Abstract
Background:
Congenital cataract (CC) is the leading cause of visual impairment or blindness in children worldwide. Because of highly genetic and clinical heterogeneity, a molecular diagnosis of the lens disease remains a challenge.
Methods:
In this study, we tested a three-generation Chinese family with autosomal dominant CCs by targeted sequencing of 45 CC genes on next generation sequencing and evaluated the pathogenicity of the detected mutation by protein structure, pedigree validation, and molecular dynamics (MD) simulation.
Results:
A novel 15 bp deletion on GJA8 (c.426_440delGCTGGAGGGGACCCT or p. 143_147delLEGTL) was detected in the family. The deletion, concerned with an in-frame deletion of 5 amino acid residues in a highly evolutionarily conserved region within the cytoplasmic loop domain of the gap junction channel protein connexin 50 (Cx50), was in full cosegregation with the cataract phenotypes in the family but not found in 1100 control exomes. MD simulation revealed that the introduction of the deletion destabilized the Cx50 gap junction channel, indicating the deletion as a dominant-negative mutation.
Conclusions:
The above results support the pathogenic role of the 15 bp deletion on GJA8 in the Chinese family and demonstrate targeted genes sequencing as a resolution to molecular diagnosis of CCs.
Keywords: Congenital Cataract, GJA8, Next Generation Sequencing, Novel In-frame Deletion, Targeted Genes Capture
INTRODUCTION
Congenital cataract (CC) refers to cataract observable at early year of life[1] and is the leading cause of visual losses in children worldwide.[2,3] The incidence of CC varies from 0.01% to 0.06% in developed countries[4] to 0.05–0.15% in less developed areas of the world.[2,3,5,6] CC can occur in isolation forms (nonsyndromic CC) or as part of a syndrome of ocular or systemic anomalies (syndromic CC).[7] Major causing factors of CC include metabolic disorders, intrauterine infectious, and genetic defects (chromosomal abnormalities or gene defects). Moreover, approximately 8.3–25.0% of the CC cases are thought to have a genetic basis of etiology, a majority of which are Mendelian diseases caused by monogenic mutations.[8,9]
CC is a group of clinically and genetically heterogeneous diseases. Clinically, in addition to other abnormalities in syndromic CC, cataract itself includes a variety of morphologies, such as sutural, pulverulent, whole lens, nuclear, lamellar (also referred to as perinuclear), cortical, polar, cerulean, coralliform, and others.[9,10] Genetically, over 110 genes have been found associated with CC,[11] including more than 20 genes involved in nonsyndromic CC (hitherto the initial of the study).[9] Inherited patterns of nonsyndromic CC includes autosomal dominant (AD), autosomal recessive, and X-linked dominant patterns, with AD as the most common forms.[1,10] Of the more than 29 known nonsyndromic CC genes, at least 22 are involved in autosomal dominant congenital cataract (ADCC), including BEST1, BFSP2, CHMP4B, CRYAA, CRYAB, CRYBA1, CRYBA4,CRYBB1, CRYBB2, CRYBB3, CRYGC, CRYGD, CRYGS, EPHA2, GJA3, GJA8, HSF4, MAF, MIP, PITX3, SLC16A12, and VIM. Most of the genes mainly involve in specific processes in lens development, intercellular communication in the lens, or the organization of lens fibers.[9] Of note, the relation between cataract phenotypes and mutant genes is even more complex for nonsyndromic CC, i.e., mutations in different genes can cause similar cataracts phenotypes, mutations in a single gene can cause different types of cataracts, and one single mutation in a gene can cause different cataract phenotypes in patients.
Identifying the precise genetic cause of CC is essential for providing accurate diagnostics for medical management, prognostics, and recurrence risk counseling for the patient and family.[12] However, due to the highly clinical and genetic heterogeneity, clinical genetic diagnostic practice of CC, especially for nonsyndromic CC, is greatly limited with the traditional sequencing method which sequences a few candidate genes at each time. Despite this, recently, the next generation sequencing (NGS) combined with targeted genomic enrichment has proved to be a cost-effective resolution to the genetic test of genetically heterogeneous diseases and provide a new opportunity for genetic diagnostics of CC.
In this study, we performed parallel sequencing of 45 CC genes by combined NGS and targeted genomic enrichments to determine the genetic mutations in a three-generation Chinese family with congenital nuclear cataracts. We identified a novel c.426_440delGCTGGAGGGGACCCT in GJA8 that segregates with the disease phenotype in the family and evaluated potential pathogenicity of the deletion and modeled the functional impacts of the deletion on the structure of the connexin 50 (Cx50) protein. Our result demonstrates that the targeted gene sequencing using NGS can be used as an effective tool for molecular diagnosis of CC.
METHODS
Participants recruitment, blood sampling, and DNA extraction
This study was approved by the Institutional Review Boards of Beijing Genomic Institute (BGI)-Shenzhen. Nine members of a three-generation Chinese family from Guangdong with ADCCs were recruited in this study [Figure 1]. Careful ophthalmological examinations and hospital medical record reviews were performed for each affected member to confirm the clinical diagnosis. After obtained written informed consent, peripheral venous blood samples were collected for all participants. Genomic DNA was extracted using a QIAamp DNA Blood MiNi kit (Qiagen, Germany).
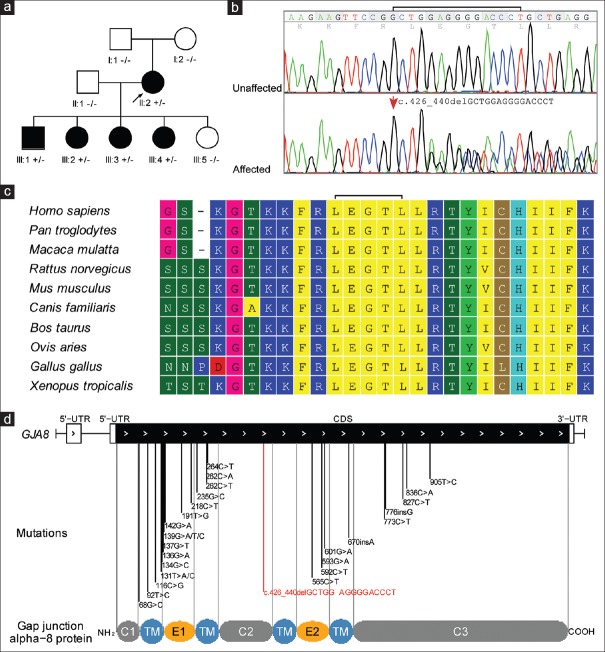
Figure 1.
