Abstract
We demonstrate that dynamic nuclear polarization (DNP) of membrane proteins in lipid bilayers may be achieved using a novel polarizing agent: pairs of spin labels covalently bound to a protein of interest interacting at an intermolecular interaction surface. For gramicidin A, nitroxide tags attached to the N-terminal intermolecular interface region become proximal only when bimolecular channels forms in the membrane. We obtained signal enhancements of 6-fold for the dimeric protein. The enhancement affect was comparable to that of a doubly tagged sample of gramicidin C, with intramolecular spin pairs. This approach could be a powerful and selective means for signal enhancement in membrane proteins, and for recognizing intermolecular interfaces.
Keywords: Dynamic Nuclear Polarization, DNP, membrane proteins, solid-state NMR, gramicidin, DEER spectroscopy
Introduction
Since membrane proteins are notoriously difficult to crystallize and sometimes perturbed by detergents, the recent progress of studying membrane proteins in bilayer environments (Opella and Marassi 2004; McDermott 2009; Warschawski et al. 2011; Hong et al. 2012) by solid-state nuclear magnetic resonance (SSNMR), has been very encouraging. There has been important progress regarding sensitivity, the principal challenge for SSNMR of membrane proteins, in that recent Dynamic Nuclear Polarization (DNP) studies indicate improvements of one or two orders of magnitude for favorable cases (Carver and Slichter 1953; Becerra et al. 1995; Hall et al. 1997; Bajaj et al. 2007; Barnes et al. 2008) including some membrane proteins (Rosay et al. 2003; Bajaj et al. 2009; Linden et al. 2011). In these studies, a protein in a lipid bilayer is mixed with a solvent containing an exogenous biradical, for example TOTAPOL (Song et al. 2006) or bTbK (Matsuki et al. 2009). DNP has been used to illustrate mechanistic aspects of membrane proteins such as bacteriorhodopsin (Bajaj et al. 2009), and to enhance spectra of oriented membrane samples (Salnikov et al. 2012).
Covalently bound radicals have been used previously as structural probes for SSNMR studies of proteins (Nadaud et al. 2007; Jaroniec 2012), and there has been a previous example of covalently bound biradicals in the DNP literature (Vitzthum et al. 2011). Here we demonstrate that doubly spin label tagged proteins and spin pairs that form at intermolecular interfaces can also be used as enhancement agents in cryogenic DNP-SSNMR, and that spin pairs that come together at intermolecular contact surfaces can be powerful sources of polarization. We illustrate this technique using spin-labled samples of gramicidin reconstituted into proteoliposomes. Specifically N-terminal labeled gramicidin A (GALN) and double-labeled gramicidin C (GCDL) were reconstituted into phospholipid bilayers, DNP enhancements were measured and evaluated based upon molecular geometry, and the overall impact of the spin label to the sample T1 was evaluated. It is hoped this work will create a new route toward membrane protein DNP enhancements and act as a means to further understand the role of radical-sample distances, bleaching, and PREs in the DNP of membrane proteins. In addition, because the protein is directly spin labeled, there is no need for an extensive glassy matrix to introduce and stabilize the DNP polarizing agents, allowing larger amounts of sample to be packed into the NMR rotor.
Materials and Methods
GALN
N-terminally labeled Gramicidin A (GALN) (Dzikovski et al. 2011) was prepared as described previously, and analysis was performed using the previously reported structure.
