Abstract
Aim:
Salmonella is an important zoonotic pathogen responsible for animal and human diseases. The aim of the present study was to determine the prevalence and stereotyping of Salmonella isolates isolated from apparently healthy poultry. Furthermore, the clonal relatedness among the isolated Salmonella serovars was assessed.
Materials and Methods:
A total of 150 cloacal swab samples from apparently healthy chickens were collected, and were subjected for the isolation and identification of associated Salmonella organisms. The isolated colonies were identified and characterized on the basis of morphology, cultural characters, biochemical tests, slide agglutination test, polymerase chain reaction, and pulsed-field gel electrophoresis (PFGE). Antibiotic sensitivity patterns were also investigated using commonly used antibiotics.
Results:
Of the 150 samples, 11 (7.33%) produced characteristics pink colony with black center on XLD agar medium, and all were culturally and biochemically confirmed to be Salmonella. All possessed serovar-specific gene SpeF and reacted uniformly with group D antisera, suggesting that all of the isolates were Salmonella Enterica serovar Gallinarum, biovar Pullorum and/or Gallinarum. Antimicrobial susceptibility testing revealed that 54.54% of the isolated Salmonella Enterica serovars were highly sensitive to ciprofloxacin, whereas the 81.81% isolates were resistant to amoxycillin, doxycycline, kanamycin, gentamycin, and tetracycline. Pulsed-field gel electrophoresis of the XbaI-digested genomic DNA exhibited identical banding patterns, suggesting that the multidrug resistant Salmonella Enterica serovars occurring in commercial layers are highly clonal in Bangladesh.
Conclusion:
The present study was conducted to find out the prevalence of poultry Salmonella in layer chicken and to find out the clonal relationship among them. The data in this study suggest the prevalence of Salmonella Enterica, which is multidrug resistant and highly clonal for commercial layers of Bangladesh.
Keywords: polymerase chain reaction, pulsed field gel electrophoresis, Salmonella, SpeF gene
Introduction
The genus Salmonella is phylogenically clustered in the family of Enterobacteriaceae [1,2]. The most Salmonella is motile, with the exception of the poultry-specific serovars of Salmonella Gallinarium and Salmonella Pullorum [2]. Salmonella is considered one of the most common causes of foodborne human infections worldwide [3]. The most characteristic feature of Salmonella is its wide host range, which comprises most animal species including all mammals, birds, and cold-blooded animals in addition to humans. Salmonella is one of the most important pathogens responsible for human food poisoning in the developed world, where chicken and chicken products are widely considered to be significant sources for this organism. Therefore, Salmonella has been isolated from a range of foods in almost all country in which it has been investigated [4]. Annual estimation of the incidence of nontyphoidal salmonellosis in the world is 1.3 billion, and annual death is estimated to 3 million cases [5]. The poultry population in Bangladesh is estimated to be 221.39 million of chickens and 41.23 million ducks [6]. About 50,000 chicken farms and 26,000 duck farms have already been set up in the private sector in addition to the Government farms. At present, there are more than 130 hatcheries producing 0.476 million day-old-chicks a week and about 1 million commercial layer and broiler farms supplying 0.6 million kg of poultry meat and 9.9 million table eggs per weeks [7]. However, the advancement of the poultry industry is seriously hampered due to the outbreak of various infectious and non-infectious diseases. Among the bacterial diseases, Salmonellosis is of the major problem [8]. Several environmental factors including air, dirty litter, feed, water and vectors, such as insects, humans, and rodents are responsible for Salmonella infection in poultry farms [9-11]. Salmonella infection remains as a serious problem to public health significance in worldwide and causes substantial economic loss resulting from mortality, morbidity, and poor growth with the hazard of transmitting food poisoning with gastroenteritis to human and represents a serious problem for the food industry [12] specially poultry meat. The possible source of Salmonella in poultry meat and eggs is due to cross contamination with feces. On the other hand, the emergence of antibiotic-resistant Salmonella has become a serious problem worldwide. Antimicrobials are generally applied to treat diseases as well as growth promoter in poultry, exposing a large number of birds to frequently subtherapeutic concentrations [13] and leading to the development of antimicrobial resistant pathogens that are subsequently transferred to humans through the food chain. Recently, the emergence of antibiotic-resistant Salmonella has also led to the ineffective treatment of salmonellosis by several antibiotics [14].