Pedigree and mutation analysis. (a) Pedigree of a three-generation Chinese family with congenital cataracts. The proband is indicated with an arrow. Squares and circles symbolize male and female individual, respectively. Black symbol indicates cataract affected status and white symbol indicates unaffected status. (b) DNA sequence chromatogram analysis. DNA sequence chromatograms of the unaffected members (top) and affected members (bottom) in the pedigree. Fifteen bases deletion in exon 2 causes a conservative deletion of LEGTL from codon 143–147 (p.143_147del). (c) Evolutionary sequence conservation analysis. Multiplex sequence alignment of connexin 50 from different species reveals that codon 143–147, where the mutation (p.143_147del) occurred, is located within a highly conserved region. (d) Allelic spectrum of reported disease-associated mutations. Schematic diagram of genomic structure of human GJA8 gene and allelic spectrum of diseases mutations are shown. The identified mutation in this study is marked with red line. CL (C1/C2/C3): cytoplasmic loop domain; TM: transmembrane domain; E1:extracellular domain 1; E2: extracellular domain 2.
Capture probes design, targeted capture, and next generation sequencing
Forty-five genes implicated in the CC, including 29 nonsyndromic cataract genes (AGK, BEST1, BFSP1, BFSP2, CHMP4B, CRYAA, CRYAB, CRYBA1, CRYBA4, CRYBB1, CRYBB2, CRYBB3, CRYGC, CRYGD, CRYGS, EPHA2, FYCO1, GJA3, GJA8, HSF4, P3H2, LIM2, MAF, MIP, NHS, PITX3, SLC16A12, TDRD7, and VIM) and 16 syndromic cataract genes (ABHD12, CNBP, CTDP1, EYA1, FTL, GALK1, GCNT2, GFER, GJA1, JAM3, OPA3, PAX6, RAB3GAP2, SIL1, SIX6, and SLC33A1), were collected from careful literature and database search. The 45 genes selected contain almost all of the genes related to CC.[1,13] Moreover, genes related to ADCC, which are interested, were all included. Targeted sequencing capture DNA Probes were designed for exons and the flanking 30 bp intronic sequences using the Nimblegen SeqCap EZ Choice system (Roche NimbleGen, Madison, WI, USA). Targeted sequences capture and sequencing library preparation were performed as previously described.[14] Paired-end sequencing (PE100) was performed on the Illumina HiSeq2000 platform (Illumina, San Dieago, CA, USA).
Short-reads mapping, variant detection, and annotation
After filtering of reads of low-quality and potential adaptor contamination,[15] the clean reads were mapped to the reference human genome (hg19) using BWA software package (Burrows Wheeler Aligner http://sourceforge.net/projects/bio-bwa/).[16] Single nucleotide variants (SNVs) were identified using SOAPsnp software (http://soap.genomics.org.cn/),[17] and small insertion and deletions (InDels) were identified using the GATK InDel Genotyper (The Genome Analysis Toolkit, http://www.broadinstitute.org/gsa/wiki/index.php/).[18] The variants were annotated using a BGI in-house developed annotation pipeline.
Sequence conservation analysis and estimation of deleteriousness
Multiple alignments of Cx50 protein sequences between species were directly obtained from the UCSC genome browser home (http://genome.ucsc.edu/) and realigned with the Molecular Evolutionary Genetics Analysis (MEGA 6.06, http://www.megasoftware.net). The online free tool Combined Annotation-Dependent Depletion (CADD, http://cadd.gs.washington.edu/),[19] a newly developed framework that integrates diverse annotations into a single quantitative score (C-score) to measure the deleteriousness of SNVs and small InDels, was used to evaluate the pathogenicity of the deletion.
Construction of connexin 50 three-dimensional structure model
In the absence of Cx50 experimental structures, the comparative modeling methods based on high sequence identity can be employed to predict the Cx50 protein structure, which can subsequently be used to aid in the understanding of protein functional mechanisms. The amino acid sequences of wild type (WT) Cx50 and mutant Cx50 above were used to perform the sequence similarity searches using the NCBI BLAST in SWISS-MODEL server (http://swissmodel.expasy.org/).[20] The newly disclosed structure of human Cx26 (Protein Data Bank [PDB] code: 2ZW3)[21] was selected as the template to construct the Cx50 structure that shares a sequence identity of 57% with WT Cx50 sequence. The modeling result is shown in Supplementary Figure 1 (564.4KB, tif) .
(a and b) The overall structure of the connexin 50 chennal model in hydrophocipy surface representation.
The system of generating lipid bilayer structures as well as membrane-bound protein structures, CHARMM-GUI Lipid Builder[22] was used to build a Cx50 protein/membrane complex for molecular dynamics (MD) simulations. The Cx50 models (WT and mutant) were inserted into a fully hydrated palmitoyl-oleyl phosphatidylcholine (POPC) lipid bilayer of 400 molecules (set 200 on the lower leaflet and 200 on the upper leaflet) [Supplementary Figure 2 (876KB, tif) ]. The water model used was three-point model (TIP3P). To remove the net charge of the system, an ionic concentration of 150 mM KCl by transmuting random water molecules into K+ and Cl− was added as counterions.
The illustration of homo-6-mer connexin 50 protein/membrane complex models.
Molecular dynamics simulation
MD simulations were carried out using the parallel MD program GROMACS 4.6.5 for all protein molecules, POPC lipid molecules, and ions along with the TIP3P model for water molecules. A cutoff of 10 Å for van der Waals interactions was imposed. The particle mesh Ewald[23] technique with a short-range cutoff of 10 Å was employed to calculate long-range electrostatic forces. The periodic boundary conditions were introduced on all simulations with an integration time step of 2 fs to employ a multiple time stepping algorithms. The simulations were equilibrated as an NPT ensemble, using the Langevin dynamics[24] method to keep the pressure at 1 atm, and the temperature was maintained at 303 K using Langevin dynamics with a very weak friction coefficient. The MD simulation protocol is detailed description as follows: To remove the largest strains in the system, all simulation systems were first subjected to 5000 cycles of steepest descent, while position restraints were applied to the residues of the Cx50 models as well as the molecules of POPC lipid bilayer. Afterward, MD with position restraints applied only to the protein was performed for 0.1 ns. All the positional restraints were eliminated in the third round, and the systems were allowed to preequilibrate for 5 ns. The production simulations were finally conducted for 10 ns for all systems.
Sanger sequencing
Polymerase chain reaction (PCR) primer sets were designed to sequence the first exons of MAF,EPHA2, and NHS genes not covered in targeted sequencing, as well as to analyze the segregation of the GJA8 deletion in the family [Supplementary Table 1]. The PCR amplification was conducted in a 25 μl reaction volume containing 1X PCR buffer with 1.5 mM MgCl2, 200 μM each dNTP, 0.25 U Taq DNA polymerase (Takara, Dalian, China), 1.5 μM primers, and 50 ng genomic DNA. The PCR condition was 94°C for 4 min, 35 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 45 s, and a final 72°C for 10 min. The PCR products were purified and sequenced at both ends on an ABI3730xl DNA sequencer (PE Applied Biosystems, Forest City, CA, USA).
Supplementary Table 1.