Synthesis of double labeled gramicidin C (GCDL)
Naturally occurring gramicidin (gramicidin D) is a mixture of three isoforms, gramicidins A, B and C in a ratio of approximately 80% A, 5% B and 15% C. Gramicidins B and C have either phenylalanine or tyrosine, respectively, replacing tryptophan at position 11 of gramicidin A. Separation of gramicidin C (GC) from the mixture was carried out in a procedure similar to an earlier report (Pepinsky and Feigenson 1978). GC was then introduced into the DCC-promoted coupling reaction with 2,2,5,5 – tetramethyl – 3 – pyrrolin –1 - oxyl – 3 – carboxylic acid, yielding a double labeled product (GCDL), with spin labels attached to the hydroxyl groups at the C-terminus and 11Tyr. To 94 mg dry GC 16 mg of 4-Dimethylaminopryidine (DMAP) was added. 116 mg of 2,2,5,5–tetramethyl –3 –pyrrolin –1 - oxyl –3 –carboxylic acid was dissolved in 0.83 ml DMF and mixed with a 0.33 ml of 1M N,N'-Dicyclohexylcarbodiimide (DCC) in CH2Cl2. The solution was then added to the dry GC/DMAP mixture and the resulting solution was allowed to stir for 96 hrs. After DMF was removed in vacuum overnight, the reaction mixture was initially washed by suspending/spinning down with several portions of water until the triplet EPR signal of the supernatant disappeared, then dried and applied to a column. The column was packed with 230–400 mesh silica gel 60. Chromatography was carried out under nitrogen pressure in 95:5 CHCl3/MeOH. The fractions were collected; each fraction was checked by EPR and TLC. 81 mg of dry, slightly yellowish powder was collected. Rf in CHCl3/MetOH 9:1 is 0.47, ESI MS: M/z =2192.5
Vesicle Sample Preparation
In each case protein was dissolved in a mixture of 5:1 CHCl3:MeOH and lipids were added in chloroform. The solvent was initially removed by nitrogen gas flow, then the samples were placed under vacuum for 3–12 hrs to remove residual solvent. Protonated vesicles were obtained using a mixture of DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) and DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine) at a 9:1 ratio and by adding a buffer composed of 50 mM KCl, 50 mM Tris (pH 7.5) followed by 2–3 minutes of vortexing and then a period of 10–15 minutes in a sonic bath after the sample began to turn cloudy. Each sample was then stored in a refrigerator for two hours, after which time vesicles were collected using centrifugation. Samples with different ratios of lipid:protein were prepared as summarized in Table 1. Samples with deuterated lipids and solvent were made by a similar protocol, but using 50 mM KCl in D2O and 2H-lipids. The samples were prepared using a protein:lipid molar ratio of 1:6, with a lipid mixture composed of a zwitterion:anionic lipid ratio of 9:1 ( 9 DSPE D70 (1,2-distearoyl(d70)-sn-glycero-3-phosphoethanolamine) lipids: 1 DS PS D70 (1,2-distearoyl(d70)-sn-glycero-3- phospho-L-serine lipid). Samples were packed into 3.2 mm sapphire SSNMR rotors (Bruker Biospin, Billerica, MA).
Table 1.
DNP signal enhancements
| Number | Protein | Protein:Lipid | ε (±0.5) |
|---|---|---|---|
| 1 | GCDLb | 1:6, 2H-lipids | 3.5 |
| 2 | GCDLb | 1:3.5 | 3.5 |
| 3 | GCDLb | 1:6 | 3 |
| 4 | GCDLb | 1:17 | 3.5 |
| 5 | GCDLb | 1:17/1:6 | 3 |
| 6 | GALNc | 1:6, 2H-lipids | 5 |
| 7 | GALNc | 1:3.5 | 6 |
| 8 | GALNc | 1:6 | 5.5 |
| 9 | GALNc | 1:17 | 5 |
| 10 | GALNc | 1:17/1:6 | 5 |
ε measured at peak with highest signal intensity, corresponding to peak 4 in Figure 3. ε values appeared to be relatively uniform for other peaks.
Average interspin distance is 9 Å.
Interspin distance is 7.5 Å.
