Abstract
Contaminated water is a serious concern in many developing countries with severe health consequences particularly for children. Current methods for monitoring waterborne pathogens are often time consuming, expensive, and labor intensive, making them not suitable for these regions. Electrochemical detection in a microfluidic platform offers many advantages such as portability, minimal use of instrumentation, and easy integration with electronics. In many parts of the world, however, the required equipment for pathogen detection through electrochemical sensors is either not available or insufficiently portable, and operators may not be trained to use these sensors and interpret results, ultimately preventing its wide adoption. Counterintuitively, these same regions often have an extensive mobile phone infrastructure, suggesting the possibility of integrating electrochemical detection of bacterial pathogens with a mobile platform. Toward a solution to water quality interventions, we demonstrate a microfluidic electrochemical sensor combined with a mobile interface that detects the sequences from bacterial pathogens, suitable for rapid, affordable, and point-of-care water monitoring. We employ the transduction of DNA hybridization into a readily detectable electric signal by means of a conformational change of DNA stem-loop structure. Using this platform, we successfully demonstrate the detection of as low as 100 nM E. coli sequences and the automatic interpretation and mapping of the detection results via a mobile application.
Keywords: Biosensors, electrochemical devices, biomedical telemetry, mobile computing, cellular phones
I. Introduction
More than one billion people have no access to any type of safe source of water, and as a direct consequence, 1.6 million people die every year from waterborne diarrheal diseases [1]. This mortality rate is greater than that from all forms of violence combined including war, and children younger than five years of age are the most severely affected [2]. Conventional methods for detecting bacterial pathogens are culture-based, requiring traditional laboratory settings using bacterial surrogates or indicators, such as total coliform bacterium, fecal coliform bacterium or Escherichia coli. These tests point to the possible presence of human microbial pathogens, but the results are not precise enough to confirm the contamination from certain pathogens. Many publications report that outbreaks of waterborne disease have occurred in spite of the absence of these indicators in source water and treated water [3]. These methods are also time-consuming and impractical for tracing outbreaks and routine monitoring [4].
To replace the culture-based methods, several methods have been reported for rapid detection of bacterial pathogens, and among those techniques, nucleic acid based assays have been employed in a wide range of biosensing technologies, due to impressive selectivity and affinity [5]–[7]. To develop point-of-care (POC) diagnostics for the developing world, the World Health Organization (WHO) has established a set of criteria, and any proposed solution should be: 1) affordable, 2) sensitive, 3) specific, 4) user-friendly, 5) rapid and robust, 6) equipment-free, and 7) deliverable to those in need, abbreviated as “ASSURED” [8]. Toward these requirements, microfluidic platforms implementing electrochemical detection offer strong advantages such as facile integration of multiple components into a single chip and no need for bulky optical equipment required for detection [9], [10]. Aptamers are nucleic acids that selectively bind to low-molecular weight organic and inorganic substrates [11], and can be stored and transported at room temperature while antibodies are sensitive to temperature and humidity changes [12]. Aptamers also provide selective, reusable biosensors and their small size and versatility allow efficient immobilizations and high-density monolayers, which are critical in miniaturized systems [13], [14]. Owing to these advantages, electrochemical detection of bioanalytes using aptamer-based sensors has become an increasingly attractive method for POC diagnostics in a wide range of applications in recent years [15]–[19].
However, in many parts of the world, the required equipment for pathogen detection is either not available or insufficiently portable, and operators may not be well trained to use the sensors and interpret their results, ultimately preventing wide adoption of pathogen detection through electrochemical sensors at POC. Counterintuitively, these same regions often have an extensive mobile phone infrastructure, suggesting the possibility of integrating electrochemical detection of bacterial pathogens with a mobile platform. There are currently 6 billion mobile phone subscriptions worldwide, which corresponds to 86% of the world's population, with a continued growth projected in the next 3–5 years [20]. Note that mobile phones are essentially computers that can be connected to portable sensors to take measurements, interpret results, and store and send records for a global analysis. Therefore, utilizing mobile phones in POC diagnostics offers a compelling advantage of increased accessibility, especially for those in remote areas. Owing to this advantage, mobile platform assisted medical technology has begun to spread in recent years [21], [22].
Motivated by these clear advantages, we present a microfluidic electrochemical DNA-based sensor capable of continuous, real-time detection of pathogens, which we have combined with a mobile interface (Fig. 1). We have presented brief, preliminary results at a conference [23], and here we present a detailed study of our system. Our electrochemical sensor offers an attractive platform for point-of-care diagnostics because it is readily integrated with microelectronics and it requires minimal instrumentation. The detection methods developed in this work are applicable not only to hybridization-based nucleic acid detection, but also to other aptamer-based protein and small molecule detection. Considering that the communities that need these sensors the most lack internet access and the ability (personnel and equipment) to analyze the data obtained by the sensor locally, we have adapted the sensor to a mobile device, e.g., a cellular telephone, to enable the instant interpretation of the data obtained and the mapping of the test results from different places, indicating safe vs. not-safe water sources. This aspect of our platform could resolve the issues associated with the lack of trained personnel, which is a major and severe constraint in deploying health technology in these regions.
Fig. 1.