Primers for PCR amplification of exons of candidate genes and the size of the PCR products
| Gene | Exon | Primers sequence (5’–3’) | Fragment size (bp) |
|---|---|---|---|
| GJA8 | 2 | F*-GTGCACTACGTCCGCATG | 298 |
| R†-CGAAGCAGTCCACCACATTG | |||
| MAF | 1 | F-AGCTGGTGACCATGTCTGTG | 407 |
| R-AGAACTAGCAAGCCCACACC | |||
| F-AACTGGCAATGAGCAACTCC | 548 | ||
| R-GTGGTGGTGGTGGTGGTAGT | |||
| F-GAGCGAGGGAGCACATTG | 352 | ||
| R-CCGGTTCCTTTTTCACTTCA | |||
| F-CCGCACTACCACCACCAC | 432 | ||
| R-CTGGTTCTTCTCCGACTCCA | |||
| EPHA2 | 1 | F-GACCAAGCTGAAACCGCTTA | 684 |
| R-TACCAGGCTCAGAGATCCCT | |||
| NHS | 1 | F-AGGCAAGGTGAGCAGAGAAG | 764 |
| R-CGCAGAAACCCATAGCCTG |
*F: Forward primers; †R: Reverse primers; PCR: Polymerase chain reaction.
RESULTS
Pedigree and clinical features
The family includes 5 affected and 4 unaffected members in a three-generation pedigree [Figure 1a]. The transmission of cataract from the female founder (also the proband) to four out of five of her offsprings with both genders supports a dominant inheritance pattern of the lens diseases in the family. All affected members have nuclear cataracts with no other ocular or systemic abnormalities [Figure 1a and Table 1]. The presences of cataracts at birth were confirmed by hospital records, and nonsyndromic CC was diagnosed for all affected members.
Table 1.
Summary of clinical evaluations for the Chinese cataract family
| Member | Gender | Age at onset | Age at diagnosis | Cataract phenotypes | Other abnormalities | Diagnosis |
|---|---|---|---|---|---|---|
| I:1 | Male | No | 65 | Normal | No | Normal |
| I:2 | Female | No | 67 | Normal | No | Normal |
| II:1 | Male | No | 48 | Normal | No | Normal |
| II:2 | Female | On birth | 42 | Nuclear, bilateral | No | Congenital nuclear cataract |
| III:1 | Male | On birth | 13 | Nuclear, bilateral | No | Congenital nuclear cataract |
| III:2 | Female | On birth | 22 | Nuclear, bilateral | No | Congenital nuclear cataract |
| III:3 | Female | On birth | 25 | Nuclear | No | Congenital nuclear cataract |
| III:4 | Female | On birth | 15 | Nuclear | No | Congenital nuclear cataract |
| III:5 | Female | On birth | 18 | Normal | No | Normal |
Targeted sequencing of 45 cataract genes
The coding sequence-exons and adjacent intronic sequences of 45 cataract genes, consisting of 366 exons and 63003 bp, were captured and sequenced for the proband DNA (see Materials and Methods). A total of 224,437 clean reads in 15,940,353 bp length (or ~15.94 Mb data) mapped to targeted regions were generated on Hiseq2000 [Table 2]. This formed a mean coverage depth of 218-fold on the targeted regions of the 45 cataract genes, with a general sequence coverage rate of 99.23% and a high-quality genotype assignment (>×10) rate of 97.29% [Table 2]. The uncovered targeted regions were all the first coding exons with extreme GC content [mean CC% >69%, Supplementary Table 2]. Supplementary sequencing was performed to cover the sequences of three of the uncovered first coding exons (see Materials and Methods) while the others were excluded for the analysis considering a syndromic form or a nondominant inheritance pattern of cataracts associated with the related genes [Supplementary Table 2].
Table 2.
Summary statistics for targeted sequencing of 45 cataracts genes in the proband
| Gene | OMIM diseases | Transcript | Number of coding exons | Sizes (bp) | Sequencing depth (X) | Coverage rate (%) | ||
|---|---|---|---|---|---|---|---|---|
| >1X | >4X | >10X | ||||||
| ABHD12 | #612674 (AR, Sa) | NM_015600.4 | 13 | 1215 | 192 | 99.26 | 84.28 | 84.28 |
| AGK | #614691 (AR, NSa) | NM_018238.3 | 16 | 1269 | 270 | 100.00 | 100.00 | 100.00 |
| BEST1 | #193220 (AD, NS) | NM_001139443.1 | 9 | 1815 | 320 | 100.00 | 100.00 | 100.00 |
| BFSP1 | #611391 (AR, NS) | NM_001195.3 | 8 | 1998 | 272 | 99.95 | 97.00 | 85.44 |
| BFSP2 | #611597 (AD, NS) | NM_003571.2 | 7 | 1248 | 164 | 100.00 | 100.00 | 100.00 |
| CHMP4B | #605387 (AD, NS) | NM_176812.4 | 5 | 675 | 214 | 100.00 | 100.00 | 100.00 |
| CNBP | #602668 (AD, S) | NM_001127192 | 4 | 513 | 155 | 97.87 | 94.92 | 93.94 |
| CRYAA | #604219 (AD, NS) | NM_000394.2 | 3 | 522 | 154 | 100.00 | 100.00 | 100.00 |
| CRYAB | #613763 (AD/AR, NS) | NM_001885.1 | 3 | 528 | 313 | 100.00 | 100.00 | 100.00 |
| CRYBA1 | #600881 (AD, NS) | NM_005208.4 | 6 | 648 | 245 | 100.00 | 100.00 | 100.00 |
| CRYBA4 | #610425(AD/AR, NS) | NM_001886.2 | 6 | 591 | 141 | 100.00 | 100.00 | 100.00 |
| CRYBB1 | #611544 (AD/AR, NS) | NM_001887.3 | 6 | 759 | 145 | 100.00 | 100.00 | 100.00 |
| CRYBB2 | #601547 (AD, NS) | NM_000496.2 | 6 | 618 | 194 | 100.00 | 100.00 | 100.00 |
| CRYBB3 | #609741 (AD/AR, NS) | NM_004076.3 | 6 | 636 | 144 | 100.00 | 100.00 | 100.00 |
| CRYGC | #604307 (AD, NS) | NM_020989.3 | 3 | 525 | 179 | 100.00 | 100.00 | 100.00 |
| CRYGD | #115700 (AD, NS) | NM_006891.3 | 3 | 525 | 179 | 100.00 | 100.00 | 100.00 |
| CRYGS | #116100 (AD, NS) | NM_017541.2 | 3 | 537 | 364 | 100.00 | 100.00 | 100.00 |
| CTDP1 | #604168 (AR, S) | NM_004715.4 | 13 | 2886 | 126 | 92.31 | 89.12 | 89.12 |
| EPHA2 | #116600 (AD, NS) | NM_004431.3 | 17 | 2931 | 150 | 100.00 | 97.10 | 97.10 |