DNP Spectra
All DNP spectra were acquired on site at Bruker Biospin (Billerica, MA) using a 400 MHz Avance spectrometer equipped with a low-temperature HCN triple channel DNP probe, a 263 GHz gyrotron, and a μ-waveguide. Set temperatures for all experiments were maintained at 100–105 K, and MAS rates were maintained at 9 kHz (estimated true sample temperature was 115–120 K). All 13C one-dimensional spectra were acquired with 1 ms 1H to 13C cross polarization and 100 kHz of SPINAL-64 decoupling. All 13C cross polarization one-dimensional spectra spectra of fully 1H samples were acquired with 32 transients and a 6.5 s pulse delay. One-dimensional 13C spectra of GALN reconstituted into vesicles composed of 2H-lipids were acquired with 1024 transients and a pulse delay of 10 s. One-dimensional spectra of GCDL with 2H-lipids were acquired with 3840 transients and a 3 s pulse delay. The discrepancy is signal averaging time arose from sample quantity limitations. 1H-lipid samples were prepared with ~4 mg of protein, while 2H-lipid samples contained ~1 mg of protein per sample. All reported 1H T1 values were measured using inversion recovery
Results and Discussion
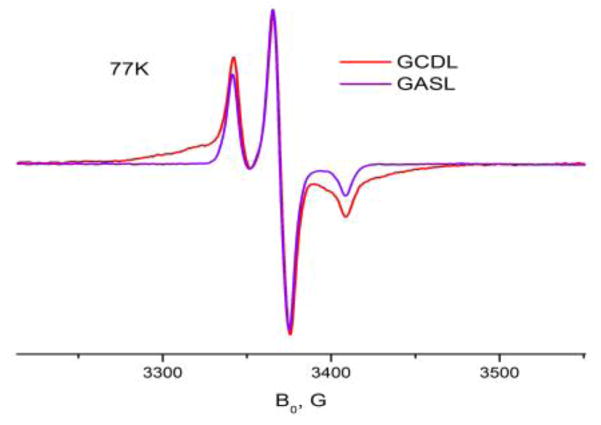
A new spin-labeled gramicidin was introduced for this study, GCDL. GCDL was synthesized from gramicidin C (GC) using DCC-promoted Steglich esterification similar to C-terminal labeling of gramicidin A (GA) (Dzikovski et al. 2004) as described above. The GC molecule has two OH groups that participate in this reaction, one is at the C-terminal end (as in GA), and another one is the phenyl hydroxide of Y11. Figure 1 shows a 77 K ESR spectrum of double-labeled GCDL in 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) in comparison with single-labeled gramicidin A (GASL), which is single-labeled at the C-terminus. A detailed analysis of ESR spectra for GCDL indicates that in the channel form these two spin labels are located close to each other in the polar area of the bilayer, near the membrane interface with an average distance between them ~ 9 Å, but with a relatively broad distribution of distances and relative orientations. One can see a broad singlet-like feature corresponding to interaction between spins separated by a short distance. GCDL has a free N-terminus. It allows for formation of head-to-head dimers (HHDs) in lipid bilayers of matching thickness (Dzikovski et al. 2004), such as 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and DLPC. In these lipids there is little difference in spectra between pure GCDL and a mixture 1:19 GCDL/GA. It indicates that spins separated by the short interspin distance are located on the same GC monomer. The short distance and the presence of a broad signal is consistent with the results of molecular modeling for HHD (Figure 2) which give ~ 9Å. Analysis of the CW spectrum, similar to (Rubenstein and Shin, 1995) also gives an interspin distance estimate of ~ 9 Å, with presence of a minor fraction (< 30%) of spins separated by a larger distance. Pulse Dipolar ESR (DQC-ESR) shows in DLPC and DMPC a short interspin distance (<10Å), along with distances of ~ 22–30 Å. These longer distances correspond to separations between spin labels (C-terminal label to 11Tyr label and C-terminal label to C-terminal label) located on different monomers of the HHD dimer. Because the distance between spin labels on neighboring molecules is greater than 25 Å and so these interactions are likely not to be relevant for the DNP polarization.
Figure 1.
ESR spectra of 0.6% GCDL (red line) and GASL (blue line) in DLPC at 77K. The broad feature of the GCDL spectrum is due to two spin labels attached to the same GC molecule in close proximity to each other. Analysis according to (Rabenstein and Shin, 1995) gives an average interspin distance of ~ 9Å with a relatively broad distance distribution.
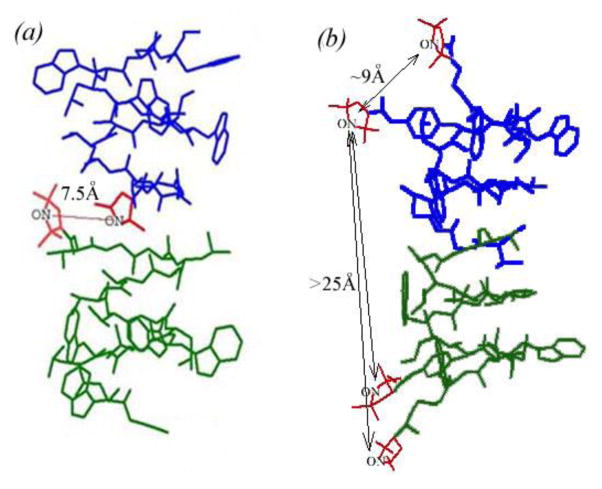
FIGURE 2.