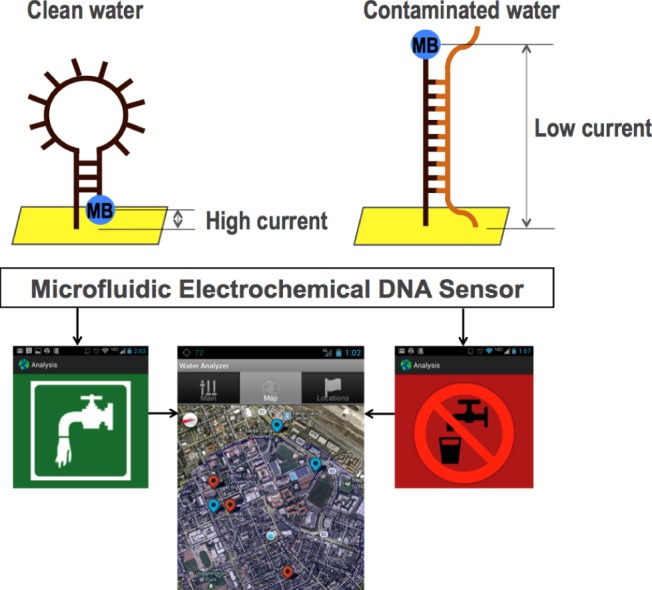
Overview of our microfluidic DNA sensor integrated with a mobile interface: an electrochemical sensor provides an electrochemical signal when hybridized to its target pathogen sequence. The mobile application inputs the data, interprets the test results, and maps the locations of the test results obtained from different locations.
II. System Overview
A. Electrochemical DNA Sensors
Our electrochemical DNA sensor offers an attractive platform for POC diagnostics due to its compact size, portability, low cost, high sensitivity and specificity. In addition, the simple instrumentation required for electrochemical detection leads to low electrical power requirements ideal for POC use. Our electrochemical sensor implements two working electrodes, a platinum counter electrode, and a platinum reference electrode (Fig. 2, bottom). The working electrodes contain an immobilized DNA probe in the stem-loop structure, holding methylene blue (MB) in proximity to the electrode surface, which enables efficient electron transfer with high reduction peak current (Fig. 1, top left, and Fig. 3, yellow peak from green curve). When a target sequence is introduced into the chamber, it hybridizes with the immobilized probe, opening up the stem-loop structure. This specific target-binding induced conformational change generates a large decrease in the reduction peak current of MB (Fig. 1, top right, and Fig. 3, cyan peak from pink curve). Once the pathogen detection is complete, a simple distilled water wash disrupts the hybridization of the target and resets the sensor for future measurements.
Fig. 2.
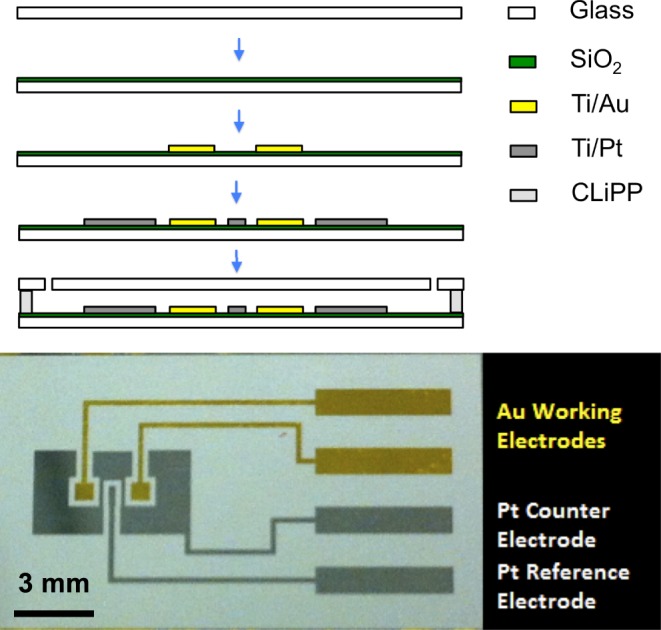
Process flow of microfluidic DNA sensors (top) and fabricated sensor (bottom): electrochemical DNA sensors are fabricated via standard metallization techniques. The polymeric microfluidic chambers are fabricated via contact liquid photolithographic polymerization (CLiPP).
Fig. 3.
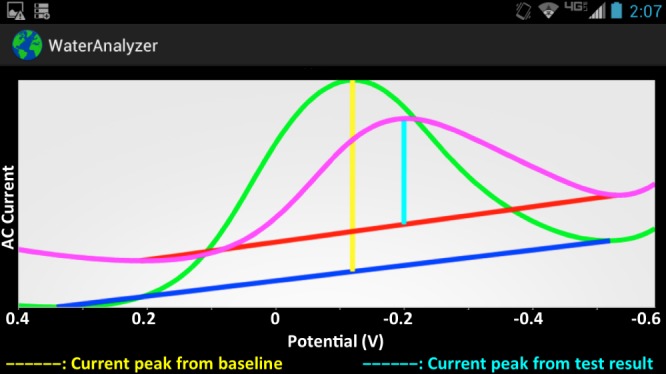
Snapshot of mobile app displaying results from baseline and measurement curves: the mobile app displays the results obtained from the sensor, and executes calculations to decide whether the water is contaminated or not.
B. Mobile Interface
The mobile app is the interface to the device. Its main goal is to show the end result (safe or not safe to drink) instantly to the user by automatically interpreting the results from the data obtained and making a decision based on water safety guidelines. We decided to implement the interface for an Android phone because of its recent spread through under-served communities around the world [24]. It communicates with the device through a USB cable through which, after each experiment, the data are uploaded to the phone as a table of voltage measurements. The app processes the data and shows the result to the user immediately. The result can be presented in two formats: as a curve in a graph (Fig. 3) or as a safe/not-safe guideline (Fig. 5). The app also keeps a log of previous results, which can be shown in a map (Fig. 6), in which red spots represent places with unsafe water, and blue spots represent places with safe water.
Fig. 5.
