC3 growth and improves the in vivo efficacy of bortezomib. (A) Growth of indicated PC3 lines after 3 and 5 days of culture (n = 3). (B) Quantification of clonogenic growth of the indicated PC3 lines 12 days after seeding in 6-well plates at 200 cells/well. Crystal violet staining of representative wells (top) and mean colony numbers (bottom) are shown. (C) PC3 cells were incubated for 72 hrs in 0.5 μg/ml doxycycline prior to treatment with the indicated concentrations of bortezomib for 24 hrs. Quantification of cell viability by Trypan blue dye exclusion is shown (left). Representative phase contrast images show shNTV and shCEBPB PC3 cells that had been treated with 50 nM bortezomib for 24 hrs (right). (D) Quantification of cell viability by Trypan blue dye exclusion in PC3 cultures that had been treated with the indicated drugs for 24 hrs. Bort – 25 nM bortezomib; CQ – 50 uM chloroquine. (E) Quantification of PC3 tumor growth in NSG mice treated with vehicle or bortezomib (1 mg/kg) on days 1, 4 and 8. (n = 7). Image to the right of the figure shows representative tumors from the indicated groups. Statistically significant differences were determined by linear regression analysis. Alpha adjusted by Holm–Bonferroni correction. Bar graphs represent the average of three experiments, error bars represent SEM (*p < 0.05; **p < 0.025; ***p < 0.0167).
Discussion
Although clinical trials investigating bortezomib in PCa have produced lackluster results, a recent study suggests that the proteasome may still be a valid target in a subset of prostate cancer patients [4]. In patients younger than 65 with localized cancer, proteasome and protein catabolism genes were shown to be strong predictors of metastatic progression after radical prostatectomy. Understanding the response of PCa cells to proteasome inhibition is therefore of potential clinical utility.
The results of our study suggest that C/EBPβ regulates fusion of lysosomes and autophagosomes in PCa cells. The effects of C/EBPβ on autophagy were dependent upon the expression ratio of the C/EBPβ translational isoforms, with higher relative levels of LIP : LAP expression suppressing autophagy in LNCaP cells and vice versa in PC3 cells. The observed effect of LIP down-regulating autophagic gene expression is consistent with its role as a transcriptional repressor owing to its lack of N-terminal trans-activation domains. Our data are consistent with previous reports that show a link between C/EBPβ and autophagy. C/EBPβ was reported to control circadian autophagy in mouse liver by broadly activating autophagy gene transcription [23]. Guo and colleagues showed that C/EBPβ activates autophagy by directly regulating ATG4B in 3T3-L1 pre-adipocytes to promote differentiation to mature adipocytes [24]. However, the specific defect in the autophagosome–lysosome fusion that we have revealed in PCa cells is a distinct mechanism from that reported in these earlier studies (Fig. 6). Analogous to what we have found, however, C/EBPβ(−/−) macrophages exhibit defects in bactericidal activity and fusion of phagosomes with lysosomes [47]. Overall, our findings and those of others show that the effect of C/EBPβ on autophagy is cell type and context specific.
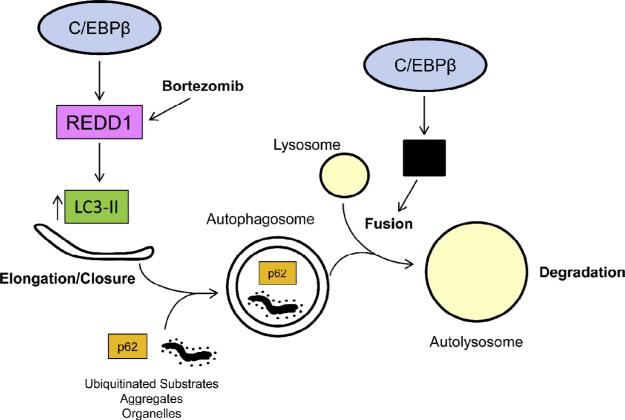
Fig. 6.