Spin labeled gramicidin compounds used in this study; nitroxide moieties are shown in red. (a) N-terminally labeled gramicidin A (GALN). Two spin labels, each from a different monomer, form a membrane embedded spin pair in the dimer, with an experimentally characterized interspin distance of 7.5 Å. (b) Doubly labeled gramicidin C (GCDL) has spin pairs on each gramicidin monomer, located close to the membrane surface and separated by an average interspin distance of ~ 9 Å. The intermolecular interspin distance between spin labels on neighboring GCDL molecules is greater than 25 Å. The Right-handed, b6.3-helical structure (PDB: 1MAG) is depicted for both compounds, as confirmed by ESR.
The current and previously reported DEER and DQC ESR studies were used to constrain the fold of GALN and GCDL as described previously (Dzikovski et al. 2004; Dzikovski et al. 2011) and the proposed structures are presented in Figure 2. Upon gramicidin channel formation (dimerization), spin labels on two different GALN molecules form a rigid biradical pair with an interspin distance of 7.5 Å located at the middle of the membrane hydrophobic core. This interspin distance is shorter than typical interspin distances in biradicals commonly used for DNP (~13 Å) (Hu et al. 2004; Song et al. 2006; Hu et al. 2008). The efficiency of the cross effect using radicals at shorter than a 11 Å distance has not been evaluated in detail, but has been postulated to be efficient (Hu et al. 2004). Analysis of the molecular coordinates in Hyperchem suggests relative orientations of the two nitroxide tags forming the GALN pair show a relative orientation of ~26° between the X-tensors, ~21° between the Y tensors, and 27° between the Z tensors. It is known that the optimal DNP effect is observed when the tensors are closer to perpendicular relative to parallel (Hu et al. 2004). Thus, the combination of tensor orientation and distance could intensify the observed DNP enhancement once optimized.
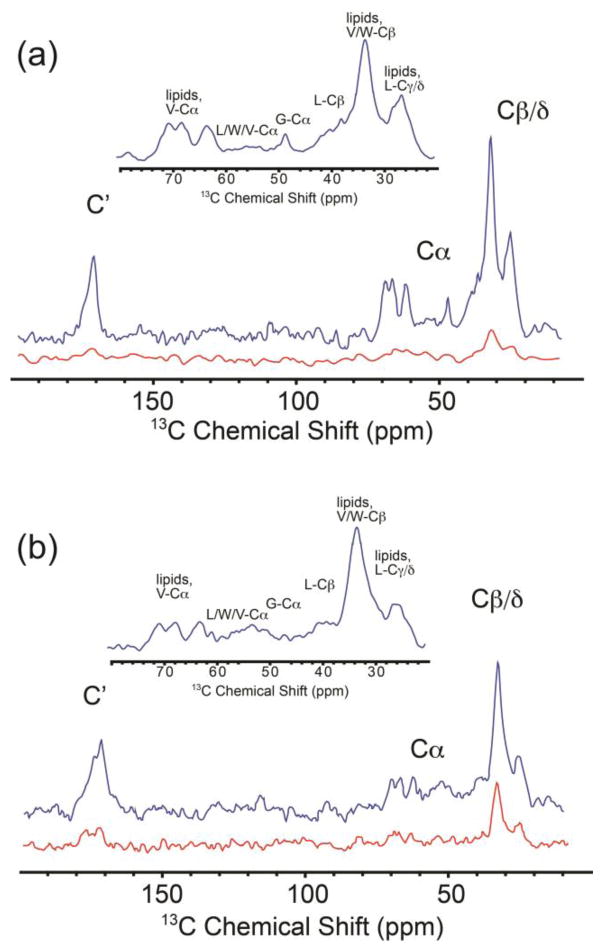
Because all materials used in this study contain 15N and 13C resonances in natural abundance, in order to evaluate the relative enhancements of protein signals the lipid signals must be minimized. We found lipid signals can be dramatically suppressed in 1H- 13C cross polarized 1D spectra of samples of GALN and GCDL by creating proteoliposmes composed of lipids with 2H enriched tails. 13C (Figure 3) and 15N (Figure S1) spectra of these samples allowed us to better resolve protein signals for this natural abundance membrane protein and these spectra were utilized to determine the overall signal enhancement of the protein peaks (Figure 3). The samples were prepared using a protein:lipid molar ratio of 1:6, with a lipid mixture composed of a zwitterion:anionic lipid ratio of 9:1 ( 9 DSPE D70 (1,2-distearoyl(d70)-sn-glycero-3-phosphoethanolamine) lipids: 1 DS PS D70 (1,2-distearoyl(d70)-sn-glycero-3- phospho-L-serine lipid), a composition that provides ample lipid to ensure complete channel formation. It is important to note here only the lipids and buffer were deuterated, but the protein sample remained protonated. Thus, we were unable to determine the impact protein deutration might have upon the signal enhancement from the current data. Protein peaks were clearly visible in the aliphatic and carbonyl regions, especially between 35 and 60 ppm where W, V, L, and G Cα resonances and L Cβ resonances were observed (Figure 3 a,b inserts) (Quist 1998); intensity from V and W Cβ and L Cδ resonances are also evident in the methylene region (along with lipid signals). The effect of the paramagnetic tag on the spectral quality and lineshape was subtle or absent in most cases; however signals from the W side chain (which are expected to be approximately 12 Å from the radicals) are broadened and shifted compared to unlabeled gramicidin D (Figure S2). 