Abstract
DNA methyltransferase 1 (DNMT1) plays a pivotal role in maintaining DNA methylation status. Polymorphisms of DNMT1 may modify the role of DNMT1 in prognosis of gastric cancer (GC). Our aim was to test whether polymorphisms of DNMT1 gene were associated with overall survival of GC. Four hundred and forty-seven GC patients who underwent radical tumorectomy were enrolled in the study. Five tagging SNPs (rs10420321, rs16999593, rs2228612, rs2228611, and rs2288349) of the DNMT1 gene were genotyped by TaqMan assays. Kaplan-Meier survival plots and Cox proportional hazard regression were used to analyze the associations between SNPs of DNMT1 and survival of GC. Patients carrying rs2228611 GA/AA genotype tended to live longer than those bearing the GG genotype (HR 0.68, 95% CI: 0.51–0.91, P = 0.007). Further multivariate Cox regression analysis showed that rs2228611 was an independent prognostic factor (GA/AA versus GG: OR 0.67, 95% CI 0.49–0.91, P = 0.010). Nevertheless, other SNPs did not show any significant associations with survival of GC. Polymorphisms of the DNMT1 gene may affect overall survival of GC. The SNP rs2228611 has the potentiality to serve as an independent prognostic marker for GC patients.
1. Introduction
Gastric cancer (GC) is the most common gastrointestinal malignancy and the third leading cause of cancer-related death worldwide [1]. Although radical gastrectomy has been regarded as an effective treatment, prognosis for GC patients remains poor, with a 5-year overall survival (OS) rate of only 27.4% in China [2]. GC patients have different outcomes even when they are diagnosed at the same clinical stage and receiving the same treatment [3], which suggests that the host characteristics, specifically the genetic factors, play crucial roles in GC prognosis. Single nucleotide polymorphisms (SNPs) are the most common genetic variations and reportedly influence the patients' clinical outcomes [4–6].
DNA methylation, which is established and maintained by DNA methyltransferases (DNMTs), is the major direct modification of eukaryotic DNA and is known to have profound effects on the regulation of gene expression [7–10]. DNMT1 is the primary enzyme that maintains the level of DNA methylation [11]. Aberrant expression of DNMT1 is correlated with progression and prognosis of various cancers, such as lung cancer [12], hepatocellular carcinoma [13], pancreatic cancer [14], and bladder cancer [15]. In GC, overexpression of DNMT1 correlates with worse differentiation, advanced stage, and increased risk of death [16–18]. Meanwhile, inhibition or blockade of DNMT1 can arrest cell cycle, increase cell apoptosis, decrease invasion, and enhance treatment sensitivity [15, 16, 19].
Previous studies have reported that SNPs of DNMT1 gene are associated with susceptibility to cancers such as ovarian cancer [20] and breast cancer [21]. In GC, though Khatami et al. found no association in an Iranian population [22], Yang et al. observed a 45% increased risk in individuals bearing rs16999593 C allele in Southern Chinese population [23]. Our previous study showed that rs10420321 GG or rs8111085 CC genotype was associated with lower risk of Helicobacter pylori infection but higher risk of gastric atrophy [24].
Given the role of DNMT1 in cancer progression and prognosis, we hypothesized that SNPs of DNMT1 may have prognostic value of cancer. As far as we know, there was no study on it. The aim of our study was to explore the role of SNPs of DNMT1 in the prognosis of gastric cancer [23].
2. Material and Methods
2.1. Subject Selection
Subjects of the study were described elsewhere [24, 25]. From August 2008 to December 2010, a total of 447 GC patients who underwent tumorectomy in the Department of Gastric and Colorectal Surgery of the First Hospital of Jilin University were recruited in this study. All of them were histologically diagnosed with gastric cancer and none of them received chemotherapy or radiotherapy before surgery. All patients were Han descent from the area of Changchun. Clinical and pathological data were collected including age, gender, tumor size, pathological type, differentiation, depth of invasion, lymph metastasis, distant metastasis, lymph-vascular invasion, and therapy. The clinical stage of gastric cancer was evaluated by the TNM classification according to American Joint Committee on Cancer (AJCC) in 2010, the seventh edition [26]. Written informed consent was obtained from each patient before sample collection. This study was approved by the ethics committee of the First Hospital of Jilin University.
The follow-up of patients was performed by telephone calls three months, six months, and one year after the tumorectomy and every one year thereafter until death or being lost to follow-up. The following data were recorded during each follow-up: the health status of patients, therapy after operation, and date of death if the patients died. About one-third of the patients received chemotherapy after surgery. The chemotherapy mainly consisted of three regimens: FOLFOX-4 regimen (combination with 5-fluorouracil, leucovorin, and oxaliplatin); XELOX regimen (capecitabine and oxaliplatin); and other chemotherapies such as capecitabine or 5-fluorouracil alone. A patient would be considered to have postoperative chemotherapy only if they received the therapy for at least 3 cycles.
2.2. Selection of Tagging SNPs
The detailed description of tagging SNPs selection was depicted in our previous work [24]. Briefly, tagging SNPs were selected from the Chinese Han data (CHB) in the HapMap Project (06-02-2009 HapMap) using the SNP browser Software v4.0 to capture SNPs with minimum minor allele frequency (MAF) > 0.05 and pairwise r 2 ≥ 0.8 [27]. There were 27 SNPs with MAF > 0.05 in DNMT1 gene region in the Chinese on HapMap. Three of them lie in the coding regions, including two nonsynonymous SNPs of rs16999593 (His97Arg) in exon 4 and rs2228612 (Ile327Val) in exon 12 and one synonymous SNP rs2228611 in exon 17. The remaining 24 SNPs reside in introns of DNMT1. Five linkage disequilibrium (LD) blocks were identified. The 3 SNPs in coding region could tag the blocks they belonged to (D′ = 1 and r 2 = 0.8) and were selected. Finally, five SNPs, rs10420321, rs16999593, rs2228612, rs2228611, and rs2288349, were selected as tagging SNPs for further study.