Salmonella serovars of poultry have been studied by most authors worldwide [10,11,15] and in Bangladesh [16-19]. However, most of the research works on Salmonella in poultry have been limited to phenotypic characterization. Data on molecular typing, such as polymerase chain reaction (PCR) and pulsed field gel electrophoresis (PFGE) are useful for epidemiological studies and is of value in identifying the potential risk to public health associated with poultry meat, eggs and other poultry products.
The aim of the present study was to determine the prevalence and stereotyping (serotypes, PCR, antimicrobial resistance patterns) of Salmonella isolates from apparently healthy looking poultry. In addition, the clonal relatedness among the isolated Salmonella serovars was assessed.
Materials and Methods
Ethical approval
All samples were collected as per standard sample collection procedure without harming or giving stress to any birds.
Sample collection and processing
To isolate the Salmonellla spp. the samples were collected from commercial poultry farms in Mymensingh region of Bangladesh. The droppings and cloacal swabs collected from birds were transferred to the laboratory at the Department of Microbiology and Hygiene, BAU, Mymensingh. The cloacal swabs were collected in a test tube containing nutrient broth followed by incubation at 37°C for 2 h. A total of 150 samples were collected and analyzed. After taking necessary permission from the poultry farm owners, the sample collection was performed as per the guidelines set by the Department of Microbiology and Hygiene, BAU.
Isolation and identification of Salmonella
The isolation and identification of Salmonella from the samples were performed according to the conventional bacteriological methods [20-24]. In short, after 2 h of incubation 10 fold dilutions was performed using nutrient broth (Himedia, India), vortexed and 0.1 ml was spread onto Salmonella-Shigella agar, followed by incubation at 37°C for overnight, and the plates were examined for characteristics colony produced by Salmonella spp.
The isolated colonies were identified and characterized on the basis of morphology, cultural characters, biochemical tests, slide agglutination test, PCR, and PFGE. The isolated organisms were identified by Gram’s staining method to determine their staining characteristics, morphology, arrangement, etc. Five basic sugar fermentation (glucose, maltose) tests were performed to identify the organism. The pure colony was identified by indole test, Methyl red, VP test and with API kit. Then the motility test was performed by hanging drop slide method and by culturing onto motility indole urea (MIU) media. Slide agglutination was performed using group D antisera (S and A Reagent Lab, Bankok, Thailand).
Amplification of Salmonella specific gene by PCR
To perform PCR bacterial DNA was extracted by boiling method, in brief, 100 µl distilled water was taken in an eppendorf tube, a pure bacterial colony from overnight cultured was mixed with the distilled water, boiled for 10 min and then immediately cooled on ice followed by centrifugation at 10,000 rpm for 10 min. The supernatant was collected and used as DNA template for PCR.
PCR was performed using the primer pair targetting SpeF forward 5´-TTAGCCGTCATTGCCCGGATT-3´ and SpeF reverse 5´-ACGAGGTTTAATGACGTAGC-3´. For the detection of Salmonella spp. PCR mixture was prepared by the following method. To make 25 µl of PCR master mixture, 12.5 µl of 2 X Master mixtures (Promega, USA), 1 µl of each primer (10 pmol/µl), 4 µl of DNA template and remaining 6.5 µl deionized water were added in PCR tube. To amplify the SpeF, PCR reaction was performed in 25cycles with initial denaturation 92°C for 5 min, denaturation 92°C for 30 sec, annealing 50°C for 1 min, extension at 72°C for 3 min and a final extension at 72°C for 5 min. The amplified products were resolved in 1% agarose gel stained with ethidium bromide, electrophoresed at 100 volt and examined under UV transilluminator.