15N amide signals were observed using low temperature 15N DNP SSNMR (Figure S1). The DNP signal enhancements, ε (ε=signal intensity with microwave irradiation/signal intensity without microwaves), were uniform across all observed peaks within the observed signal-to-noise ratio. For GCDL ε ≥3.5, and for GALN ε ≥6. For a mono-radical under these conditions, no detectible enhancement is seen; to our knowledge this is the first example of the use of a bimolecular biradical at a molecular recognition interface for polarization studies and it suggests a broad range of applications where polarization would be diagnostic of radical pair formation at an interface. The enhancement of six-fold results in a 36-fold improvement in data collection times, and remarkably allowed for detection of membrane proteins using natural abundance 13C. These results are promising, considering the opportunity to further optimize the radicals’ relative geometry (Song et al. 2006; Matsuki et al. 2009).
FIGURE 3.
1H-13C Cross Polarization 1D 13C NMR spectra of GALN and GCDL. Blue traces are DNP-enhanced spectra upon microwave irradiation while red traces are the same experiment without microwave irradiation. Spectra for 2H lipid vesicle bound GALN (a) and GCDL (b) reveal protein peaks, despite the fact that the protein has natural abundance 13C. Aliphatic regions are highlighted in the insert. All spectra were acquired at 400 MHz 1H Larmor frequency with a sample temperature of 105 K (uncorrected for microwave heating and MAS. True sample temperature is estimated to be 115 K.). Spectra were processed with 30 Hz of Gaussian line broadening. Observed signal enhancements for GALN (a) was 6, and 3.5 in the case of GCDL. To obtain comparable S/N in spectra acquired without DNP enhancement would require ~25 hours of signal averaging.
We also considered the possibility that a radical tagged protein could be used to serve as a magnetization source to enhance the NMR of nearby molecules (lipid or protein). While the standard DNP recipe has been useful for a number of different kinds of samples, it is possible that embedding radicals directly into the membrane might be advantageous in some sample preparations, and could, provide greater flexibility in the sample preparation of membrane proteins for DNP, in part because it would not require the introduction of a matrix holding the polarizing agent allowing for a higher final sample concentration. For this purpose, GCDL and GALN were reconstituted into vesicles with unsaturated and protonated (natural isotopic abundance) lipids, DOPE and DOPS (9:1 ratio of DOPE:DOPS), and protein:lipid ratios ranging from 1:3.5 to 1:17, and the enhancement of lipid signals was recorded. In addition, another sample was prepared with a 1:3.5 protein:lipid ratio, but with only 17% of the protein spin-labeled (5:1 ratio of gramicidin D:GALN) and enhancement of the protein NMR spectrum was recorded for this co-reconstituted system.