Model for C/EBPβ driven autophagy in PCa cells. C/EBPβ promotes autophagy in PCa cells by promoting autophagosome–lysosome fusion and by driving REDD1 gene expression. REDD1 suppresses LC3-II delipidation to LC3-I to increase autophagosome formation. Bortezomib may promote autophagy by increasing REDD1 protein levels. In addition, autophagosome–lysosome fusion is directly driven by C/EBPβ through an as of yet undescribed mechanism (indicated by the black box).
Our results also demonstrate that C/EBPβ further promotes autophagy through direct induction of REDD1 gene expression (Fig. 6). Previous reports had shown that C/EBPβ cooperates with ATF4 by binding to adjacent half-sites at −1004 bp in the REDD1 promoter in cells challenged by oxidative stress [39,40]. Our ChIP data and over-expression/knockdown studies suggest that C/EBPβ can also bind to a consensus site centered at −603 bp and a near consensus site at −99 bp in the REDD1 proximal promoter in the absence of oxidative stress to induce REDD1 expression. REDD1 indirectly promotes autophagosome turnover by inhibiting LC3-II delipidation, thereby preventing LC3 recycling [11]. The latter function is consistent with our findings that show decreased LC3-II and p62 levels and increases in core autophagy regulators in PC3 cells over-expressing C/EBPβ. Because REDD1 is not known to directly mediate or control autophagosome–lysosome fusion, it is likely that there are additional genes regulated by C/EBPβ whose protein products promote this function independent of REDD1. And as noted, shCEBPB expression in PC3 cells only reduced REDD1 2-fold, likely allowing autophagosome formation, albeit at a slower rate. However, we were able to partly rescue the defect in autophagy in shCEBPB cells by ectopic expression of REDD1. In particular, LC3-II was not increased by shCEBPB in the presence of exogenous REDD1, suggesting increased flux through autophagy. The latter effect suggests that in the presence of shCEBPB, where autophagosome–lysosome fusion is suppressed, ectopic expression of REDD1 may have a positive feedback on the autophagy pathway or additional activities beyond its ability to induce LC3-II.
During periods of metabolic stress such as exercise, starvation, or hypoxia, REDD1 is critical for autophagosome maturation by inducing LC3-II and regulating proper autophagosome size [11]. REDD1 transcript levels were not increased 16 hrs after 25 nM bortezomib treatment despite elevation of C/EBPβ LAP, indicating that bortezomib interferes with C/EBPβ induction of REDD1 transcription, and protein levels were increased more than two-fold at this time point, suggesting stabilization of REDD1 protein. Consistent with the idea that bortezomib affects REDD1 protein levels independent of C/EBPβ, knockdown of CEBPB did not prevent up-regulation of REDD1 protein by bortezomib, though basal levels were reduced. We also found that LC3-II proteins were up-regulated within 4 hours of bortezomib treatment. However, in cells expressing shCEBPB, LC3-II levels showed only modest changes relative to control cells. These findings suggest that regulation of basal REDD1 transcript and protein levels by C/EBPβ is crucial for LC3B processing early after proteasome inhibitor treatment.
Proteasome inhibition increases cytosolic and endoplasmic reticulum (ER) protein content and promotes amino acid deprivation [42,48]. These events activate the amino acid sensitive protein kinase GCN2 and the unfolded protein response in the ER, leading to phosphorylation of eIF2α and suppression of protein translation [49]. Autophagy is thought to promote cell survival in cells treated with proteasome inhibitors by restoring amino acid homeostasis and suppressing ER stress [42]. We found that PCa cells treated with bortezomib show a dynamic regulation of autophagy that was dependent upon C/EBPβ expression. Suppression of autolysosome catabolism has been reported in ovarian and breast cancer cell lines treated with bortezomib, owing to a decrease in cathepsins D and B [50,51]. The conclusions of these studies are consistent with our findings which showed strong up-regulation of p62 and ULK1 but suppression of Cathepsin L (not shown) in PC3 cells treated with bortezomib.