| EYA1 | #113650 (AD, S) | NM_000503.4 | 18 | 1779 | 314 | 100.00 | 100.00 | 100.00 |
| FTL | #600886 (AD, S) | NM_000146.3 | 4 | 528 | 131 | 100.00 | 100.00 | 100.00 |
| FYCO1 | #610019 (AR, NS) | NM_024513.3 | 18 | 4437 | 194 | 100.00 | 100.00 | 100.00 |
| GALK1 | #230200 (AR, S) | NM_000154.1 | 8 | 1179 | 80 | 100.00 | 100.00 | 100.00 |
| GCNT2 | #110800 (AR, S) | NM_145649.4 | 5 | 1209 | 421 | 100.00 | 100.00 | 100.00 |
| GFER | #613076 (AR, S) | NM_005262.2 | 3 | 618 | 120 | 100.00 | 95.63 | 73.46 |
| GJA1 | #257850 (AR, S) | NM_000165.3 | 2 | 1149 | 371 | 100.00 | 100.00 | 100.00 |
| GJA3 | #601885 (AD, NS) | NM_021954.3 | 2 | 1308 | 127 | 100.00 | 100.00 | 100.00 |
| GJA8 | #116200 (AD, NS) | NM_005267.4 | 2 | 1302 | 270 | 100.00 | 100.00 | 100.00 |
| HSF4 | #116800 (AD, NS) | NM_001040667.2 | 15 | 1479 | 125 | 100.00 | 100.00 | 98.24 |
| JAM3 | #613730 (AR, S) | NM_032801.4 | 9 | 933 | 275 | 100.00 | 100.00 | 100.00 |
| LEPREL1 | #614292 (AR, NS) | NM_018192.3 | 15 | 2127 | 191 | 100.00 | 99.76 | 92.57 |
| LIM2 | #615277 (AR, NS) | NM_030657.3 | 5 | 648 | 166 | 100.00 | 100.00 | 100.00 |
| MAF | #610202 (AD, NS) | NM_005360.4 | 2 | 1212 | 100 | 79.04 | 76.32 | 73.10 |
| MIP | #615274 (AD, NS) | NM_012064.3 | 4 | 792 | 204 | 100.00 | 100.00 | 100.00 |
| NHS | #302200 (XD, NS and S) | NM_198270.2 | 8 | 4893 | 355 | 97.06 | 95.26 | 90.88 |
| OPA3 | #165300 (AD, S) | NM_001017989.2 | 2 | 543 | 75 | 100.00 | 100.00 | 100.00 |
| PAX6 | #106210 (AD, S) | NM_001258462.1 | 14 | 1311 | 283 | 100.00 | 100.00 | 100.00 |
| PITX3 | #610623 (AD, NS) | NM_005029.3 | 4 | 909 | 59 | 100.00 | 100.00 | 100.00 |
| RAB3GAP2 | #212720 (AR, S) | NM_012414.3 | 35 | 4182 | 327 | 100.00 | 100.00 | 100.00 |
| SIL1 | #248800 (AR, S) | NM_001037633.1 | 11 | 1386 | 193 | 100.00 | 100.00 | 100.00 |
| SIX6 | #212550 (AR, S) | NM_007374.2 | 2 | 741 | 206 | 100.00 | 100.00 | 100.00 |
| SLC16A12 | #612018 (AD, NS) | NM_213606.3 | 8 | 1551 | 334 | 100.00 | 100.00 | 100.00 |
| SLC33A1 | #614482 (AR, S) | NM_004733.3 | 6 | 1650 | 391 | 100.00 | 100.00 | 100.00 |
| TDRD7 | #613887 (AR, NS) | NM_014290.2 | 17 | 3297 | 368 | 100.00 | 100.00 | 100.00 |
| VIM | #116300 (AD, NS) | NM_003380.3 | 10 | 1401 | 201 | 100.00 | 100.00 | 100.00 |
| Total | 366 | 63,003 | 218 | 99.23 | 98.43 | 97.29 | ||
OMIM: Online Mendelian Inheritance in Man; AR: Autosomal recessive; AD: Autosomal dominant; XD: X-linked dominant; S: Syndromic; NS: Non-syndromic.
Supplementary Table 2.
Gene (exons) with poor sequencing coverage and the associated diseases
| Gene | Related cataracts | Exon | Size (bp) | GC content (%) | Sequencing depth (x) | Coverage rate (%) | Uncovered sequences | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Length (bp) | Percentage of all exons | |||||||||
| EPHA2 | #116600, cataract 6, multiple types, AD | EX1 | 240 | 74.17 | 29.70 | 54.12 | c.1_85 | 85 | 2.14 | ||
| BFSP1 | #611391, cataract 33, AR | EX1 | 417 | 76.13 | 100.89 | 99.73 | c.1_377 | 377 | 18.87 | ||
| ABHD12 | #612674, polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract, AR | EX1 | 470 | 78.13 | 96.43 | 95.29 | c.1_191 | 191 | 15.72 | ||
| MAF | #610202, cataract, pulverulent or cerulean, with or without microcornea, AD | EX1 | 1941 | 69.65 | 72.14 | 77.28 | c.1_1118 | 1118 | 42.24 | ||
| CTDP1 | #604168, congenital cataracts, facial dysmorphism, and neuropathy, AR | EX1 | 461 | 78.09 | 5.30 | 29.30 | c.1_314 | 314 | 8.37 | ||
| NHS | #302200, cataract 40, X-linked, XD | EX1 | 903 | 70.99 | 79.55 | 74.51 | c.1_565 | 565 | 6.45 | ||
AR: Autosomal recessive; AD: Autosomal dominant; XD: X-linked dominant.
Identification of potential causal mutation
Mapped against the human reference genome sequences, a total of 43 variants, including forty substitutes and 3 small InDels, were identified on the 45 genes [Table 3 and Supplementary Table 3]. A vast majority of these variants (29/43) were either synonymous substitutes or from untranslated regions [Table 3]. These variants are thought to be of less potentially pathogenicity, and we put them aside first. Of the remaining variants with potential function importance (including ten nonsynonymous, 3 splice acceptor and donor site mutations, and 1 coding InDels), only the heterozygous in-frame coding on GJA8 was novel meeting the criteria as nonpolymorphic with an allele frequency <5% in any of the single nucleotide polymorphism database, HapMap, or 1000 genome project database. Multiplex protein sequences alignment shows that the deleted sequence was evolutionarily conserved among ten species with high-quality reference genome sequences available on UCSC genomes database [Figure 1c]. More importantly, the deletion was found in full cosegregation with cataract phenotypes in Sanger sequencing analysis of the mutation in the pedigree [Figure 1a and 1b] but was not detected in 1100 BGI in-house control exomes [Supplementary Table 3], genetically suggesting a potential pathogenic role of the deletion. Intriguingly, the deletion was not detected on both unaffected parents of the proband in Sanger sequencing validation analysis [Figure 1a and 1b], supporting a de novo occurrence of the deletion in the proband.
Table 3.