2.3. Genotyping
Blood samples of each subject were collected in EDTA tubes and stored in −80°C until DNA extraction. Genomic DNA was extracted using commercial kits following the protocol provided by the manufacturer (AxyPrep Blood Genomic DNA Miniprep kit, Axygen Biosciences, USA). Genotypes of each SNP were determined by TaqMan SNP genotyping assays in 384-well plates (Applied Biosystems, USA) and the detailed process of polymerase chain reaction (PCR) was described elsewhere [24]. The PCR products were read on ABI PRISM 7900 Sequence Detector in the end-point mode and genotypes were identified using the Allelic Discrimination Sequence Detector Software V2.3. Ten randomly selected samples were genotyped twice in all SNPs to verify genotyping and sample-handling accuracy and the concordance rate was 100%. The call rates for rs10420321, rs16999593, rs2228612, rs2228611, and rs2288349 were 91.7%, 100%, 100%, 99.5%, and 99.3%, respectively.
2.4. Statistical Analysis
Overall survival was defined as the time from the date of surgery to the date of death (if the patients were dead) or to the date of final follow-up (if the patients were alive) or to the date of the last follow-up (if the patients were lost to follow-up). Survival time would be right-censored in case that the patients were alive, were lost to follow-up, or died of causes other than GC. Patients would be excluded from survival analysis if they were lost to follow-up at the first interview or they died of postoperative complications within 30 days such as uncontrollable bleeding during perioperative period.
Survival curves of the GC patients within each SNP were plotted by Kaplan-Meier method and compared by log-rank test. The median survival time was estimated by the Kaplan-Meier method. If median OS could not be calculated, mean OS would be used instead. Hazard ratios (HRs) with their 95% confidence intervals (CIs) were calculated by the Cox proportional hazards model after adjusting for age (scale variable), sex (nominal variable), clinical stage (scale variable), and postoperational chemotherapy (nominal variable). Stepwise Cox regression analysis was conducted to determine independent predictive factors of prognosis, with a significance level of P < 0.05 for entering and P > 0.05 for removing the variables. All analyses were performed using SPSS18.0 Software (SPSS Inc., USA). Two-tailed P values < 0.05 were considered to be statistically significant.
3. Results
3.1. Characteristics of Patients
A total of 447 patients with histologically confirmed GC were enrolled in the study. Twelve patients (2.7%) could not be connected any more after they left the hospital and follow-up information was available for 435 patients (97.3%) until March 2016. Thirteen patients (2.9%) died of postoperative complications. These 25 cases (12 + 13 = 25, 5.6%) were excluded from the analysis of effects of SNPs on survival. The remaining 422 patients were enrolled in further analysis. During the follow-up period, 197 (46.2%) patients died of gastric cancer, 19 (3.3%) died of other causes such as cerebral hemorrhage, 17 (2.4%) were lost to follow-up, and 189 (48.1%) were alive. The duration of follow-up ranged from 1.1 to 85.0 months with a median of 59.9 months (95% CI: 59.4–60.5 months).
The baseline characteristics and clinical features of these patients are detailed in Table 1. Of these patients, 72.3% were male gender, 87.4% were tubular adenoma type, 62.9% were poorly differentiated, 43.1% were at advanced stage (TNM III or IV), 96.4% received radical gastrectomy, and 32.7% received postoperational chemotherapy. Survival analysis shows that patients in male gender, with tumor size > 5 cm, with deeper invasion, with presence of lymph node metastasis or distant metastasis, with advanced TNM stage, positive for lymph-vascular or neural invasion, and receiving palliative surgery only had shorter postoperational survival compared to patients without (log-rank P < 0.05, Table 1).
Table 1.
Characteristics and clinical features of gastric cancer patients.