Finally, our findings also show decreased cell and tumor growth rates in PC3 cells lacking C/EBPβ coincident with a decline in basal autophagy. This decline in autophagy could explain the poor growth rates of cells lacking C/EBPβ as autophagy maintains cellular fitness by turnover of damaged organelles and long-lived proteins [44]. We also found that suppression of C/EBPβ increased PC3 cell and tumor susceptibility to bortezomib. Further, CQ did not additively increase PC3 cell death following bortezomib treatment, indicating that bortezomib-induced cell death was related to inhibition of autophagy. These results are in accordance with previous studies wherein combined autophagy and proteasome blockade additively increased tumor cell killing [52–54]. In particular, our results implicate C/EBPβ as a key mediator of autophagy in PCa and validate the utility of targeting C/EBPβ in combination with bortezomib or other proteasome inhibitors as a novel approach to PCa therapy.
Supplementary Material
Acknowledgements
This work was supported by Department of Defense Post-Doctoral Research Grant PC131609 (to DJB), by NIH training grant T32 CA60441 (to TB), by the Patrick Walsh Cancer Research Fund (to SKK, IPP, and ADF), by NIH Cancer Center grant P30 CA006973, by an AEGON International Scholarship (to JM), by FAMRI (to SKK), by NIH Prostate SPORE grant P50 CA58236 (to SKK), and by the Giant Food Children's Cancer Research Fund.
Abbreviations
- bZIP
basic region-leucine zipper
- C/EBPβ
CCAAT/enhancer binding protein β
- ChIP
chromatin immunoprecipitation
- CQ
chloroquine
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- KD
knockdown
- NOD/SCID
non-obese diabetic/severe-combined immuno-deficient
- NSG
IL2Rγ−/−
- PE
phosphatidylethanolamine
- PCa
prostate cancer
- qPCR
quantitative polymerase chainreaction
- PB
PiggyBac
- mRFP
monomeric red fluorescent protein
- shRNA
short hairpin RNA
- TSS
transcription start site
Footnotes
Authors’ contributions
DJB, SKK, IPP, and ADF conceived and designed the research. TB and JZ generated shCEBPB and shNTV constructs and cell lines. JM conducted the autophagosome–lysosome fusion analysis. DJB conducted the balance of the experiments. DJB and ADF wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest
None to declare.
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.canlet.2016.03.005.
References
- 1.Morris MJ, Kelly WK, Slovin S, Ryan C, Eicher C, Heller G, et al. A phase II trial of bortezomib and prednisone for castration resistant metastatic prostate cancer. J. Urol. 2007;178:2378–2383. doi: 10.1016/j.juro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Dreicer R, Petrylak D, Agus D, Webb I, Roth B. Phase I/II study of bortezomib plus docetaxel in patients with advanced androgen-independent prostate cancer. Clin. Cancer Res. 2007;13:1208–1215. doi: 10.1158/1078-0432.CCR-06-2046. [DOI] [PubMed] [Google Scholar]
- 3.Hainsworth JD, Meluch AA, Spigel DR, Barton J, Jr., Simons L, Meng C, et al. Weekly docetaxel and bortezomib as first-line treatment for patients with hormone-refractory prostate cancer: a Minnie Pearl Cancer Research Network phase II trial. Clin. Genitourin. Cancer. 2007;5:278–283. doi: 10.3816/CGC.2007.n.004. [DOI] [PubMed] [Google Scholar]
- 4.Zhao SG, Jackson WC, Kothari V, Schipper MJ, Erho N, Evans JR, et al. High-throughput transcriptomic analysis nominates proteasomal genes as age-specific biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2015;18:229–236. doi: 10.1038/pcan.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 8.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao S, Dennis M, Song X, Vadysirisack DD, Salunke D, Nash Z, et al. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat. Commun. 2015;6:7014. doi: 10.1038/ncomms8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 13.