Lipid signals dominated the spectra when the protein was reconstituted into fully 1H DOPE/DOPS bilayers. DNP enhancements of the lipid peaks exhibited by these samples were comparable to the protein enhancements observed in 2H lipids, i.e. ranged from 3- to 6-fold (Table 1). In the case of the GCDL samples (Samples 1–5, Table 1), ε values were ~ 3.5 and relatively unaffected by protein concentration. In the GALN samples (Samples 6–10, Table 1), where, as noted above, the intermolecular intra-radical distance determined by ESR is 7.5 Å (Dzikovski et al. 2011), ε was > 5 fold (Figure 4). There is no notable effect on the DNP enhancement or on the NMR lineshape as the effective radical concentration is increased (e.g. the samples prepared with a 1:3.5 protein:lipid ratio vs. with a 1:17 lipid:protein molar ratio). Based upon the gramicidin area and bilayer thickness, the effective concentration for the dilute samples in bilayers is estimated to be 27 mM for GALN (with 1:17 protein:lipid molar ratio, estimated treating the lipids in the bilayer as solvent, ignoring the presence of bulk water, since the radicals are localized to the bilayer). These concentrations are comparable to saturated solutions of TOTAPOL and to concentrations used in prior studies with exogenously added TOTAPOL. Presumably the proximity of the NMR observed site to the radical is consistent throughout the series, and is the important parameter, whereas the average concentrations appear not to be relevant.
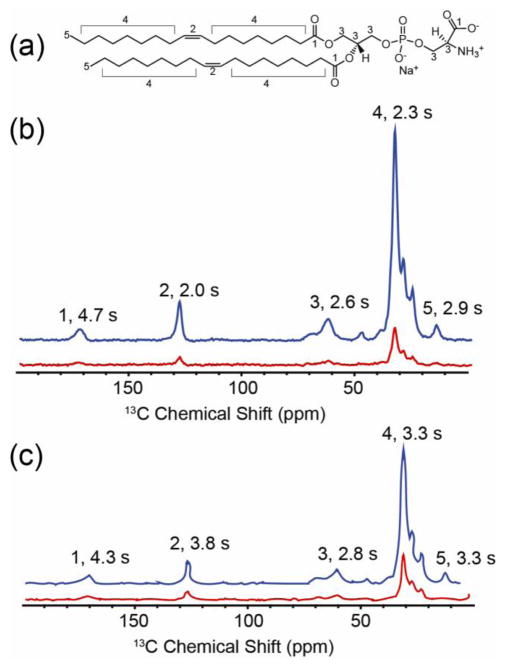
FIGURE 4.
1H-13C Cross Polarization 1D 13C NMR spectra of GALN and GCDL in protonated lipids. (a) 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine (DOPE), the dominant lipid in the vesicles. Numerical labels correspond to resonances identified in spectra of (b) GALN and (c) GCDL prepared with 1:3.5 protein:lipid ratio in vesicles of 9:1 DOPE:DOPS. Blue traces are DNP-enhanced spectra upon microwave irradiation while red traces are the same experiment without microwave irradiation. Spectra appear similar because they are dominated by lipid signals. In the case of GALN (b) the patterns of T1 obtained from a 3D inversion recovery array, peaks 2, 4, and 5 exhibit shorter T1 relaxation times compared to peaks 1 and 3, suggesting the nitroxide spin labels are situated in the middle of the bilayer, as predicted by the IMAG crystal structure. In the case of GCDL, the opposite trend is observed. All spectra were processed with 30 Hz of Gaussian line broadening. t
In the next step in our investigation, we measured 1H T1 relaxation for various lipid sites in the fully protonated samples. Relaxation effects are important in determining the overall enhancement and the quality of the spectra. Interestingly, the two samples GCDL and GALN exhibited very different relaxation profiles. We recorded the lipid 1H T1s for vesicles containing GALN and GCDL; the proton relaxation is of interest since the DNP effect was based on the proton polarization (and subsequently transferred to the heteronuclei). Lipid NMR spectra of GCDL vesicles showed 1H relaxation times of 2.8–3.8 seconds for sites within the membrane (peaks 2, 4, 5, Figure 4). This may be consistent with the formation of a channel structure where each spin label is situated near the membrane head groups, and distant from the membrane interior, consistent with the results from EPR. The 1H T1 of the lipid head group was 4.3 s. In the case of GALN, however, T1 relaxation rates for these internal resonances were considerably shorter. The peaks originating from the transmembrane lipid double bond exhibit a T1 half of that observed in the GCDL samples. This is consistent with the formation of the head-to head dimer (PDB: 1MAG) (Ketchem et al. 1996) with the nitroxide pair located in the middle of the bilayer (Dzikovski et al. 2011). However, this