| Variables | Number of patients | Number of deaths | Mean OS (months) | Log-rank P | HR (95% CI) |
|---|---|---|---|---|---|
| All | 422 | 195 | 54.5 | ||
| Gender | |||||
| Female | 117 (27.7) | 45 | 57.8 | 0.044 | 1.00 |
| Male | 305 (72.3) | 152 | 48.8 | 1.41 (1.01–1.96) | |
| Age | |||||
| ≤60 | 209 (49.5) | 91 | 52.5 | 0.218 | 1.00 |
| >60 | 213 (50.5) | 106 | 50.2 | 1.19 (0.90–1.58) | |
| Tumor size | |||||
| ≤5 cm | 231 (55.8) | 75 | 62.6 | <0.001 | 1.00 |
| >5 cm | 183 (44.2) | 116 | 39.5 | 2.63 (1.96–3.52) | |
| Pathological type | |||||
| Tubular adenocarcinoma | 368 (87.4) | 170 | 52.2 | 0.167 | 1.00 |
| Signet ring cell carcinoma | 22 (5.2) | 8 | 49.1 | 0.75 (0.37–1.53) | |
| Others | 31 (7.4) | 19 | 38.8 | 1.49 (0.93–2.40) | |
| Differentiation | |||||
| Well to moderate | 155 (37.1) | 70 | 55.7 | 0.161 | 1.00 |
| Poor | 263 (62.9) | 124 | 49.4 | 1.23 (0.92–1.65) | |
| Depth of invasion | |||||
| T1/T2 | 97 (23.0) | 13 | 74.2 | <0.001 | 1.00 |
| T3/T4 | 322 (77.0) | 182 | 44.8 | 5.96 (3.39–10.46) | |
| Lymph metastasis | |||||
| N0 | 119 (29.1) | 15 | 75.5 | <0.001 | 1.00 |
| N1/N2/N3 | 300 (70.9) | 180 | 42.0 | 7.16 (4.22–12.14) | |
| Distant metastasis | |||||
| M0 | 392 (92.9) | 174 | 53.9 | <0.001 | 1.00 |
| M1 | 30 (7.1) | 23 | 25.4 | 2.70 (1.75–4.18) | |
| TNM stage | |||||
| I/II | 240 (56.9) | 57 | 69.5 | <0.001 | 1.00 |
| III/IV | 182 (43.1) | 140 | 28.6 | 5.97 (4.37–8.16) | |
| Lymph-vascular invasion | |||||
| Absent | 112 (26.5) | 20 | 73.4 | <0.001 | 1.00 |
| Present | 305 (73.5) | 173 | 43.9 | 4.39 (2.76–6.99) | |
| Neural invasion | |||||
| Absent | 199 (47.2) | 65 | 63.0 | <0.001 | 1.00 |
| Present | 218 (52.8) | 128 | 42.4 | 2.42 (1.80–3.27) | |
| Radical surgery | |||||
| Yes | 407 (96.4) | 182 | 53.6 | <0.001 | 1.00 |
| Noa | 15 (3.6) | 15 | 10.8 | 5.61 (3.28–9.60) | |
| Chemotherapyb | |||||
| None | 284 (67.3) | 132 | 51.7 | 0.157 | 1.00 |
| XELOX | 29 (6.9) | 9 | 62.8 | 0.55 (0.28–1.08) | |
| FOLFOX-4 | 77 (18.2) | 36 | 51.3 | 0.98 (0.68–1.41) | |
| Others | 32 (7.6) | 20 | 39.6 | 1.34 (0.84–2.15) |
OS: overall survival; HR: hazard ratio; CI: confidence interval.
aFifteen patients received palliative surgery.
bXELOX (capecitabine and oxaliplatin); FOLFOX-4 (5-fluorouracil, leucovorin, and oxaliplatin); other chemotherapies included 5-fluorouracil; Xeloda alone; paclitaxel plus leucovorin and tegafurum; LV5-FU2 (leucovorin plus 5-fluorouracil); and FOLFIRI (irinotecan, 5-fluorouracil, and leucovorin).
3.2. Association between Genotypes and Survival of Gastric Cancer
As reported by our previous work [24], no association was found between the SNPs and pathological type, tumor differentiation, TNM stage, and lymph-vascular or neural invasion (Table 2).
Table 2.
Associations between the SNPs and clinical parameters of gastric cancer patients.
| rs10420321 | P | rs16999593 | P | rs2228612 | P | rs2228611 | P | rs2288349 | P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG + GG | TT | TC + CC | TT | TC + CC | GG | GA + AA | GG | GA + AA | ||||||
| N | 130 | 257 | 268 | 154 | 137 | 285 | 221 | 199 | 237 | 182 | |||||
| Gender | |||||||||||||||
| Male | 31.9 | 68.1 | 0.334 | 62.4 | 37.6 | 0.532 | 31.4 | 68.6 | 0.650 | 53.3 | 46.7 | 0.885 | 55.2 | 44.8 | 0.538 |
| Female | 36.9 | 63.1 | 65.6 | 34.4 | 33.6 | 66.4 | 54.0 | 46.0 | 58.4 | 41.6 | |||||
| Age | |||||||||||||||