Summary statistics for variants detected in targeted sequencing of 45 cataract genes in the proband
| Mutation type | Number |
|---|---|
| SNVs | 40 |
| NS | 10 |
| Synonymous | 28 |
| SS | 2 |
| InDels | 3 |
| Coding (I) | 1 |
| SS | 1 |
| UTR | 1 |
| NS/SS/I | 14 |
| Allele frequency ≤0.05 in dbSNP (snp137), HapMap or 1000 genomes project | 2 |
| Allele frequency ≤0.05 in 1100 BGI in-house control exomes | 1 |
| Frequency ≤0.05 in either | 1 |
NS: Nonsynonymous; SS: Spicing sites; UTR: Untranslated regions; dbSNP: Single nucleotide polymorphism database; BGI: Beijing Genomic Institute; SNVs: Single nucleotide variants; InDels: Insertion and deletions.
Supplementary Table 3.
Variants detected in the 45 cataract genes in the proband
| Gene | Transcript | Variant | Hom/Het | Reads | Mutation type | Gene region | Functional region | rsID* | Frequency in dbSNP† | Frequency in HapMap‡ | Frequency in 1000 genomes§ | Frequency in 1100 exomes|| |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLC16A12 | NM_213606 | c.49 T>G | Het | C99/103A;202 | SNV | CDS1 | Missense | rs3740030 | 0.163 | 0.38 | 0.1538 | 0.3641 |
| PITX3 | NM_005029 | c.285 C>T | Het | A25/22G;47 | SNV | CDS2 | Synonymous | rs2281983 | 0.43 | 0 | 0.424 | 0.3795 |
| BEST1 | NM_001139443 | c.39 C>A | Hom | A142;142 | SNV | CDS1 | Synonymous | rs1109748 | 0.075 | 0.547 | 0.1474 | 0.5333 |
| BEST1 | NM_001139443 | c.1428 T>C | Hom | C249/1T;250 | SNV | CDS8 | Synonymous | rs1800009 | 0 | 0 | 0.3553 | 0.7638 |
| JAM3 | NM_032801 | c.978−3 T>C | Het | C106/142T;248 | SNV | Intron7 | Splice | rs610382 | 0.498 | 0.434 | 0.402 | 0.4342 |
| GJA3 | NM_021954 | c.1017 G>A | Het | T41/19C;60 | SNV | CDS1 | Synonymous | rs11617415 | 0.371 | 0 | 0.3736 | 0.1949 |
| GJA3 | NM_021954 | c.895 C>A | Hom | T103;103 | SNV | CDS1 | Missense | rs968566 | 0.966 | 0 | 0.967 | 1 |
| CNBP | NM_001127192 | c.156 C>T | Het | A108/86G | SNV | CDS2 | Synonymous | rs4303883 | 0.381 | 0.277 | 0.2115 | 0.2308 |
| CTDP1 | NM_004715 | c.978 G>A | Het | A127/122G;249 | SNV | CDS7 | Synonymous | rs599554 | 0.326 | 0.175 | 0.283 | 0.1128 |
| CTDP1 | NM_004715 | c.1461 G>A | Het | A54/28G;82 | SNV | CDS8 | Synonymous | rs2126082 | 0.304 | 0 | 0.2756 | 0.1026 |
| CTDP1 | NM_004715 | c.2817 T>C | Hom | C33;33 | SNV | CDS13 | Synonymous | rs626169 | 0.846 | 0.993 | 0.8672 | 0.9949 |
| EPHA2 | NM_004431 | c.2874 C>T | Het | A42/49G;91 | SNV | CDS17 | Synonymous | rs3754334 | 0.355 | 0.226 | 0.326 | 0.241 |
| EPHA2 | NM_004431 | c.1983 C>T | Het | A77/94G;171 | SNV | CDS11 | Synonymous | rs10907223 | 0.267 | 0.206 | 0.2024 | 0.1949 |
| EPHA2 | NM_004431 | c.987 C>T | Het | A27/35G;62 | SNV | CDS5 | Synonymous | rs2230597 | 0.433 | 0.184 | 0.4158 | 0.2564 |
| BFSP1 | NM_001195 | c.1749 A>G | Hom | C250;250 | SNV | CDS8 | Synonymous | rs6080718 | 0 | 0.358 | 0.5403 | 0.4154 |
| BFSP1 | NM_001195 | c.1500 G>A | Het | T109/141C;250 | SNV | CDS8 | Synonymous | rs6136118 | 0.431 | 0.453 | 0.4295 | 0.4615 |
| BFSP1 | NM_001195 | c.1033 G>A | Het | T102/108C;210 | SNV | CDS7 | Missense | rs6080719 | 0.378 | 0.489 | 0.3608 | 0.4564 |
| BFSP1 | NM_001195 | c.90 G>A | Het | T2/3C;5 | SNV | CDS1 | Synonymous | snp105 | 0 | 0 | 0 | 0 |
| ABHD12 | NM_015600 | c.1068 T>C | Hom | G211;211 | SNV | CDS12 | Synonymous | rs10966 | 0.31 | 0 | 0.3581 | 0.516 |
| CRYAA | NM_000394 | c.6 C>T | Hom | T149;149 | SNV | CDS1 | Synonymous | rs872331 | 0.276 | 0.03 | 0.315 | 0.2278 |
| CRYBB3 | NM_004076 | c.337 C>G | Hom | G144;144 | SNV | CDS4 | Missense | rs9608378 | 0.456 | 0.867 | 0.4908 | 0.8718 |
| CRYBB2 | NM_000496 | c.449+9 G>A | Hom | A96;96 | SNV | Intron5 | Splice | rs4049505 | 0 | 0 | 0.6145 | 0.9026 |
| CRYBB2 | NM_000496 | c.483 G>A | Het | A67/79G;146 | SNV | CDS5 | Synonymous | rs8140949 | 0.412 | 0 | 0.3819 | 0.4923 |
| CRYBA4 | NM_001886 | c.171 T>C | Hom | C128;128 | SNV | CDS3 | Synonymous | rs5761637 | 0.844 | 0 | 0.848 | 0.9949 |
| CRYGD | NM_006891 | c.285 A>G | Hom | C249;249 | SNV | CDS3 | Synonymous | rs2305430 | 0.626 | 0.453 | 0.6392 | 0.3846 |
| CRYGD | NM_006891 | c.51 T>C | Het | G37/40A;77 | SNV | CDS2 | Synonymous | rs200375285 | 0 | 0.518 | 0.435 | 0.2974 |
| FYCO1 | NM_024513 | c.3924 C>T | Hom | A194;194 | SNV | CDS13 | Synonymous | rs1463680 | 0.739 | 0.956 | 0.7473 | 0.8114 |
| FYCO1 | NM_024513 | c.2036 C>T | Het | A68/68G;136 | SNV | CDS7 | Missense | rs3796375 | 0.489 | 0.519 | 0.4203 | 0.3808 |
| FYCO1 | NM_024513 | c.1335 G>A | Het | T101/140C;241 | SNV | CDS7 | Synonymous | rs3796376 | 0.315 | 0.562 | 0.3168 | 0.3203 |
| FYCO1 | NM_024513 | c.962 G>C | Het | G60/84C;144 | SNV | CDS7 | Missense | rs3733100 | 0.499 | 0.489 | 0.457 | 0.3879 |
| FYCO1 | NM_024513 | c.749 G>A | Hom | T219;219 | SNV | CDS7 | Missense | rs4683158 | 0 | 1 | 0.8498 | 0.8932 |