result contrasts from what might be expected near 100 K, where molecular motions are significantly attenuated and the corresponding 1H-1H spin diffusion should be fast enough that measured 1H T1 values appear to reach uniformity. It appears with the present data that the fast spin diffusion T1 limit is reached for the water accessible sites (The T1 for the lipid head group being nearly identical between the two samples), but it is significantly different in the case of the lipids. This likely results from a weaker network of 1H-1H couplings within the bilayer, arising from an arrangement where, unlike water, there is no hydrogen bonding network and the relative distances between many of the 1H resonances is slightly longer. This is an interesting result and suggests that embedded radicals in lipid environments might prove useful for two reasons. First, once the molecular geometry is better optimized compared to GALN, where the nitroxides are slightly more distal and the relative tensor orientations are more perpendicular, the polarization enhancement might be greater as less polarization will be lost to spin diffusion. Secondly, the measured T1 gradient could also be utilized to detect both the formation of intermolecular interfaces and the depth of insertion of transmembrane proteins. Both of these observations could enhance the study of membrane proteins using DNP.
Conclusions
In summary, we illustrate that small proteins such as gramicidin, or other paramagnetically tagged molecules could be developed to provide efficient polarization enhancements of other molecules embedded into the bilayer. These results highlight a promising future for performing DNP upon membrane samples with covalently bound spin labels. Covalent attachment of the radical to the molecules of interest avoids issues of solubility of the radicals, and could therefore have many advantages over exogenously added radical polarizing agents. We further illustrate an intriguing possibility that radical pairs that form at intermolecular interaction surfaces can serve as polarization sources. Such approaches could play a significant role in future efforts to apply DNP to membrane proteins.
Supplementary Material
References
- Bajaj VS, Hornstein MK, Kreischer KE, Sirigiri JR, Woskov PP, Mak-Jurkauskas ML, Herzfeld J, Temkin RJ, Griffin RG. 250 GHz CW gyrotron oscillator for dynamic nuclear polarization in biological solid state NMR. J Magn Reson. 2007;189:251–279. doi: 10.1016/j.jmr.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. P Natl Acad Sci USA. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AB, De Paepe G, van der Wel PCA, Hu KN, Joo CG, Bajaj VS, Mak-Jurkauskas ML, Sirigiri JR, Herzfeld J, Temkin RJ, Griffin RG. High-field dynamic nuclear polarization for solid and solution biological NMR. Appl Magn Reson. 2008;34:237–263. doi: 10.1007/s00723-008-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra LR, Gerfen GJ, Bellew BF, Bryant JA, Hall DA, Inati SJ, Weber RT, Un S, Prisner TF, Mcdermott AE, Fishbein KW, Kreischer KE, Temkin RJ, Singel DJ, Griffin RG. A Spectrometer for Dynamic Nuclear-Polarization and Electron-Paramagnetic-Resonance at High-Frequencies. J Magn Reson Ser A. 1995;117:28–40. [Google Scholar]
- Carver TR, Slichter CP. Polarization of Nuclear Spins in Metals. Phys Rev. 1953;92:212–213. [Google Scholar]
- Dzikovski BG, Borbat PP, Freed JH. Spin-labeled gramicidin A: Channel formation and dissociation. Biophys J. 2004;87:3504–3517. doi: 10.1529/biophysj.104.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikovski BG, Borbat PP, Freed JH. Channel and Nonchannel Forms of Spin-Labeled Gramicidin in Membranes and Their Equilibria. J Phys Chem B. 2011;115:176–185. doi: 10.1021/jp108105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Maus DC, Gerfen GJ, Inati SJ, Becerra LR, Dahlquist FW, Griffin RG. Polarization-enhanced NMR spectroscopy of biomolecules in frozen solution. Science. 1997;276:930–932. doi: 10.1126/science.276.5314.930. [DOI] [PubMed] [Google Scholar]