| ≤60 | 34.5 | 65.5 | 0.600 | 64.7 | 35.3 | 0.571 | 34.0 | 66.0 | 0.392 | 54.0 | 46.0 | 0.837 | 52.6 | 47.4 | 0.154 |
| >60 | 32.0 | 68.0 | 62.1 | 37.9 | 30.2 | 69.8 | 53.0 | 47.0 | 59.3 | 40.7 | |||||
| Tumor size | |||||||||||||||
| ≤5 cm | 33.6 | 66.4 | 0.982 | 64.8 | 35.2 | 0.668 | 33.1 | 66.9 | 0.807 | 50.6 | 49.4 | 0.400 | 57.0 | 43.0 | 0.933 |
| >5 cm | 33.5 | 66.5 | 62.8 | 37.2 | 31.9 | 68.1 | 54.7 | 45.3 | 56.6 | 43.4 | |||||
| Pathological type | |||||||||||||||
| Tubular adenocarcinoma | 32.2 | 67.8 | 0.393 | 62.8 | 37.2 | 0.889 | 31.3 | 68.7 | 0.414 | 54.6 | 45.4 | 0.507 | 55.6 | 44.4 | 0.853 |
| Signet ring cell carcinoma | 36.8 | 63.2 | 65.2 | 34.8 | 30.4 | 69.6 | 47.8 | 52.2 | 56.5 | 43.5 | |||||
| Others | 43.8 | 56.3 | 66.7 | 33.3 | 42.4 | 57.6 | 45.5 | 54.5 | 60.6 | 39.4 | |||||
| Differentiation | |||||||||||||||
| Well to moderate | 37.6 | 62.4 | 0.191 | 61.1 | 38.9 | 0.501 | 34.4 | 65.6 | 0.458 | 57.7 | 42.3 | 0.152 | 54.5 | 45.5 | 0.646 |
| Poor | 31.1 | 68.9 | 64.4 | 35.6 | 30.9 | 69.1 | 50.5 | 49.5 | 56.8 | 43.2 | |||||
| TNM stage | |||||||||||||||
| I/II | 29.6 | 70.4 | 0.065 | 62.9 | 37.1 | 0.710 | 29.4 | 70.6 | 0.183 | 52.0 | 48.0 | 0.618 | 58.5 | 41.5 | 0.354 |
| III/IV | 38.4 | 61.6 | 64.6 | 35.4 | 35.4 | 64.6 | 54.4 | 45.6 | 54.1 | 45.9 | |||||
| Lymph-vascular invasion | |||||||||||||||
| Absent | 33.0 | 67.0 | 0.849 | 60.7 | 39.3 | 0.499 | 31.6 | 68.4 | 0.800 | 49.6 | 50.4 | 0.446 | 57.8 | 42.2 | 0.829 |
| Present | 34.0 | 66.0 | 64.2 | 35.8 | 32.9 | 67.1 | 53.7 | 46.3 | 56.6 | 43.4 | |||||
| Neural invasion | |||||||||||||||
| Absent | 32.8 | 67.2 | 0.699 | 62.6 | 37.4 | 0.794 | 33.0 | 67.0 | 0.848 | 50.0 | 50.0 | 0.305 | 60.0 | 40.0 | 0.215 |
| Present | 34.7 | 65.3 | 63.8 | 36.2 | 32.1 | 67.9 | 55.0 | 45.0 | 54.1 | 45.9 | |||||
| Radical surgery | |||||||||||||||
| Yes | 33.2 | 66.8 | 0.995 | 63.7 | 36.3 | 0.415 | 31.9 | 68.1 | 0.910 | 53.5 | 46.5 | 0.991 | 56.2 | 43.8 | 0.827 |
| No | 33.3 | 66.7 | 53.3 | 46.7 | 33.3 | 66.7 | 53.3 | 46.7 | 53.3 | 46.7 | |||||
| Chemotherapy | |||||||||||||||
| No | 30.2 | 69.8 | 0.056 | 59.9 | 40.1 | 0.028 | 28.7 | 71.3 | 0.026 | 56.4 | 43.6 | 0.069 | 55.1 | 44.9 | 0.530 |
| Yes | 39.8 | 60.2 | 70.7 | 29.3 | 39.3 | 60.7 | 47.1 | 52.9 | 58.3 | 41.7 | |||||
Survival curves within each SNP of DNMT1 were plotted and compared using the codominant, dominant, and recessive model (Table 3). More than half of patients bearing the GG genotype of rs2228611 (117/221 = 52.9%) died of GC until the final follow-up, whereas only 39.7% of patients with the GA or AA genotype died (P = 0.007). Comparing to patients carrying the GG genotype of rs2228611, patients with GA or AA genotype had reduced risk of death after surgery (GA: HR 0.66, 95% CI 0.48–0.89; AA: HR 0.81, 95% CI 0.46–1.44). Result from the dominant model combining patients with GA or AA genotype showed that the A allele bearers of rs2228611 had 32% lower risk of death comparing to the GG carriers (HR 0.68, 95% CI 0.51–0.90, log-rank P = 0.007, Figure 1). Further subgroup analysis in patients receiving radical gastrectomy showed similar correlation where A allele reduced the death risk by 35% (HR 0.65, 95% CI 0.48–0.88, log-rank P = 0.005; adjusting for age, sex, TNM stage, and chemotherapy).
Table 3.
Associations between the SNPs and overall survival of gastric cancer patients.
| Genotypes | Overall patients N = 422 | Mean OS | HR (95% CI) | P | Adjusted HR (95% CI)a | Adjusted P | |