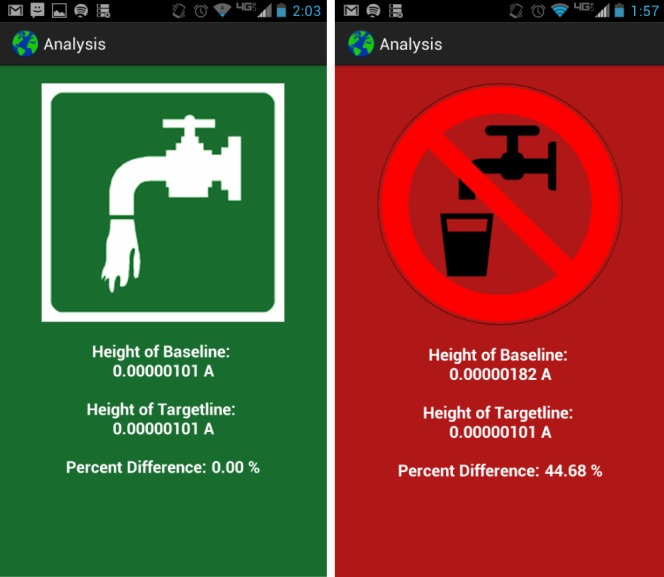
Snapshot of the mobile app displaying safe and not-safe indicators: once the percentage change of current signals is calculated in the app, the end result appears as safe or not-safe to drink based on water safety guidelines.
Fig. 6.
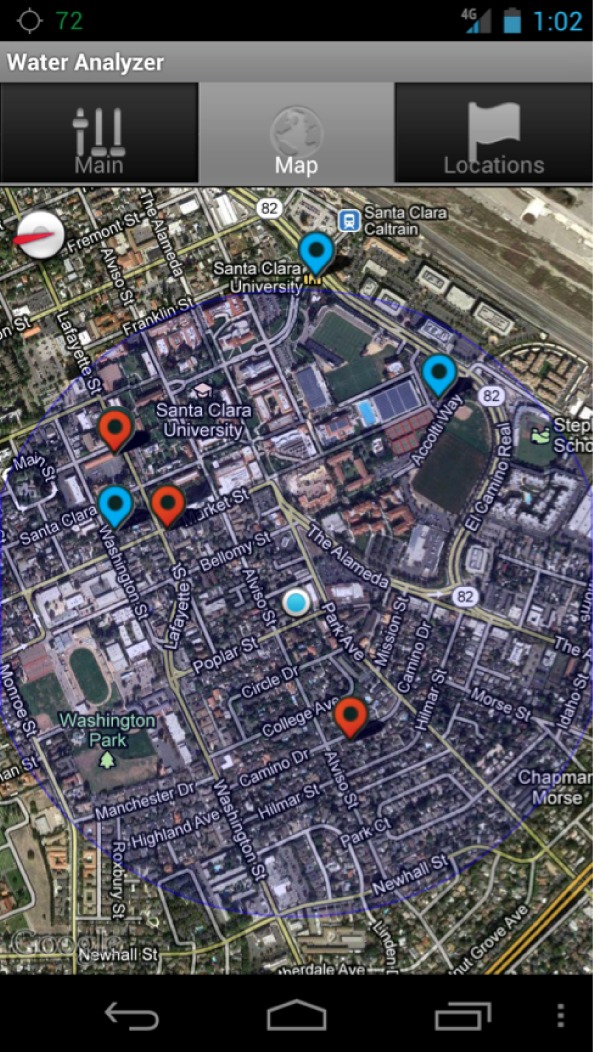
Snapshot of the mobile app showing a water safety map: data taken from different locations were processed in the mobile app to interpret the results, and the end results (safe or not-safe) were mapped.
III. Methods and Procedures
A. Chemical Reagents and Sensor Design
The single stranded probes and targets were synthesized and purified by Biosearch Technologies (Novato, CA). DTT, SSC and TRIS buffers were purchased from Promega (San Luis Obispo, CA). 6-mercapto-1-hexanol was purchased from Sigma Aldrich (Saint Louis, MO). We chose a 21-mer target sequence specific to 16S ribosomal RNA adapted from Liao et al. [25] (target sequence: 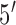 -ACA TCA TGG CAG ACA AAC AAA-
-ACA TCA TGG CAG ACA AAC AAA-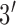 ). We added 5 base-pairs to both sides of the probe to form the molecular beacon stem. The
). We added 5 base-pairs to both sides of the probe to form the molecular beacon stem. The  terminus was labeled with thiol to be immobilized on gold electrodes via thiol-gold chemistry, and the
terminus was labeled with thiol to be immobilized on gold electrodes via thiol-gold chemistry, and the 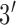 terminus was labeled with methylene blue (MB) (probe sequence:
terminus was labeled with methylene blue (MB) (probe sequence: 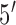 -/thiol/GCT AG [TTT GTT TGT CTG CCA TGA TGT] CTA GC/MB/-
-/thiol/GCT AG [TTT GTT TGT CTG CCA TGA TGT] CTA GC/MB/-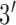 ).
).