| FYCO1 | NM_024513 | c.267 C>A | Hom | T249;249 | SNV | CDS3 | Synonymous | rs4682801 | 0.622 | 1 | 0.6667 | 0.8114 |
| BFSP2 | NM_003571 | c.603 G>A | Het | A84/129G;213 | SNV | CDS3 | Synonymous | rs2276737 | 0.495 | 0.519 | 0.4277 | 0.4513 |
| SLC33A1 | NM_004733 | c.512 A>G | Het | C113/137T;250 | SNV | CDS1 | Missense | rs3804769 | 0.174 | 0.226 | 0.1722 | 0.2278 |
| EYA1 | NM_172058 | c.1755 T>C | Het | G51/42A;93 | SNV | CDS16 | Synonymous | rs10103397 | 0.497 | 0.407 | 0.4267 | 0.388 |
| EYA1 | NM_172058 | c.1278 C>T | Het | A135/115G;250 | SNV | CDS12 | Synonymous | rs4738118 | 0.399 | 0.455 | 0.2811 | 0.4614 |
| EYA1 | NM_172058 | c.813 A>G | Hom | C248;248 | SNV | CDS7 | Synonymous | rs1445398 | 0.034 | 0.224 | 0.0632 | 0.1747 |
| TDRD7 | NM_014290 | c.33 A>G | Hom | G249;249 | SNV | CDS1 | Synonymous | rs1381532 | 0.322 | 0.299 | 0.3114 | 0.2811 |
| TDRD7 | NM_014290 | c.449 T>C | Hom | C243/5T;248 | SNV | CDS3 | Missense | rs2045732 | 0.322 | 0.281 | 0.3114 | 0.2776 |
| NHS | NM_198270 | c.3955 T>C | Het | C128/121T;249 | SNV | CDS6 | Missense | rs3747295 | 0.469 | 0.25 | 0.1374 | 0.1692 |
| MAF | NM_005360 | c.–1637_−1639 delGGC | Het | W34/M14;48 | Deletion | 5-UTR | 5-UTR | _ | No_frequency | Not_in_HapMap | 0 | 0.4744 |
| GJA8 | NM_005267 | c.426_440 delGCTG GA GGGGACCCT | Het | W169/M78;247 | Deletion | CDS1 | CDS | _ | No_frequency | Not_in_HapMap | 0 | 0 |
| RAB3GAP2 | NM_012414 | c.812−6 delT | Het | W67/M68;135 | Deletion | Intron9 | Splice | _ | No_frequency freq | Not_in_HapMap | 0 | 0.3154 |
*NCBI dbSNP ID; †Allele frequency in NCBI dbSNP; ‡Allele frequency in HapMap; §Allele frequency in 1000 genome project; ||Allele frequency in BGI in-house control exomes. dbSNP: Single nucleotide polymorphism database; SNV: Single nucleotide variant; UTR: Untranslated regions; CDS: Coding sequence; BGI: Beijing Genomic Institute.
The pathogenicity of the deletion was further evaluated with CADD. The C-score of 15.52 supports a pathogenic role of the deletion (mutations with C-score >15 were supposed to be pathogenic).[19]
This novel heterozygous deletion was concerned with a sequence deletion of 5 amino acids (or 15 bp) at the intracellular loop domain (cytoplasmic loop [CL]) of the Cx50 protein (or GJA8) [Figure 1d], which connects two transmembrane domains and was suspected to serving as binding sites of Ca2+/CaM in gap junction channel function.[25,26] Mutations on GJA8 have caused multiplex ADCCs (Cataract 1, multiple types, OMIM#116200) including nuclear cataracts.
Potential functional impact of the deletion
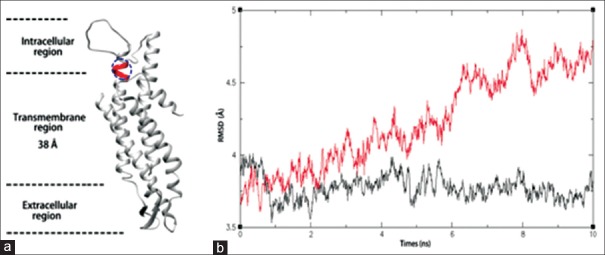
To illustrate the dysfunction of Cx50 mutant, we performed MD simulation. At first, the best hit in PDB of Cx50 protein was used to predict the three-dimensional structure (see Materials and Methods). The overall structure of Cx50 represents a typical channel protein that contains a hydrophobic surface and hydrophilic channel [Supplementary Figure 1 (564.4KB, tif) ]. Then, the protein/membrane complex with lipid bilayer system and water was built to simulate the environment of Cx50 in cell. Both WT and mutant of Cx50 were inserted into the lipid bilayer system [Figure 2a and Supplementary Figure 2 (876KB, tif) ]. Finally, MD simulation was performed on both models (see Methods). As shown in Figure 2b, WT protein molecules were very stable in our system. However, the mutant molecules were more dynamic and the root-mean-square deviation increased when mutant molecules stay in the system for a longer time. Thus, the mutant Cx50 becomes unstable in the simulated system, which may also happen in real cellular environment.
Figure 2.
Molecular dynamics simulation. (a) The illustration of mono connexin 50 protein/membrane complex models. The blue circle represents the lost region in connexin 50 mutant. (b) Simulation of molecular dynamics. The black curve denotes the structure dynamics of wild type and the red curve denotes mutants. X-axis is the time of simulation and Y-axis is the root-mean-square deviation of atoms.
DISCUSSION
The lens of human eye is an avascular structure, and gap junction channels encoded by Cxs function as the passage of intercellular transport of the ions and low molecular weight biomolecules. Cx50 (GJA8) and Cx46 (GJA3) are the major components of mammalian lens fiber cells, and mutations of these two genes account for approximately 20% of nonsyndromic familial cataract cases.[7] The typical structure of Cx includes a cytoplasmic N-terminal domain (NT), four transmembrane domains (TM1 to TM4), two extracellular loops (E1 and E2), a CL between TM2 and TM3, and a C-terminal domain (CT). In this study, we identified a novel in-frame deletion of 15 bp in the coding region of GJA8 gene (c.426_440delGCTGGAGGGGACCCT) associated with CCs in a three-generation Chinese family by targeted genes sequencing.