|---|---|---|---|---|---|---|---|
| n (%) | Deaths | ||||||
| rs10420321 | |||||||
| AA | 130 (33.6) | 62 | 50.0 | 1.00 | 1.00 | ||
| AG | 189 (48.8) | 85 | 53.6 | 0.90 (0.65–1.24) | 0.510 | 0.91 (0.65–1.26) | 0.553 |
| GG | 68 (17.6) | 32 | 52.1 | 0.97 (0.63–1.49) | 0.890 | 0.98 (0.64–1.50) | 0.917 |
| Dominant | |||||||
| AA | 130 (33.6) | 62 | 50.0 | 1.00 | 1.00 | ||
| AG/GG | 257 (66.4) | 117 | 53.3 | 0.90 (0.67–1.24) | 0.490 | 0.92 (0.68–1.26) | 0.618 |
| Recessive | |||||||
| AA/AG | 319 (82.4) | 147 | 52.4 | 1.00 | 1.00 | ||
| GG | 68 (17.6) | 32 | 52.1 | 1.04 (0.71–1.52) | 0.730 | 1.04 (0.70–1.52) | 0.857 |
| rs16999593 | |||||||
| TT | 268 (63.5) | 127 | 51.3 | 1.00 | 1.00 | ||
| TC | 135 (32.0) | 63 | 52.3 | 0.98 (0.72–1.32) | 0.891 | 0.99 (0.73–1.34) | 0.986 |
| CC | 19 (4.5) | 7 | 50.5 | 0.80 (0.37–1.70) | 0.555 | 0.80 (0.37–1.72) | 0.802 |
| Dominant | |||||||
| TT | 268 (63.5) | 127 | 51.3 | 1.00 | 1.00 | ||
| TC/CC | 154 (36.5) | 70 | 52.9 | 0.96 (0.72–1.28) | 0.767 | 0.96 (0.72–1.29) | 0.801 |
| Recessive | |||||||
| TT/TC | 403 (95.5) | 190 | 51.8 | 1.00 | 1.00 | ||
| CC | 19 (4.5) | 7 | 50.5 | 0.80 (0.38–1.70) | 0.471 | 0.81 (0.38–1.72) | 0.576 |
| rs2228612 | |||||||
| TT | 137 (32.4) | 65 | 50.1 | 1.00 | 1.00 | ||
| TC | 197 (46.7) | 90 | 52.9 | 0.92 (0.67–1.27) | 0.629 | 0.92 (0.67–1.27) | 0.614 |
| CC | 88 (20.9) | 42 | 51.3 | 1.00 (0.68–1.47) | 1.000 | 1.03 (0.70–1.53) | 0.864 |
| Dominant | |||||||
| TT | 137 (32.4) | 65 | 50.1 | 1.00 | 1.00 | ||
| TC/CC | 285 (67.6) | 132 | 52.5 | 0.95 (0.70–1.27) | 0.720 | 0.96 (0.71–1.29) | 0.760 |
| Recessive | |||||||
| TT/TC | 334 (79.1) | 155 | 52.0 | 1.00 | 1.00 | ||
| CC | 88 (20.9) | 42 | 51.3 | 1.05 (0.75–1.47) | 0.790 | 1.09 (0.77–1.53) | 0.638 |
| rs2228611 | |||||||
| GG | 221 (52.6) | 117 | 47.8 | 1.00 | 1.00 | ||
| GA | 169 (40.3) | 66 | 57.5 | 0.66 (0.48–0.89) | 0.006 | 0.65 (0.48–0.88) | 0.006 |
| AA | 30 (7.1) | 13 | 45.9 | 0.81 (0.46–1.44) | 0.469 | 0.80 (0.45–1.42) | 0.444 |
| Dominant | |||||||
| GG | 221 (52.6) | 117 | 47.8 | 1.00 | 1.00 | ||
| GA/AA | 199 (47.4) | 79 | 56.7 | 0.68 (0.51–0.90) | 0.007 | 0.67 (0.51–0.90) | 0.007 |
| Recessive | |||||||
| GG/GA | 390 (92.9) | 183 | 52.1 | 1.00 | 1.00 | ||
| AA | 30 (7.1) | 13 | 45.9 | 0.96 (0.55–1.69) | 0.894 | 0.95 (0.54–1.68) | 0.860 |
| rs2288349 | |||||||
| GG | 237 (56.6) | 102 | 54.7 | 1.00 | 1.00 | ||
| GA | 151 (36.0) | 77 | 47.4 | 1.27 (0.95–1.71) | 0.1105 | 1.26 (0.94–1.70) | 0.128 |
| AA | 31 (7.4) | 17 | 46.0 | 1.39 (0.84–2.33) | 0.204 | 1.37 (0.82–2.30) | 0.228 |
| Dominant | |||||||
| GG | 237 (56.6) | 102 | 54.7 | 1.00 | 1.00 | ||
| GA/AA | 182 (43.4) | 94 | 47.7 | 1.29 (0.98–1.71) | 0.072 | 1.28 (0.97–1.70) | 0.086 |
| Recessive | |||||||
| GG/GA | 388 (92.6) | 179 | 52.4 | 1.00 | 1.00 | ||
| AA | 31 (7.4) | 17 | 46.0 | 1.27 (0.77–2.08) | 0.353 | 1.25 (0.76–2.06) | 0.376 |
OS: overall survival; HR: hazard ratio; CI: confidence interval.
aAdjusted for age, sex, TNM stage, and postoperational chemotherapy.
Figure 1.
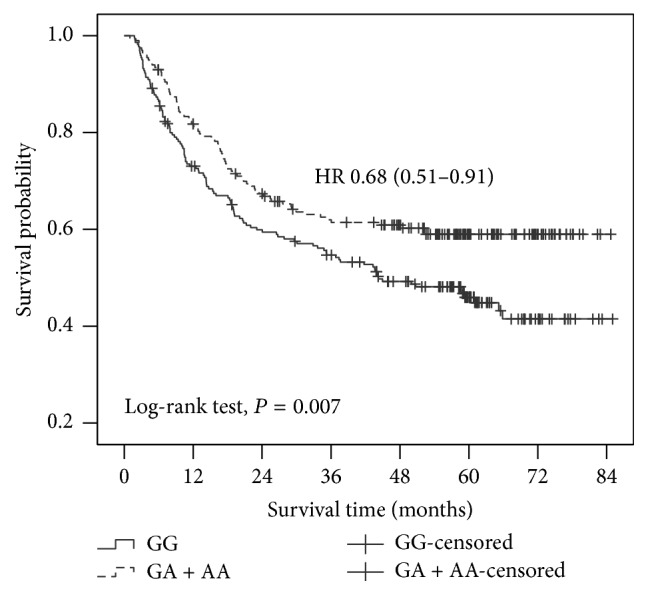
Association of DNMT1 rs2228611 with overall survival in gastric cancer patients.
Other four SNPs, rs10420321, rs16999593, rs2228612, and rs2288349, were not associated with the overall survival of GC (Table 3). Further haplotype analysis did not show associations between the haplotype and survival in GC (Table 3).