B. Sensor Fabrication
The devices were processed using a modular fabrication architecture. The electrodes were fabricated via standard metallization techniques. First, borofloat glass wafers were passivated with a 100-nm-thick layer of 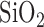 via sputtering. Then, the working electrodes (20 nm Ti/200 nm Au) were patterned via lift-off process after standard photolithography. The reference and counter electrodes (20 nm Ti/200 nm Pt) were patterned using the same process. Next, the wafer was diced and cleaned with a piranha solution, and the molecular beacon probes were allowed to self-assemble on the gold working electrodes via gold-thiol bonds. The polymeric microfluidic chambers were fabricated with contact liquid photolithographic polymerization (CLiPP) [26]. The electrodes were cleaned within the sensor before the immobilization of the thiolated DNA probes, by performing cyclic voltammetry (CV) in 0.1 M H2SO4 with a voltage sweep between 0 and 1.8 V for 5 min at a rate of 500 mV/s as described previously [27]. After cleaning, the 2
via sputtering. Then, the working electrodes (20 nm Ti/200 nm Au) were patterned via lift-off process after standard photolithography. The reference and counter electrodes (20 nm Ti/200 nm Pt) were patterned using the same process. Next, the wafer was diced and cleaned with a piranha solution, and the molecular beacon probes were allowed to self-assemble on the gold working electrodes via gold-thiol bonds. The polymeric microfluidic chambers were fabricated with contact liquid photolithographic polymerization (CLiPP) [26]. The electrodes were cleaned within the sensor before the immobilization of the thiolated DNA probes, by performing cyclic voltammetry (CV) in 0.1 M H2SO4 with a voltage sweep between 0 and 1.8 V for 5 min at a rate of 500 mV/s as described previously [27]. After cleaning, the 2 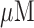 probe DNA was immobilized onto the gold electrodes by incubating for 1 hour, which produces probe densities of
probe DNA was immobilized onto the gold electrodes by incubating for 1 hour, which produces probe densities of 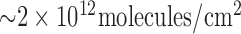 on the working electrode [28], followed by passivation in 2 mM mercapto-1-hexanol (C6) for 2 hours.
on the working electrode [28], followed by passivation in 2 mM mercapto-1-hexanol (C6) for 2 hours.
C. AC Voltammetry
The sensor is connected to an electrochemical analyzer (CH Instruments, Austin, TX). AC voltammetry is performed with the following parameters: an initial potential of 0.4 V and a final potential of  with an amplitude of 0.025 V at a frequency of 50 Hz. All scans were performed in triplicate to verify the reliability of signals.
with an amplitude of 0.025 V at a frequency of 50 Hz. All scans were performed in triplicate to verify the reliability of signals.
D. On-Chip Measurements
To obtain the baseline, a 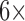 SSC buffer was introduced, and ACV scans were established. Next, the sample containing the target to the sensor was introduced into the chamber and incubated for 30 min, and the ACV scans were obtained. Another ACV scan was performed after the regeneration of the working electrodes with deionized water. The measurements were repeated with the samples containing different concentrations of target and non-target DNA to determine the sensitivity and specificity of the sensor.
SSC buffer was introduced, and ACV scans were established. Next, the sample containing the target to the sensor was introduced into the chamber and incubated for 30 min, and the ACV scans were obtained. Another ACV scan was performed after the regeneration of the working electrodes with deionized water. The measurements were repeated with the samples containing different concentrations of target and non-target DNA to determine the sensitivity and specificity of the sensor.
E. Mobile Interface Development
The app is designed for Android phones or tablets. Once data are uploaded to the phone, the result can be displayed as a curve in a graph or as a safe/not-safe guideline. The app processes the data to determine the safety of the water, which is given by the concentration of pathogens. The data are represented as a curve measured from our electrochemical sensor. The app compares the measurement curve (Fig. 3, pink curve) with the baseline data (Fig. 3, green curve) as follows. For each curve, it determines the minimum line, which connects the two minimum points of the curve (Fig. 3, blue and red lines), and the height line between the maximum of the curve and the minimum line, at the 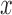 value where it provides the maximum current signal (Fig. 3, yellow and cyan lines). The result is given by the difference between the length of the height line calculated for the baseline curve and the one calculated for the measurement curve (Fig. 4). The difference needs to be below the threshold determined by health guidelines. After processing the data, the app provides the user with the curves (Fig. 3) and the final result, which is given by either the safe or the not-safe screen (Fig. 5). We used the AChartEngine library [29] to graph these results on the Android phone.
value where it provides the maximum current signal (Fig. 3, yellow and cyan lines). The result is given by the difference between the length of the height line calculated for the baseline curve and the one calculated for the measurement curve (Fig. 4). The difference needs to be below the threshold determined by health guidelines. After processing the data, the app provides the user with the curves (Fig. 3) and the final result, which is given by either the safe or the not-safe screen (Fig. 5). We used the AChartEngine library [29] to graph these results on the Android phone.
Fig. 4.
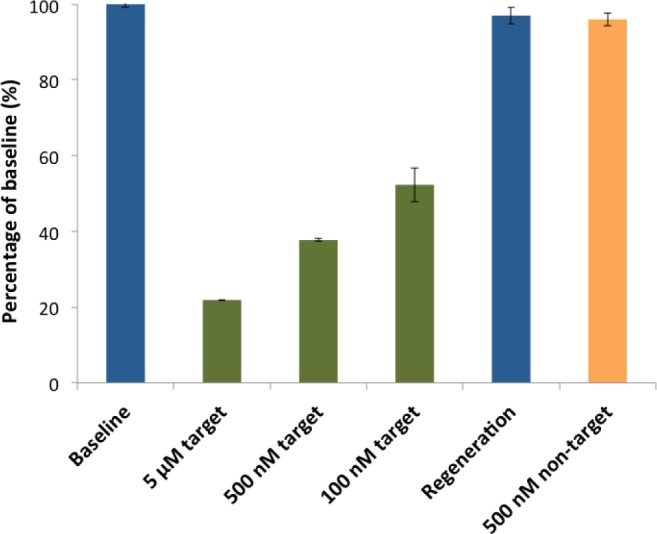
Electrochemical detection of a specific target sequence: the sensor was interrogated via AC voltammetry. First, the baseline was established from the sample without any sequences, and measurements were taken after the addition of complementary target of 5 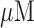 , 500 nM, and 100 nM, showing the percentage changes from baseline increase with increasing target concentrations. Additional measurements were taken for regeneration of the sensor and specificity against other sequences. Each error bar represents the standard deviation of three separate measurements.