PFGE
Whole agarose-embedded genomic DNA from the Salmonella isolates was prepared. PFGE was carried out using a contour-clamped homogeneous electrical field-DRII apparatus (Bio-Rad), according to procedures described elsewhere [25]. Genomic DNA of the test strains was digested by XbaI restriction enzyme (Gibco-BRL, Gaithersburg, MD), and Salmonella Enterica serovar Braenderup was digested using XbaI, with fragments employed as molecular size markers. Restriction fragments were separated in 1% pulsed-field-certified agarose in 0.5X TBE (Tris-borate-ethylenediaminetetraacetic acid) buffer. Post-electrophoresis gel-treatment included gel stained and de-stained. The DNA was visualized using a UV-transilluminator, and images were digitized via a one-dimensional gel documentation system (Bio-Rad). The fingerprint pattern in the gel was analyzed using a computer software package, Bionumeric (Applied Maths, Belgium). The fingerprint patterns were subjected to typing based on banding similarity and dissimilarity, using the dice similarity coefficient and unweighted pair group method employing average linkage (UPGMA) clustering, as recommended by the manufacturer. The results were graphically represented as a dendrogram.
Antimicrobial susceptibility test
The antimicrobial susceptibility tests of the isolates were performed by disc diffusion method according to CLSI [26]. The antibiotic discs impregnated with ciprofloxacin (5 mcg), amoxycillin (10 mcg), doxycycline (30 mcg), kanamycin (30 mcg), gentamycin (10 mcg), tetracycline (30 mcg) (Oxoid, UK) were used in this study.
Results
In the present study out of 150 samples collected, eleven samples were identified as positive for Salmonella by conventional techniques, API 20E system, serotyping and molecular methods. On S-S agar the Salmonella isolates produced translucent, black, smooth, small round colonies. Microscopic examination of Gram stain revealed Gram-negative, pink colored short rod-shaped bacteria, arranged either single or paired. All of the test isolates were indole and V-P negative and Methyl Red test positive, fermented glucose and maltose and produced both acid and gas. The isolates were agglutinated with group D antiserum, indicated that all 11 isolates were Salmonella Pullorum and/or Salmonella Gallinarum. All isolates were found to be non-motile characterized by forming the stab line without producing turbidity in the MIU medium. All the isolated Salmonella were confirmed as Salmonella Pullorum and/or Salmonella Gallinarum by PCR targeting SpeF gene. The SpeF genes specific 2 kb amplicon sizes (Figure-1) were successfully amplified in all isolates. In this study out of 150 cloacal swabs, Salmonella Pullorum and Gallinarum were isolated from 5 (3.33%) and 6 (4.00%) birds and differntiated based on biochemical characters. The biovars Salmonella Gallinarum and Pullorum were differentiated based on different sugar fermentation tests particularly dulcitol fermentation test. The biovar, Gallinarum can ferment galactose and dulcitol. In our study total 6 of the biovars fermented galactose and dulcitol and 5 isolates were found non-fermenter. All 5 isolates were fermented glucose and confirmed as Pullorum. All the isolated 11 Salmonella spp. was subjected to PFGE to determine their clonal relatedness. The XbaI- restriction enzyme digested the genomic DNA and produced 16-20 fragments (Figure-2) ranged from 550 to 20-kb of molecular sizes. Cluster analysis, which was performed with dendrograms (prepared by dice similarity coefficient and UPGMA clustering methods), revealed a single cluster with 100% similarity coefficient (Figure-2). Antimicrobial susceptibility test using six drugs revealed that 54.54% of the isolated Salmonella Enterica serovars were highly sensitive to ciprofloxacin, whereas 81.81% isolates were resistant to amoxycillin, doxycycline, kanamycin, gentamycin, and tetracycline.