- Hong M, Zhang Y, Hu FH. Membrane Protein Structure and Dynamics from NMR Spectroscopy. An Rev Phys Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KN, Song C, Yu HH, Swager TM, Griffin RG. High-frequency dynamic nuclear polarization using biradicals: A multifrequency EPR lineshape analysis. J Chem Phys. 2008;128 doi: 10.1063/1.2816783. [DOI] [PubMed] [Google Scholar]
- Hu KN, Yu HH, Swager TM, Griffin RG. Dynamic nuclear polarization with biradicals. J Am Chem Soc. 2004;126:10844–10845. doi: 10.1021/ja039749a. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP. Solid-state nuclear magnetic resonance structural studies of proteins using paramagnetic probes. Solid State Nucl Mag. 2012;43–44:1–13. doi: 10.1016/j.ssnmr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Ketchem RR, Lee KC, Huo S, Cross TA. Macromolecular structural elucidation with solid-state NMR-derived oreintational constraints. J Biomol NMR. 1996;8:1. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- Linden AH, Lange S, Franks WT, Akbey U, Specker E, van Rossum BJ, Oschkinat H. Neurotoxin II Bound to Acetylcholine Receptors in Native Membranes Studied by Dynamic Nuclear Polarization NMR. J Am Chem Soc. 2011;133:19266–19269. doi: 10.1021/ja206999c. [DOI] [PubMed] [Google Scholar]
- Matsuki Y, Maly T, Ouari O, Karoui H, Le Moigne F, Rizzato E, Lyubenova S, Herzfeld J, Prisner T, Tordo P, Griffin RG. Dynamic Nuclear Polarization with a Rigid Biradical. Angew Chem Int Ed Engl. 2009;48:4996–5000. doi: 10.1002/anie.200805940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott A. Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- Nadaud PS, Helmus JJ, Hofer N, Jaroniec CP. Long-range structural restraints in spin-labeled proteins probed by solid-state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2007;129:7502. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepinsky RB, Feigenson GW. Purification of Gramicidin C. Anal Biochem. 1978;86(2):512–8. doi: 10.1016/0003-2697(78)90776-5. [DOI] [PubMed] [Google Scholar]
- Quist PO. C-13 solid-state NMR of gramicidin A in a lipid membrane. Biophys J. 1998;75:2478–2488. doi: 10.1016/S0006-3495(98)77692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosay M, Lansing JC, Haddad KC, Bachovchin WW, Herzfeld J, Temkin RJ, Griffin RG. High-frequency dynamic nuclear polarization in MAS spectra of membrane and soluble proteins. J Am Chem Soc. 2003;125:13626–13627. doi: 10.1021/ja036898k. [DOI] [PubMed] [Google Scholar]
- Rabenstein MD, Shin YK. Determination of a distance between two spin labels attached to a micromolecule. Proc Natl Acad Sci USA. 1995;92:8239– 8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov ES, Ouari O, Koers E, Sarrouj H, Franks T, Rosay M, Pawsey S, Reiter C, Bandara P, Oschkinat H, Tordo P, Engelke F, Bechinger B. Developing DNP/Solid-State NMR Spectroscopy of Oriented Membranes. Appl Magn Reson. 2012;43:91–106. [Google Scholar]
- Song CS, Hu KN, Joo CG, Swager TM, Griffin RG. TOTAPOL: A biradical polarizing agent for dynamic nuclear polarization experiments in aqueous media. J Am Chem Soc. 2006;128:11385–11390. doi: 10.1021/ja061284b. [DOI] [PubMed] [Google Scholar]
- Vitzthum V, Borcard F, Jannin S, Morin M, Mieville P, Caporini MA, Sienkiewicz A, Gerber-Lemaire S, Bodenhausen G. Fractional Spin-Labeling of Polymers for Enhancing NMR Sensitivity by Solvent-Free Dynamic Nuclear Polarization. Chemphyschem. 2011;12:2929–2932. doi: 10.1002/cphc.201100630. [DOI] [PubMed] [Google Scholar]
- Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand E, Marcotte I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Bba-Biomembranes. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.