, 500 nM, and 100 nM, showing the percentage changes from baseline increase with increasing target concentrations. Additional measurements were taken for regeneration of the sensor and specificity against other sequences. Each error bar represents the standard deviation of three separate measurements.
To implement the mapping feature, we keep a log of previous measurements in a local database, which contains, for each measurement, the coordinates of the place (provided by the GPS feature of the Android phone) where the measurement was obtained. This log is used to generate a map, in which safe spots are marked blue and unsafe spots are marked red (Fig. 6). The map is created using Google Maps API [30].
IV. Results
A. Detection of Targets
Our electrochemical sensing mechanism is particularly suited for POC applications because it enables real-time, continuous monitoring of targets with high sensitivity and specificity. First, the sample without any target sequences is infused into the microfluidic sensor, and after the incubation, the MB redox current is measured to obtain a baseline current. Then, the sample containing target sequences is loaded into the sensor, and after the incubation, the MB redox current is measured and recorded to detect the presence of the targets. One can define a baseline current from the clean water sample without any target sequences, and determine the concentration of the target from the measured current. We selected the detection of the target sequence from the 16S ribosomal RNA of Escherichia coli as a model system because E. coli is one of the major enteric pathogens affecting children and causing deaths mainly through diarrhea in developing countries [31]. 16S rRNA is a 1542 nucleotide-long bacterial specific biosignature, and due to its abundance (5.5% by weight), it is commonly used in molecular assays in bacterial cells [32].
To validate our sensor, we performed AC voltammetry for detection against different concentrations of the target and non-target sequences (Fig. 4). AC voltammetry is very sensitive to changes in electron transfer kinetics, and therefore it is ideally suited to monitor electrochemical sensors in this class. The responses of the sensor are specific and rapid. First, in the absence of the target, the immobilized DNA probe maintained its intact stem-loop structure, holding methylene blue (MB) in proximity to the electrode surface. This methylene blue (MB) reduction current peak was observed from the working electrode, providing the baseline (Fig. 4, first blue bar). In the presence of 5 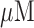 of the target strands, we observed a robust 78% decrease in our current signal (Fig. 4, first green bar). As we decreased the target concentrations down to 500 nM and 100 nM, we observed smaller 62% and 48% decreases, respectively, as expected (Fig. 4, second and third green bars), indicating that our sensor is able to distinguish different concentrations of the target sequence. Furthermore, we confirmed that the reduction in current values is indeed caused by hybridization between the target strands and the sensor probes by regenerating the sensor, which returns the current value to within 97% of the initial baseline (Fig. 4, second blue bar). To further verify the signal, the sensor was challenged with 500 nM non-target DNA, generating little change from the regeneration signal, with a signal drop of 4% (Fig. 4, orange bar).
of the target strands, we observed a robust 78% decrease in our current signal (Fig. 4, first green bar). As we decreased the target concentrations down to 500 nM and 100 nM, we observed smaller 62% and 48% decreases, respectively, as expected (Fig. 4, second and third green bars), indicating that our sensor is able to distinguish different concentrations of the target sequence. Furthermore, we confirmed that the reduction in current values is indeed caused by hybridization between the target strands and the sensor probes by regenerating the sensor, which returns the current value to within 97% of the initial baseline (Fig. 4, second blue bar). To further verify the signal, the sensor was challenged with 500 nM non-target DNA, generating little change from the regeneration signal, with a signal drop of 4% (Fig. 4, orange bar).
B. Mobile Interface
The mobile interface enables the device to be used in remote areas, where testing the water is really important, but the ability to do so is limited by the lack of trained personnel to analyze the data obtained by the sensor locally. We have adapted the sensor to a mobile device to enable the instant interpretation of the data obtained and the mapping of the test results at different places, indicating safe vs. not-safe water sources. As a proof of concept of this capability, we marked different places with sampled data using our mobile interface as described in Experiments above (see Section III-E). The samples were prepared by spiking different concentrations of target sequences. We set the threshold to determine the water to be unsafe to 10% decrease from the baseline of safe water. This threshold has been determined from the infective dose of pathogenic E. coli[33] and the average number of rRNA copies per cell [32]. The end results interpreted from the sample data were used to create a map, in which safe spots are marked blue and unsafe spots are marked red (Fig. 6).
As future work, we plan to create a web-based application to coordinate the water safety program. This application will run on a server and will store measurements obtained with different phones, which will communicate information as measurements are obtained. This application will provide the user with a broader perspective on the safety of water sources. It will also enable the creation of a red-flag mechanism, which should give a warning about places in which the water might be affected by a contaminated remote source. This aspect of our mobile app could provide communities with access to quick information about water in their and in other neighborhoods, which might help them both to determine where to get safe water and to be warned in case a source becomes unsafe. This also enables the study of patterns and consequent prediction of changes in the water quality at specific spots.