Several lines of evidence support a pathogenic role of the deletion. First, the deletion was concerned with an in-frame deletion of 5 amino acids within a highly evolutionarily conserved region in the CL domain of Cx50 protein, which binds Ca2+/CaM to mediate gap junction channel in the lens of the eye.[25,26] Second, the deletion was fully cosegregated with cataracts in the family but was not found in 1100 control exomes. Third, the cataract morphology of the affected members and inheritance pattern of the cataracts in the family were compatible with those of cataract caused by GJA8 mutations. Fourth, deleteriousness evaluation with CADD supported pathogenicity of the deletion. Fifth, protein structure modeling revealed that mutant protein disrupts the structure stability of Cx50 channel.
To date, about 28 GJA8 mutations have been reported in CC patients [Figure 1d]. The majority of these mutations (26/28) are missense substitutes although rare cases of nonsense and frameshift mutations (2/28) have also been reported [Supplementary Table 4]. These mutations occur at the NT, TM1 to TM4, E1 and E2, or CT,[27] but none has been reported on the CL domain [Figure 1d and Supplementary Table 4]. No correlation between the mutation types or mutant region and the cataract phenotypes were observed. In vitro, functional analysis showed that these mutations triggered the formation of cataracts either through loss of normal channel functions (loss of function, altered gating, or reduced channel numbers) or gain of abnormal functions (gain of hemichannel function and formation of cytoplasmic accumulations).[9,27] The in-frame deletion reported here was first ever detected in the intracellular loop [Figure 1d and Supplementary Table 4] and its likely pathogenicity indicated the functional importance of the deleted sequence in the Cx50 gap junction protein. The structure modeling here showed that the deletion destabilized the structure of Cx50 protein channel. Thus, it is supposed that the mutation causes cataracts in the Chinese family as a dominant-negative function mutation due to impaired function of the channel or reduced number of normal channels.
Supplementary Table 4.
Reported connexin 50 mutants and associated cataracts
| Mutation | Amino acid change | Location | Cataract types | Inheritary | Family origin | References |
|---|---|---|---|---|---|---|
| 68G>C | R23T | NT* | Progressive dense nuclear | Autosomal dominant | Iranian | Willoughby et al. 2003 |
| 92T>C | I31T | M1† | Nuclear cataract | Autosomal dominant | Chinese | Wang et al. 2009 |
| 116C>G | T39R | M1 | Cataract and microcornea and iris hypoplasia | Autosomal dominant | Chinese | Sun et al. 2011 |
| 131T>A | V44E | M1 | Cataract and microcornea | Autosomal dominant | Indian | Devi and Vijayalakshmi 2006 |
| 131T>C | V44A | M1 | Suture-sparing nuclear cataracts | Autosomal dominant | Chinese | Zhu et al. 2014 |
| 134G>C | W45S | M1 | Jellyfish-like bilateral and microcornea | Autosomal dominant | Indian | Vanita et al. 2008b |
| 136G>A | G46R | M1 | Complete and microcornea | Autosomal dominant | Chinese | Sun et al. 2011 |
| 137 G>T | G46V | M1 | Total cataract | Autosomal dominant | Pakistani | Minogue et al. 2009 |
| 139G>A | D47N | E1‡ | Nuclear pulverulent | Autosomal dominant | British | Arora et al. 2008; Wang et al. 2011 |
| 139G>T | D47Y | E1 | Nuclear cataract | Autosomal dominant | Chinese | Lin et al. 2008 |
| 139G>C | D47H | E1 | Nuclear cataract | Autosomal dominant | Chinese | Li et al. 2013 |
| 142G>A | E48K | E1 | Zonular nuclear pulverulent | Autosomal dominant | Pakistani | Berry et al. 1999 |
| 191T>G | V64G | E1 | Nuclear | Autosomal dominant | Chinese | Ma et al. 2005 |
| 218C>T | S73F | E1 | Dense and “star-shaped,” various locations in the nucleus or the poles | Autosomal dominant | Danish | Hansen et al. 2009 |
| 235G>C | V79L | M2§ | “Full moon” Y-sutural opacity | Autosomal dominant | Indian | Vanita et al. 2006 |
| 262C>T | P88S | M2 | Zonular pulverulent | Autosomal dominant | British | Shiels et al. 1998 |
| 262C>A | P88Q | M2 | Lamellar pulverulent | Autosomal dominant | British | Arora et al. 2006 |
| 262C>A | P88Q | M2 | “Balloon-like” Y-sutural opacities | Autosomal dominant | Indian | Vanita et al. 2008a |
| 264C>T | P88T | M2 | Total cataract | Autosomal dominant | Chinese | Ge et al. 2014 |
| 565C>T | P189L | E2|| | Cataract and microcornea | Autosomal dominant | Danish | Hansen et al. 2007 |
| 593G>A | R198Q | E2 | Cataract and microcornea | Autosomal dominant | Indian | Devi and Vijayalakshmi 2006 |
| 592C>T | R198W | E2 | Cataract and microcornea | Autosomal dominant | Chinese | Hu et al. 2010 |
| 601G>A | E201K | E2 | Perinuclear cataracts | Autosomal dominant | Chinese | Su et al. 2013 |
| 670insA | 203fs | E2 | Cataract | Autosomal recessive | Indian | Ponnam et al. 2007 |
| 773C>T | S258F | CT¶ | Nuclear | Autosomal dominant | Chinese | Gao et al. 2010 |
| 776insG | fs | CT | Triangular | Autosomal recessive | Germany | Schmidt et al. 2008 |
| 836C>A | S259Y | CT | – | Autosomal dominant | Danish | Hansen et al. 2009 |
| 827C>T | S276F | CT | Nuclear pulverulent | Autosomal dominant | Chinese | Yan et al. 2008 |
| 905T>C | L281C | CT | Lamellar/zonular | Autosomal dominant | Indian | Kumar et al. 2011 |
*Cytoplasmic amino-terminal, †First transmembrane domain, ‡Extracellular loop 1, §Second transmembrane domain, ||Extracellular loop 2, ¶Cytoplasmic carboxy-terminal. NT: N-terminal; CT: C-terminal.
In recent several years, great advances have been achieved in NGS and targeted genomic enrichment technologies. The combination of these two technologies has shown considerable potential and value in clinical applications, especially in genetic diagnosis of highly heterogeneous rare genetic diseases.[28] First, targeted sequencing of a small proportion of the genome (about 1/1000–1/100 of the genome) could reduce the sequencing cost to a level acceptable in clinical context. Second, the flexibility of targeted gene panel design and the availability of deep sequencing depth on NGS allow to simultaneously sequence all known genes for certain genetic disease and to comprehensively analyze SNVs, small InDels, and copy number variations at a high accurate level. Here, we performed targeted sequencing of 45 genes involved in CC (mainly in nonsyndromic CC) to explore the utility of targeted NGS sequencing for genetic diagnosis of ADCC. The high depth and completeness of sequences coverage for these 45 genes, as well as the readily identification of potential causative mutation, indicate that the targeted genes sequencing using NGS provides a tool for genetic diagnostics of nonsyndromic CC. More importantly, this method could be expanded to all CC with the addition of more CC genes in target panel.