3.3. Stratified Analysis between Genotypes and Survival
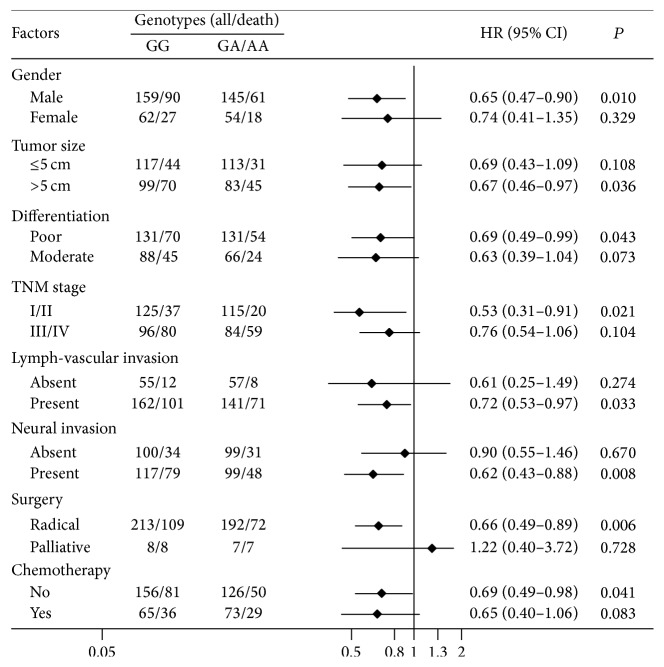
We further performed the stratified analysis for rs2228611 to investigate if this SNP on survival was modified by some important clinicopathological factors. As presented in Figure 2, patients carrying GA/AA variant genotypes had a lower death risk than those with the GG genotype. The protective effect was more pronounced among patients with relatively early clinical stage (HR 0.53, 95% CI: 0.31–0.91; P = 0.021).
Figure 2.
Stratified survival analysis for rs2228611 using the dominant model.
3.4. Stepwise Cox Regression Model for GC Survival
Further multivariate Cox regression analysis showed that rs2228611 was an independent predictive factor for GC (GA/AA versus GG, HR 0.67, 95% CI 0.49–0.91, P = 0.010). Other factors, male gender (HR 1.80, 95% CI 1.22–2.67; P = 0.003), advanced TNM stage (HR 4.76, 95% CI: 3.28–6.90; P < 0.001), tumor size ≥ 5 cm (HR 1.56, 95% CI: 1.13–2.16; P = 0.007), and lymph-vascular invasion (HR 1.52, 95% CI: 1.08–2.14; P = 0.015), were associated with shorter survival of GC. In addition, receiving palliative surgery increased risk (HR 2.55, 95% CI: 1.40–4.64; P = 0.002) while receiving postsurgery chemotherapy (HR 0.69, 95% CI: 0.50–0.96; P = 0.029) decreased risk of death (Table 4).
Table 4.
Multivariate Cox regression analysis on the survival of GC.
| Variables | HR | 95% CI | P |
|---|---|---|---|
| rs2228611 (GA/AA versus GG) | 0.67 | 0.49–0.91 | 0.010 |
| Sex (male versus female) | 1.80 | 1.22–2.67 | 0.003 |
| TNM stages (III + IV versus I + II) | 4.76 | 3.28–6.90 | <0.001 |
| Tumor sizes (≥5 cm versus <5 cm) | 1.56 | 1.13–2.16 | 0.007 |
| Lymph-vascular invasion (present versus absent) | 1.52 | 1.08–2.14 | 0.015 |
| Surgical method (palliative versus radical surgery) | 2.55 | 1.40–4.64 | 0.002 |
| Chemotherapy (yes versus no) | 0.69 | 0.50–0.96 | 0.029 |
HR: hazard ratio; CI: confidence interval.
4. Discussion
In the present study, we explored the associations of five SNPs of DNMT1 gene (rs16999593, rs10420321, rs2288349, rs2228611, and rs2228612) with postoperational survival in Chinese gastric cancer patients. We found that rs2228611 was an independent prognostic factor where A allele of rs2228611 was associated with a 33% decreased risk of death (HR 0.67, 95% CI 0.49–0.91, P = 0.010, Table 4).
To the best of our knowledge, this is the first study to evaluate the prognosis value of DNMT1 SNPs on cancer. Previous studies mainly focused on the role in tumorigenesis and they reported controversial results. Khatami et al. first assessed the associations of SNPs of DNMT1 with sporadic colorectal cancer in 208 cases and 213 controls in Iranian population and they observed no significant correlation [28]. Liu et al. expanded the sample size (1609 colon cancer cases and 1974 controls) and found that rs2228612 together with dietary factors (folate, methionine, vitamin B2, and vitamin B12 intake) could modify risk of colon cancer [29]. Similar controversial results were reported in breast cancer [21, 30–32]. In gastric cancer, Khatami et al. did not find any association between SNPs of DNMT1 (rs2228611, rs721186, rs11488, and rs13784) and disease risk in Iranian patients [22]. Yang et al. examined this association in the Southern Chinese population and found that the C allele of rs16999593 increased the GC risk by 45% (OR 1.45, 95% CI: 1.00–2.11, P = 0.05). Our previous study, however, found that rs2228612 CC genotype increased the risk of precancerous disease (gastric atrophy) but not GC [24]. In the present study, we assessed their roles in the prognosis of GC and observed that A allele of rs2228611 was associated with favorable postoperational survival (HR 0.67, P = 0.010).