V. Discussion on Potential Clinical Impacts
We have developed a microfluidic electrochemical DNA sensor capable of detecting sequence-specific bacterial pathogens rapidly, reliably, and with high selectivity. Additionally, we have demonstrated that a mobile interface can be adapted for interpreting, storing, and mapping the results. This microfluidic platform integrated with the mobile interface has a great potential to contribute to water quality monitoring and other diagnostic technologies available for global healthcare, especially in the developing world and rural areas where the mobile infrastructure is already well-established but communities suffer from lack of trained personnel, laboratory facilities, and clinical expertise on site. For instance, these electrochemical sensors operated on a mobile platform could provide a rapid, affordable, POC method to detect microbial pathogens that can cause infectious diseases in drinking water, which indicate fecal or other contamination in the water source. Feces are an important source of many pathogens, and the greatest microbial risks are associated with ingestion of water that is contaminated with feces from humans or animals [34]. Therefore, microbial monitoring for drinking-water quality is one of the most important aspects of public health. Such a detection platform presented here would support the World Health Organization's Rapid Assessment of Drinking Water Quality Guidelines (RADWQ), which were designed to help in the implementation of rapid assessment of water quality to improve the knowledge and understanding of the level of safety of water and, hence, to support the development of water supply technologies that deliver safe drinking-water [33]. A thorough meta-analysis has reported that water quality interventions, especially point-of-use interventions, indeed reduce diarrheal disease in developing countries, and the impact of the interventions has an even greater effect on children under the age of 5 [35]–[38]. With the electrochemical sensors presented here, water quality could be potentially evaluated in real time, at point-of-use, rather than requiring days as with conventional indicator methods. Although our sensors were fabricated in a standard semiconductor cleanroom, much progress has been made in recent years with the development of low cost alternative fabrication techniques, and on-chip electrochemical sensors can be purchased for as little as US 60 [39]–[41]. A truly POC platform should not have any complex and bulky peripherals and should be integrated with downstream analysis modules, including electrochemical analyzers used in this work. Miniaturized, hand-held electrochemical analyzers have been proposed and are commercially available to be easily integrated with the mobile interface presented here [42], [43].
Another advantage of the proposed system is that its electrochemical detection methods can be exploited for a wide range of analytes other than microbial pathogens. For instance, chemical contents in water such as nitrates, fluoride, and arsenic can cause health risks to young infants or serious chronic health problems when ingested over a long period of time, and thus should be monitored as core parameters for water quality assessment in addition to microbial quality [33]. Multiple sensors selective to a series of these critical analytes can be incorporated in different chambers within a single device, allowing the measurement of multiple analytes simultaneously. Aptamer-based electrochemical sensors are especially suited for such multi-target platforms as these sensors' response is based on a target-induced conformational change, rather than on a less specific signaling mechanism such as absorbed mass or charge, and they are unaffected by nonspecific contaminants [44]. Such a system could simplify current water monitoring practice, which relies on different types of technologies against each contaminant.
Mobile assisted sensor systems would not only alleviate the problems of inadequate access to water quality monitoring in rural and developing regions, but they would also enable remote access to digital record, automated result interpretation, and epidemiological monitoring. The acute lack of qualified personnel is a major and severe constraint in deploying health technology in these regions, and combining the mobile assisted sensor system with automated sample preparation systems could enable its use by minimally trained personnel [45].
VI. Conclusion
We demonstrated an integrated microfluidic system that detects sequence-specific bacterial pathogens rapidly and reliably with high specificity. We detected as low as 100 nM target sequences of bacterial samples through electrochemical sensing. The device is designed as an inexpensive disposable unit, which provides an attractive platform in resource-limited settings.
The microfluidic DNA sensor combined with a mobile interface represents one example of a growing trend toward affordable, point-of-care diagnostic devices. Our platform, capable of transmitting digital data over existing communication systems, enables detection in locations that are impractical to access by trained personnel. Though our system demonstrated the detection of E. coli for water quality monitoring as a proof of principle, it can be implemented for diagnosis of other infectious diseases as well.
Acknowledgement
The authors would like to thank S. Krishnan and R. Basu for their support.
Biographies

Unyoung Kim received the B.S. and M.S. degrees in mechanical engineering from the Korea Advanced Institute of Science and Technology, Daejeon, Korea, and the Ph.D. degree in mechanical engineering from the University of California, Santa Barbara, CA, USA. Currently, she is an Assistant Professor of bioengineering with Santa Clara University, Santa Clara, CA, USA, where she serves as the Director of the Biological Micro/Nanosystems Laboratory. Her research interests involve the investigation of integrated microfluidic systems to address challenging needs in the biomedical applications. She is a member of the SCU Frugal Innovation Laboratory. She leads several projects on developing diagnostic platforms for environment/health monitoring for under-served communities and emerging markets.

Sarah Ghanbari was born in California in 1989. She received the B.S. degree in bioengineering from Santa Clara University, Santa Clara, CA, USA, in 2011. Since 2012, she has been with Life Technologies, focusing on digital PCR. Her research interests include DNA detection and diagnostics.

Anusha Ravikumar received the B.S. degree in bioengineering from Santa Clara University, Santa Clara, CA, USA. Her current research interests include medical device, bioimaging and signal processing, and Bio-MEMS.

John Seubert received the B.S. and M.S. degrees in computer science and engineering from Santa Clara University, Santa Clara, CA, USA, where he was an Active Member and Teaching Assistant with the Frugal Innovation Laboratory. His main area of research is mobile computing, and he has experience in mobile development for emerging markets.

Silvia Figueira received the B.S. and M.S. degrees in computer science from the Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, and the Ph.D. degree in computer science from the University of California, San Diego, CA, USA. Currently, she is an Associate Professor of computer engineering with Santa Clara University, Santa Clara, CA, USA. Her research is in the area of performance evaluation and prediction, recently with a focus on energy efficiency. She is a member of the SCU Frugal Innovation Laboratory. She leads the Mobile Laboratory and advises students working on mobile applications for under-served communities and emerging markets. She has published over 50 papers and has established several collaborations with both companies in Silicon Valley, San Francisco, CA, USA, and social entrepreneurs in the U.S. and abroad.