However, a major limitation of this approach is that if the panel does not include genes responsible in tested patients, mutations will not be detected. Hence, the panel used needs to be updated when new genes are implicated in the disease. Another problem of the method is that some deep intronic mutations or mutations affecting regulatory elements may not be detected, which needs to be solved.
In conclusion, clinical molecular diagnosis of CC, particularly for ADCC, remains a challenge because of highly genetic and clinical heterogeneity. In this study, by combined NGS and targeted genomic enrichment technology, we identified a novel 15 deletion in a highly conserved region of CL domain of the GJA8 gene associated with cataracts in a three-generation Chinese family with ADCC (mutations have never reported in CL domain before). Evidence supports a pathogenic role of the deletion in the family. More importantly, protein modeling indicates a dominant-negative impact of the deletion on the protein function. Our results demonstrate targeted genes sequencing on NGS as a useful tool for molecular diagnosis of CCs.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
This work was supported by the experimental facilities and reagents of the BGI-Shenzhen, China.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
This work was supported by the experimental facilities and reagents of the BGI-Shenzhen, China. We would like to thank all of the blood donors for their uncompensated contribution to this study. We wish to thank the staff of BGI-Guangzhou for all the assistance provided in the process of sample collections. We gratefully acknowledge the support from Jiu-Cheng Liu for his assist in figures editing.
Footnotes
Edited by: Yi Cui
REFERENCES
- 1.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–49. doi: 10.1016/j.semcdb.2007.10.003. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apple DJ, Ram J, Foster A, Peng Q. Elimination of cataract blindness: A global perspective entering the new millenium. Surv Ophthalmol. 2000;45:S1–2. [PubMed] [Google Scholar]
- 3.Sun W, Xiao X, Li S, Guo X, Zhang Q. Exome sequencing of 18 Chinese families with congenital cataracts: A new sight of the NHS gene. PLoS One. 2014;9:100455. doi: 10.1371/journal.pone.0100455. doi: 10.1371/journal.pone.0100455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes JM, Leske DA, Burke JP, Hodge DO. Birth prevalence of visually significant infantile cataract in a defined U.S. population. Ophthalmic Epidemiol. 2003;10:67–74. doi: 10.1076/opep.10.2.67.13894. doi: 10.1076/opep.10.2.67.13894. [DOI] [PubMed] [Google Scholar]
- 5.Haargaard B, Wohlfahrt J, Fledelius HC, Rosenberg T, Melbye M. Incidence and cumulative risk of childhood cataract in a cohort of 2.6 million Danish children. Invest Ophthalmol Vis Sci. 2004;45:1316–20. doi: 10.1167/iovs.03-0635. doi: 10.1167/iovs.03-0635. [DOI] [PubMed] [Google Scholar]
- 6.Santana A, Waiswo M. The genetic and molecular basis of congenital cataract. Arq Bras Oftalmol. 2011;74:136–42. doi: 10.1590/s0004-27492011000200016. doi: 10.1590/S0004-27492011000200016. [DOI] [PubMed] [Google Scholar]
- 7.Shiels A, Bennett TM, Hejtmancik JF. Cat-map: Putting cataract on the map. Mol Vis. 2010;16:2007–15. [PMC free article] [PubMed] [Google Scholar]
- 8.Hejtmancik J, Kaiser-Kupfer M, Piatigorsky J. Vol. 8. New York: McGraw Hill; 2001. Molecular biology and inherited disorders of the eye lens; pp. 6033–62. [Google Scholar]
- 9.Shiels A, Hejtmancik JF. Genetics of human cataract. Clin Genet. 2013;84:120–7. doi: 10.1111/cge.12182. doi: 10.1111/cge.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–15. doi: 10.1016/j.survophthal.2004.02.013. doi: 10.1016/j.survophthal.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie RL, O’Sullivan J, Ashworth J, Bhaskar S, Williams S, Biswas S, et al. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology. 2014;121:2124–37.e1-2. doi: 10.1016/j.ophtha.2014.06.006. doi: 10.1016/j.ophtha.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Kondo Y, Saitsu H, Miyamoto T, Lee BJ, Nishiyama K, Nakashima M, et al. Pathogenic mutations in two families with congenital cataract identified with whole-exome sequencing. Mol Vis. 2013;19:384–9. [PMC free article] [PubMed] [Google Scholar]
- 13.Reis LM, Tyler RC, Muheisen S, Raggio V, Salviati L, Han DP, et al. Whole exome sequencing in dominant cataract identifies a new causative factor, CRYBA2, and a variety of novel alleles in known genes. Hum Genet. 2013;132:761–70. doi: 10.1007/s00439-013-1289-0. doi: 10.1007/s00439-013-1289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G, Wei X, Chen R, Zhou H, Li X, Sun Y, et al. A novel mutation of the SLC25A13 gene in a Chinese patient with citrin deficiency detected by target next-generation sequencing. Gene. 2014;533:547–53. doi: 10.1016/j.gene.2013.10.021. doi: 10.1016/j.gene.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Wei X, Ju X, Yi X, Zhu Q, Qu N, Liu T, et al. Identification of sequence variants in genetic disease-causing genes using targeted next-generation sequencing. PLoS One. 2011;6:e29500. doi: 10.1371/journal.pone.0029500. doi: 10.1371/journal.pone.0029500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–32. doi: 10.1101/gr.088013.108. doi: 10.1101/gr.088013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. doi: 10.1093/nar/gku340. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 22.Wu EL, Cheng X, Jo S, Rui H, Song KC, Dávila-Contreras EM, et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–93. doi: 10.1063/1.470117. [Google Scholar]
- 24.Goga N, Rzepiela AJ, de Vries AH, Marrink SJ, Berendsen HJ. Efficient algorithms for langevin and DPD dynamics. J Chem Theory Comput. 2012;8:3637–49. doi: 10.1021/ct3000876. doi: 10.1021/ct3000876. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Zhou Y, Lin X, Wong HC, Xu Q, Jiang J, et al. Molecular interaction and functional regulation of connexin50 gap junctions by calmodulin. Biochem J. 2011;435:711–22. doi: 10.1042/BJ20101726. doi: 10.1042/BJ20101726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou J, Salarian M, Chen Y, Veenstra R, Louis CF, Yang JJ. Gap junction regulation by calmodulin. FEBS Lett. 2014;588:1430–8. doi: 10.1016/j.febslet.2014.01.003. doi: 10.1016/j.febslet.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beyer EC, Ebihara L, Berthoud VM. Connexin mutants and cataracts. Front Pharmacol. 2013;4:1–14. doi: 10.3389/fphar.2013.00043. doi: 10.3389/fphar.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–5. doi: 10.1038/ng.499. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a and b) The overall structure of the connexin 50 chennal model in hydrophocipy surface representation.
The illustration of homo-6-mer connexin 50 protein/membrane complex models.