Methylation of CpG islands in promoter region can lead to transcriptional silence of genes. Silencing genes related to tumor suppressors or invasion suppressors promote tumorigenesis and metastasis [33–35]. DNMT1 is a key enzyme to maintain DNA methylation pattern [11]. Upregulation of DNMT1 downexpresses genes such as P53, P21 via increasing methylation, which could enhance proliferation of cancer cells [36]. Inhibition of DNMT1 restores expression of tumor suppression-related genes such as p16 and RGS10 and contributes to reduced cancer cell viability, decreased invasive capability, and enhanced treatment sensitivity [15, 16, 19, 37]. Aberrant expression of DNMT1 is associated with unfavorable prognosis of various cancers including GC [16]. Evaluating the prognostic value of genetic variations in DNMT1 in the prognosis of cancer makes sense. In our study, we found that the minor allele A of rs2228611 was associated with longer life-span of postoperational GC patients.
Rs2228611 is located in exon 17 of DNMT1 and the G-to-A change mediates a synonymous variation (CCG→CCA, Proline→Proline). Bioinformatics tool SNPinfo predicts that rs2228611 is in the region of exonic splicing enhancer (ESE) [38]. The G-to-A variation may alter the binding activity to serine/arginine-rich (SR) protein, change pre-RNA splicing of DNMT1, and therefore lead to alteration of normal DNMT1 expression. However, the exact mechanism of rs2228611 contributing to the prognosis of GC is still unknown and needs further investigation.
Two limitations should be noted in our study. The first one was that we did not clarify the underlying specific function of rs2228611 in the prognosis of GC due to our study design. The second one was that the follow-up time was relatively short as 50.5% of the patients are alive and the right side of the survival plot was censored, although the median follow-up time was 54.5 months. Therefore, more studies with larger-scale and longer follow-up time are needed to confirm our finding and to clarify the exact mechanism.
5. Conclusions
In summary, our study provides the first evidence that polymorphisms of the DNMT1 gene could modify the postoperational survival of gastric cancer cases. The A allele of rs2228611 contributes to favorable prognosis and has potentiality to serve as a prognostic predicting biomarker for gastric cancer.
Acknowledgments
The authors would like to thank all of those who participated in this study, especially Ying Song for her work on follow-up of subjects and Chang-Song Guo for his technical support. This work was supported by grants from National Natural Science Foundation of China (nos. 81373084 and 81273065), the Norman Bethune Program of Jilin University (no. 2013025), and the Youth Fund of the First Hospital of Jilin University (no. JDYY42013014).
Competing Interests
The authors declare that there are no competing interests regarding the publication of this paper.
Authors' Contributions
Zhifang Jia and Xing Wu contributed equally to this study and were listed as co-first authors.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Zeng H., Zheng R., Guo Y., et al. Cancer survival in China, 2003–2005: a population-based study. International Journal of Cancer. 2015;136(8):1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 3.Son S.-Y., Kim H.-H. Minimally invasive surgery in gastric cancer. World Journal of Gastroenterology. 2014;20(39):14132–14141. doi: 10.3748/wjg.v20.i39.14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S., Lv C., Jin H., et al. A common genetic variation in the promoter of miR-107 is associated with gastric adenocarcinoma susceptibility and survival. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2014;769:35–41. doi: 10.1016/j.mrfmmm.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Yu H., Wu X., Zhang Y., Jin Z., Li G., Zhao H. Genetic variability of DNA repair mechanisms influences chemotherapy outcome of gastric cancer. International Journal of Clinical and Experimental Pathology. 2015;8(4):4106–4112. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao T., Gu D., Xu Z., et al. Polymorphism in one-carbon metabolism pathway affects survival of gastric cancer patients: large and comprehensive study. Oncotarget. 2015;6(11):9564–9576. doi: 10.18632/oncotarget.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 8.Curradi M., Izzo A., Badaracco G., Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Molecular and Cellular Biology. 2002;22(9):3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X., Yan L., Davidson N. E. DNA methylation in breast cancer. Endocrine-Related Cancer. 2001;8(2):115–127. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- 10.Luczak M. W., Jagodzinski P. P. The role of DNA methylation in cancer development. Folia Histochemica et Cytobiologica. 2006;44(3):143–154. [PubMed] [Google Scholar]
- 11.Miremadi A., Oestergaard M. Z., Pharoah P. D. P., Caldas C. Cancer genetics of epigenetic genes. Human Molecular Genetics. 2007;16(1):R28–R49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- 12.Lin R.-K., Hsu H.-S., Chang J.-W., Chen C.-Y., Chen J.-T., Wang Y.-C. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55(2):205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Saito Y., Kanai Y., Nakagawa T., et al. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. International Journal of Cancer. 2003;105(4):527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 14.Wang W., Gao J., Man X.-H., Li Z.-S., Gong Y.-F. Significance of DNA methyltransferase-1 and histone deacetylase-1 in pancreatic cancer. Oncology Reports. 2009;21(6):1439–1447. doi: 10.3892/or_00000372. [DOI] [PubMed] [Google Scholar]
- 15.Wu C.-T., Wu C.-F., Lu C.-H., et al. Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer. 2011;117(22):5221–5233. doi: 10.1002/cncr.26150. [DOI] [PubMed] [Google Scholar]