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen HG, Yang JC, Kung HJ, Shi XB, Tilki D, Lara PN, Jr., et al. Targeting autophagy overcomes enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33:4521–4530. doi: 10.1038/onc.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan Q, Joshua AM, Saggar JK, Yu M, Wang M, Kanga N, et al. Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy. Br. J. Cancer. 2015;112:832–840. doi: 10.1038/bjc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Modernelli A, Naponelli V, Giovanna Troglio M, Bonacini M, Ramazzina I, Bettuzzi S, et al. EGCG antagonizes Bortezomib cytotoxicity in prostate cancer cells by an autophagic mechanism. Sci. Rep. 2015;5:15270. doi: 10.1038/srep15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, He J, Liu Z, Lu Y, Zheng Y, Li H, et al. Anti-β2-microglobulin monoclonal antibodies overcome bortezomib resistance in multiple myeloma by inhibiting autophagy. Oncotarget. 2015;6:8567–8578. doi: 10.18632/oncotarget.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogl DT, Stadtmauer EA, Tan KS, Heitjan DF, Davis LE, Pontiggia L, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu K, Dunner K, Jr., McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.B'chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L, Huang JX, Liu Y, Li X, Zhou SR, Qian SW, et al. Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Mol. Cell. Biol. 2013;33:3180–3190. doi: 10.1128/MCB.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirley CM, Chen J, Shamay M, Li H, Zahnow CA, Hayward SD, et al. Bortezomib induction of C/EBPβ mediates Epstein-Barr virus lytic activation in Burkitt lymphoma. Blood. 2011;117:6297–6303. doi: 10.1182/blood-2011-01-332379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hungness ES, Robb BW, Luo GJ, Pritts TA, Hershko DD, Hasselgren PO. Proteasome inhibitors activate the transcription factors C/EBP-beta and delta in human intestinal epithelial cells. Biochem. Biophys. Res. Commun. 2002;290:469–474. doi: 10.1006/bbrc.2001.6168. [DOI] [PubMed] [Google Scholar]
- 27.Chen JJ, Huang WC, Chen CC. Transcriptional regulation of cyclooxygenase-2 in response to proteasome inhibitors involves reactive oxygen species-mediated signaling pathway and recruitment of CCAAT/enhancer-binding protein delta and CREB-binding protein. Mol. Biol. Cell. 2005;16:5579–5591. doi: 10.1091/mbc.E05-08-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother. Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 29.Barakat DJ, Zhang J, Barberi T, Denmeade SR, Friedman AD, Paz-Priel I. CCAAT/enhancer binding protein β controls androgen-deprivation-induced senescence in prostate cancer cells. Oncogene. 2015;34:5912–5922. doi: 10.1038/onc.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topaloglu O, Hurley PJ, Yildirim O, Civin CI, Bunz F. Improved methods for the generation of human gene knockout and knockin cell lines. Nucleic Acids Res. 2005;33:e158. doi: 10.1093/nar/gni160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paz-Priel I, Houng S, Dooher J, Friedman AD. C/EBPα and C/EBPα oncoproteins regulate nfkb1 and displace histone deacetylases from NF-κB p50 homodimers to induce NF-κB target genes. Blood. 2011;117:4085–4094. doi: 10.1182/blood-2010-07-294470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 33.Narain Y, Wyttenbach A, Rankin J, Furlong RA, Rubinsztein DC. A molecular investigation of true dominance in Huntington's disease. J. Med. Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo L, Li X, Huang JX, Huang HY, Zhang YY, Qian SW, et al. Histone demethylase Kdm4b functions as a co-factor of C/EBPβ to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes. Cell Death Differ. 2012;19:1917–1927. doi: 10.1038/cdd.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen KA, Myers JT, Swanson JA. pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 2002;115:599–607. doi: 10.1242/jcs.115.3.599. [DOI] [PubMed] [Google Scholar]