References
- [1].Global water supply and sanitation assessment 2000 report, World Health Organization, Geneva, Switzerland, 2000, Tech. Rep. NLM classification: WA 675. [Google Scholar]
- [2].Esrey S. A., Potash J. B., Roberts L., and Shiff C., “Effects of improved water supply and sanitation on ascariasis, diarrhoea, dracunculiasis, hookworm infection, schistosomiasis, and trachoma,” Bull. World Health Org., vol. 69, no. 5, pp. 609–621, 1991. [PMC free article] [PubMed] [Google Scholar]
- [3].Barrell R. A., Hunter P. R., and Nichols G., “Microbiological standards for water and their relationship to health risk,” Commun. Disease Public Health, vol. 3, pp. 8–13, Mar. 2000. [PubMed] [Google Scholar]
- [4].Swaminathan B. and Feng P., “Rapid detection of food-borne pathogenic bacteria,” Annu. Rev. Microbiol., vol. 48, pp. 401–426, Oct. 1994. [DOI] [PubMed] [Google Scholar]
- [5].Tan W. H., Wang K. M., and Drake T. J., “Molecular beacons,” Current Opinion Chem. Biol., vol. 8, pp. 547–553, Oct. 2004. [DOI] [PubMed] [Google Scholar]
- [6].Tombelli S., Minunni M., Luzi E., and Mascini M., “New trends in nucleic acids based biosensors—Florence, Italy, October 25—28, 2003,” Anal. Lett., vol. 37, no. 6, pp. 1037–1052, 2004. [Google Scholar]
- [7].Rajendran M. and Ellington A. D., “Selecting nucleic acids for biosensor applications,” Combinat. Chem. High Throughput Screen., vol. 5, pp. 263–270, Jun. 2002. [DOI] [PubMed] [Google Scholar]
- [8].Mabey D., Peeling R. W., Ustianowski A., and Perkins M. D., “Diagnostics for the developing world,” Nature Rev. Microbiol., vol. 2, pp. 231–240, Mar. 2004. [DOI] [PubMed] [Google Scholar]
- [9].Wang J., “Electrochemical biosensors: Towards point-of-care cancer diagnostics,” Biosensors Bioelectron., vol. 21, pp. 1887–1892, Apr. 2006. [DOI] [PubMed] [Google Scholar]
- [10].Gooding J. J., “Electrochemical DNA hyhridization biosensors,” Electroanalysis, vol. 14, pp. 1149–1156, Sep. 2002. [Google Scholar]
- [11].Jayasena S. D., “Aptamers: An emerging class of molecules that rival antibodies in diagnostics,” Clinical Chem., vol. 45, pp. 1628–1650, Sep. 1999. [PubMed] [Google Scholar]
- [12].Mairal T., Ozalp V. C., Sanchez P. L., Mir M., Katakis I., and O'Sullivan C. K., “Aptamers: Molecular tools for analytical applications,” Anal. Bioanal. Chem., vol. 390, pp. 989–1007, Feb. 2008. [DOI] [PubMed] [Google Scholar]
- [13].Bang G. S., Cho S., and Kim B. G., “A novel electrochemical detection method for aptamer biosensors,” Biosensors Bioelectron., vol. 21, pp. 863–870, Dec. 2005. [DOI] [PubMed] [Google Scholar]
- [14].Wu Z. S., Guo M. M., Zhang S. B., Chen C. R., Jiang J. H., Shen G. L., and Yu R. Q., “Reusable electrochemical sensing platform for highly sensitive detection of small molecules based on structure-switching signaling aptamers,” Anal. Chem., vol. 79, pp. 2933–2939, Apr. 2007. [DOI] [PubMed] [Google Scholar]
- [15].Yang S. and Rothman R. E., “PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings,” Lancet Infect. Diseases, vol. 4, pp. 337–348, Jun. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Holland C. A. and Kiechle F. L., “Point-of-care molecular diagnostic systems—Past, present and future,” Current Opinion Microbiol., vol. 8, pp. 504–509, Oct. 2005. [DOI] [PubMed] [Google Scholar]
- [17].Palchetti I. and Mascini M., “Electroanalytical biosensors and their potential for food pathogen and toxin detection,” Anal. Bioanal. Chem., vol. 391, no. 2, pp. 455–471, May 2008. [DOI] [PubMed] [Google Scholar]
- [18].van Belkum A., Renders N. H., Smith S., Overbeek S. E., and Verbrugh H. A., “Comparison of conventional and molecular methods for the detection of bacterial pathogens in sputum samples from cystic fibrosis patients,” FEMS Immunol. Med. Microbiol., vol. 27, pp. 51–57, Jan. 2000. [DOI] [PubMed] [Google Scholar]
- [19].Ferguson B. S., Buchsbaum S. F., Swensen J. S., Hsieh K., Lou X., and Soh H. T., “Integrated microfluidic electrochemical DNA sensor,” Anal. Chem., vol. 81, pp. 6503–6508, Aug. 2009. [DOI] [PubMed] [Google Scholar]
- [20].The world in 2011—ICT facts and figures, Geneva, Switzerland: International Telecommunication Union, Statistics, 2011. [Google Scholar]
- [21].Tachakra S., Wang X. H., Istepanian R. S. H., and Song Y. H., “Mobile e-health: The unwired evolution of telemedicine,” Telemed. J. E-Health, vol. 9, no. 3, pp. 247–257, 2003. [DOI] [PubMed] [Google Scholar]
- [22].Zimic M., Coronel J., Gilman R. H., Luna C. G., Curioso W. H., and Moore D. A. J., “Can the power of mobile phones be used to improve tuberculosis diagnosis in developing countries?,” Trans. R. Soc. Tropical Med. Hygiene, vol. 103, pp. 638–640, Jun. 2009. [DOI] [PubMed] [Google Scholar]
- [23].Kim U., Ravikumar A., Seubert J., and Figueira S., “Detection of bacterial pathogens through microfluidic DNA sensors and mobile interface toward rapid, affordable, and point-of-care water monitoring,” in IEEE point-of-Care healthcare technologies, Bangalore, India, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Android Developers. Available, 2012, [Online]. Available: http://developer.android.com.