- 16.Mutze K., Langer R., Schumacher F., et al. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. European Journal of Cancer. 2011;47(12):1817–1825. doi: 10.1016/j.ejca.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 17.Etoh T., Kanai Y., Ushijima S., et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. The American Journal of Pathology. 2004;164(2):689–699. doi: 10.1016/s0002-9440(10)63156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Wei X., Wu Q., et al. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Molecular Medicine Reports. 2011;4(6):1139–1143. doi: 10.3892/mmr.2011.578. [DOI] [PubMed] [Google Scholar]
- 19.Chen C.-C., Chen W.-C., Wang W.-H., et al. Role of DNA methyltransferase 1 in pharyngeal cancer related to treatment resistance. Head & Neck. 2011;33(8):1132–1143. doi: 10.1002/hed.21586. [DOI] [PubMed] [Google Scholar]
- 20.Mostowska A., Sajdak S., Pawlik P., Lianeri M., Jagodzinski P. P. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Molecular Biology Reports. 2013;40(8):4893–4899. doi: 10.1007/s11033-013-2589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun M.-Y., Yang X.-X., Xu W.-W., Yao G.-Y., Pan H.-Z., Li M. Association of DNMT1 and DNMT3B polymorphisms with breast cancer risk in Han Chinese women from South China. Genetics and Molecular Research. 2012;11(4):4330–4341. doi: 10.4238/2012.september.26.1. [DOI] [PubMed] [Google Scholar]
- 22.Khatami F., Noorinayer B., Ghiasi S., Mohebi R., Hashemi M., Zali M. R. Lack of effects of single nucleotide polymorphisms of the DNA methyltransferase 1 gene on gastric cancer in Iranian patients: a case control study. Asian Pacific Journal of Cancer Prevention. 2009;10(6):1177–1182. [PubMed] [Google Scholar]
- 23.Yang X.-X., He X.-Q., Li F.-X., Wu Y.-S., Gao Y., Li M. Risk-association of DNA methyltransferases polymorphisms with gastric cancer in the Southern Chinese population. International Journal of Molecular Sciences. 2012;13(7):8364–8378. doi: 10.3390/ijms13078364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J., Jia Z., Cao D., et al. Polymorphisms of the DNA methyltransferase 1 associated with reduced risks of Helicobacter pylori infection and increased risks of gastric atrophy. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0046058.e46058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., Jia Z., Cao D., et al. Polymorphism of DNA methyltransferase 3b and association with development and prognosis in gastric cancer. PLoS ONE. 2015;10(8) doi: 10.1371/journal.pone.0134059.e0134059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Washington K. 7th edition of the AJCC cancer staging manual: stomach. Annals of Surgical Oncology. 2010;17(12):3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 27.De La Vega F. M. Selecting single-nucleotide polymorphisms for association studies with SNPbrowser™ software. In: Collins A. R., editor. Linkage Disequilibrium and Association Mapping. Vol. 376. 2007. pp. 177–193. (Methods in Molecular Biology™). [DOI] [PubMed] [Google Scholar]
- 28.Khatami F., Noorinayer B., Mohebi S. R., Ghiasi S., Hashemi M., Zali M. R. Effects of amino acid substitution polymorphisms of two DNA methyltransferases on susceptibility to sporadic colorectal cancer. Asian Pacific Journal of Cancer Prevention. 2009;10(6):1183–1188. [PubMed] [Google Scholar]
- 29.Liu A. Y., Scherer D., Poole E., et al. Gene-diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Molecular Nutrition and Food Research. 2013;57(4):721–734. doi: 10.1002/mnfr.201200180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang G., Zhenkun F., Shuang C., et al. Association of DNMT1 gene polymorphisms in exons with sporadic infiltrating ductal breast carcinoma among Chinese Han women in the Heilongjiang Province. Clinical Breast Cancer. 2010;10(5):373–377. doi: 10.3816/cbc.2010.n.049. [DOI] [PubMed] [Google Scholar]
- 31.Lin W., Cen Y. L., Lin Y., et al. Joint effects between urinary selenium and polymorphisms in methylation related genes on breast cancer risk. Neoplasma. 2015;62(3):491–499. doi: 10.4149/neo_2015_059. [DOI] [PubMed] [Google Scholar]
- 32.Tao R., Chen Z., Wu P., et al. The possible role of EZH2 and DNMT1 polymorphisms in sporadic triple-negative breast carcinoma in southern Chinese females. Tumor Biology. 2015;36(12):9849–9855. doi: 10.1007/s13277-015-3754-y. [DOI] [PubMed] [Google Scholar]
- 33.Zeng B., Li Z., Chen R., et al. Epigenetic regulation of miR-124 by Hepatitis C Virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Letters. 2012;586(19):3271–3278. doi: 10.1016/j.febslet.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 34.Yoon J. H., Choi W. S., Kim O., Park W. S. The role of gastrokine 1 in gastric cancer. Journal of Gastric Cancer. 2014;14(3):147–155. doi: 10.5230/jgc.2014.14.3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramaniam D., Thombre R., Dhar A., Anant S. DNA methyltransferases: a novel target for prevention and therapy. Frontiers in Oncology. 2014;4, article 80 doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C.-C., Lin J.-H., Hsu T.-W., et al. IL-6 enriched lung cancer stem-like cell population by inhibition of cell cycle regulators via DNMT1 upregulation. International Journal of Cancer. 2015;136(3):547–559. doi: 10.1002/ijc.29033. [DOI] [PubMed] [Google Scholar]
- 37.Cacan E., Ali M. W., Boyd N. H., Hooks S. B., Greer S. F. Inhibition of HDAC1 and DNMT1 modulate RGS10 expression and decrease ovarian cancer chemoresistance. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0087455.e87455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Z., Taylor J. A. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Research. 2009;37(2):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]