- 37.Mauvezin C, Nagy P, Juhász G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin L, Stringfield TM, Shi X, Chen Y. Arsenite induces a cell stress-response gene, RTP801, through reactive oxygen species and transcription factors Elk-1 and CCAAT/enhancer-binding protein. Biochem. J. 2005;392:93–102. doi: 10.1042/BJ20050553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin HO, Seo SK, Woo SH, Kim ES, Lee HC, Yoo DH, et al. Activating transcription factor 4 and CCAAT/enhancer-binding protein-beta negatively regulate the mammalian target of rapamycin via Redd1 expression in response to oxidative and endoplasmic reticulum stress. Free Radic. Biol. Med. 2009;46:1158–1167. doi: 10.1016/j.freeradbiomed.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Malagelada C, Ryu EJ, Biswas SC, Jackson-Lewis V, Greene LA. RTP801 is elevated in Parkinson brain substantia nigral neurons and mediates death in cellular models of Parkinson's disease by a mechanism involving mammalian target of rapamycin inactivation. J. Neurosci. 2006;26:9996–10005. doi: 10.1523/JNEUROSCI.3292-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suraweera A, Münch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fels DR, Ye J, Segan AT, Kridel SJ, Spiotto M, Olson M, et al. Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res. 2008;68:9323–9330. doi: 10.1158/0008-5472.CAN-08-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baixauli F, López-Otín C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surmacz CA, Pösö AR, Mortimore GE. Regulation of lysosomal fusion during deprivation-induced autophagy in perfused rat liver. Biochem. J. 1987;242:453–458. doi: 10.1042/bj2420453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovacs J, Karpati AP. Regression of autophagic vacuoles in mouse pancreatic cells: a morphometric study of the effect of methylamine and chloroquine followed by cycloheximide treatment. Cell Biol. Int. Rep. 1989;13:805–811. doi: 10.1016/0309-1651(89)90057-x. [DOI] [PubMed] [Google Scholar]
- 47.Pizarro-Cerdá J, Desjardins M, Moreno E, Akira S, Gorvel JP. Modulation of endocytosis in nuclear factor IL-6(−/−) macrophages is responsible for a high susceptibility to intracellular bacterial infection. J. Immunol. 1999;162:3519–3526. [PubMed] [Google Scholar]
- 48.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 49.Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 2015;40:141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao C, Chao A, Tsai CL, Chuang WC, Huang WP, Chen GC, et al. Bortezomib enhances cancer cell death by blocking the autophagic flux through stimulating ERK phosphorylation. Cell Death Dis. 2014;5:e1510. doi: 10.1038/cddis.2014.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Periyasamy-Thandavan S, Jackson WH, Samaddar JS, Erickson B, Barrett JR, Raney L, et al. Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress and induces caspase-dependent cell death in antiestrogen-sensitive and resistant ER+ breast cancer cells. Autophagy. 2010;6:19–35. doi: 10.4161/auto.6.1.10323. [DOI] [PubMed] [Google Scholar]
- 52.Chen S, Zhang Y, Zhou L, Leng Y, Lin H, Kmieciak M, et al. A Bim-targeting strategy overcomes adaptive bortezomib resistance in myeloma through a novel link between autophagy and apoptosis. Blood. 2014;124:2687–2697. doi: 10.1182/blood-2014-03-564534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding WX, Ni HM, Gao W, Chen X, Kang JH, Stolz DB, et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol. Cancer Ther. 2009;8:2036–2045. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hui B, Shi YH, Ding ZB, Zhou J, Gu CY, Peng YF, et al. Proteasome inhibitor interacts synergistically with autophagy inhibitor to suppress proliferation and induce apoptosis in hepatocellular carcinoma. Cancer. 2012;118:5560–5571. doi: 10.1002/cncr.27586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.