- [25].Liao J. C., Mastali M., Li Y., Gau V., Suchard M. A., Babbitt J., Gornbein J., Landaw E. M., McCabe E. R. B., Churchill B. M., and Haake D. A., “Development of an advanced electrochemical DNA biosensor for bacterial pathogen detection,” J. Molecular Diag., vol. 9, pp. 158–168, Apr. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hutchison J. B., Haraldsson K. T., Good B. T., Sebra R. P., Luo N., Anseth K. S., and Bowman C. N., “Robust polymer microfluidic device fabrication via contact liquid photolithographic polymerization (CLiPP),” Lab Chip, vol. 4, no. 6, pp. 658–662, 2004. [DOI] [PubMed] [Google Scholar]
- [27].Pavlovic E., Lai R. Y., Wu T. T., Ferguson B. S., Sun R., Plaxco K. W., and Soh H. T., “Microfluidic device architecture for electrochemical patterning and detection of multiple DNA sequences,” Langmuir, vol. 24, pp. 1102–1107, Feb. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ricci F., Lai R. Y., Heeger A. J., Plaxco K. W., and Sumner J. J., “Effect of molecular crowding on the response of an electrochemical DNA sensor,” Langmuir, vol. 23, pp. 6827–6834, Jun. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].AChartEngine, 2012, [Online]. Available: http://www.achartengine.org/.
- [30].Google Maps API, 2012, [Online]. Available: https://developers.google.com/maps/.
- [31].Ashbolt N. J., “Microbial contamination of drinking water and disease outcomes in developing regions,” Toxicology, vol. 198, pp. 229–238, May 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Neidhardt F. C., Curtiss R., Gross C. A., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W., Riley M., Schaechter M., and Umbarger H. E., Escherichia Coli and Salmonella Typhimurium, Washington, DC, USA: ASM, 1996, vol. 1. [Google Scholar]
- [33].Rapid Assessment of Drinking-Water Quality: A Handbook for Implementation, Geneva, Switzerland: World Health Organization, 2012. [Google Scholar]
- [34].Dufour A. B., Bartram J., Bos R., and Gannon V., Animal Waste, Water Quality and Human Health, London, U.K.: IWA, 2012. [Google Scholar]
- [35].Colwell R. R., Huq A., Islam M. S., Aziz K. M., Yunus M., Khan N. H., Mahmud A., Sack R. B., Nair G. B., Chakraborty J., Sack D. A., and Russek-Cohen E., “Reduction of cholera in Bangladeshi villages by simple filtration,” in Proc. Nat. Acad. Sci. USA, Feb. 2003, vol. 100, pp. 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jensen P. K., Ensink J. H., Jayasinghe G., van der Hoek W., Cairncross S., and Dalsgaard A., “Effect of chlorination of drinking-water on water quality and childhood diarrhoea in a village in Pakistan,” J. Health Populat. Nutrition, vol. 21, pp. 26–31, Mar. 2003. [PubMed] [Google Scholar]
- [37].Brown J. M., Proum S., and Sobsey M. D., “Escherichia coli in household drinking water and diarrheal disease risk: Evidence from Cambodia,” Water Sci. Technol., vol. 58, no. 4, pp. 757–763, 2008. [DOI] [PubMed] [Google Scholar]
- [38].Fewtrell L. and Colford J. M., “Water, sanitation and hygiene in developing countries: Interventions and diarrhoea: A review,” Water Sci. Technol., vol. 52, no. 8, pp. 133–142, 2005. [PubMed] [Google Scholar]
- [39].EcoBioServices, 2012, [Online]. Available: http://www.ebsr.it/ebsr2/?lang=en.
- [40].BVT Technologies Electrochemical Sensors and Tools, 2012, [Online]. Available: http://www.bvt.cz/.
- [41].Bergamini M. F., Santos A. L., Stradiotto N. R., and Zanoni M. V., “A disposable electrochemical sensor for the rapid determination of levodopa,” J. Pharm. Biomed. Anal., vol. 39, pp. 54–59, Sep. 2005. [DOI] [PubMed] [Google Scholar]
- [42].Chang S. Y., Jay T., Munoz J., Kim I., and Lee K. H., “Wireless fast-scan cyclic voltammetry measurement of histamine using WINCS—A proof-of-principle study,” Analyst, vol. 137, no. 9, pp. 2158–2165, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rowe A. A., Bonham A. J., White R. J., Zimmer M. P., Yadgar R. J., Hobza T. M., Honea J. W., Ben-Yaacov I., and Plaxco K. W., “CheapStat: An open-source, ‘do-it-yourself’ potentiostat for analytical and educational applications,” PLos ONE, vol. 6, no. 9, pp. e23783–e23783, 2011, doi: 10.1371/journal.pone.0023783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Baker B. R., Lai R. Y., Wood M. S., Doctor E. H., Heeger A. J., and Plaxco K. W., “An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids,” J. Amer. Chem. Soc., vol. 128, pp. 3138–3139, Mar. 2006. [DOI] [PubMed] [Google Scholar]
- [45].Yager P., Domingo G. J., and Gerdes J., “Point-of-care diagnostics for global health,” Annu. Rev. Biomed. Eng., vol. 10, pp. 107–144, Mar. 2008. [DOI] [PubMed] [Google Scholar]