Figure-1.
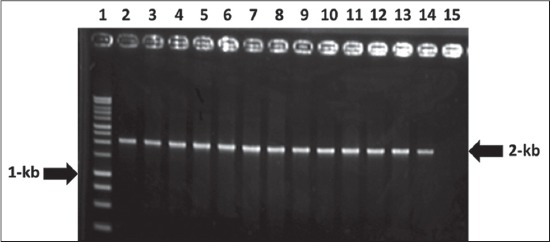
Lane 1: 1-kb DNA ladder, Lane 3-13: Tested samples positive for SpeF gene. Lane 2, 14 positive control, and Lane 15 negative control.
Figure-2.
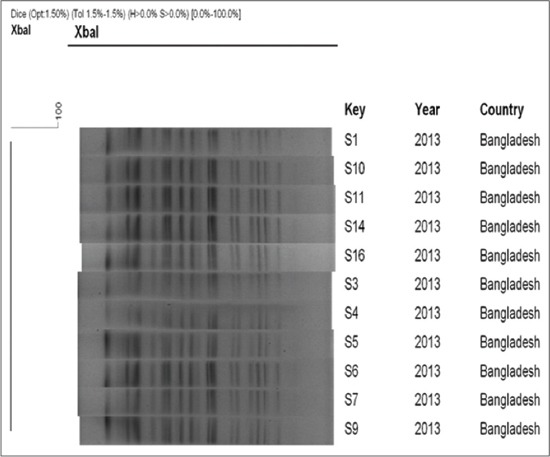
Genomic fingerprinting of Salmonella isolated from poultry farm of Bangladesh Agriculture University in 2013. The dendrogram was constructed by dice similarity coefficient and UPGMA clustering method using pulsed-field gel electrophoresis images of XbaI- restriction digested genomic DNA of the tested Salmonella strains; the scale bar at the top (left) indicates similarity coefficient.
Discussion
Salmonella Gallinarum and Salmonella Pullorum are non-motile pathogens that infect poultry and other galliform birds [27]. Salmonella Gallinarum is responsible for fowl typhoid, and Salmonella Pullorum causes Pullorum disease, which is characterized by white diarrhea in chicks [28]. SG, found mostly in adult birds, can also affect young birds. Salmonella Pullorum may infect older chickens, causing symptoms similar to those observed in fowl typhoid. Therefore, the proper identification of both Salmonella serovars is a very important from the epidemiological and control standpoints.
In the present study, we identified 7.33% Salmonella in healthy layer chicken. The results are in agreement with [29] in Basrah city who found that the overall presence of Salmonella spp. was 9.2%. On the basis of cultural, serological, biochemical and molecular characterization, i.e., the overall prevalence of Salmonella in a cloacal swab of healthy layers is 7.33% in Mymensingh region of Bangladesh. Several authors reported that unlike other Salmonella serotypes, Pullorum and Gallinarum are not excreted extensively in feces [30]. Rahaman et al. [30] observed that in the absence of clinical signs bacteriological assay of faecal swabs for Pullorum or Gallinarum will not be easy. The most important observation in this study was the excretion of Salmonella Pullorum in healthy flocks which needs to be considered when flocks are stressed due to other factors as such excretions can be a source of infection in multi-age farms. Salmonella Enterica serovar Pullorum and Gallinarum are worldwide a poultry pathogen of considerable economic importance, particularly a developing poultry industry. Salmonella Enterica serovar Pullorum colonized both the ovary and the oviduct of hens and led to 6% of laid eggs being infected by Salmonella serovar Pullorum. The colonization of several different sites of the reproductive tract suggests that serovar Pullorum may employ more than one mechanism of egg infection [18].
Although Salmonella Pullorum and Salmonella Gallinarum cause different diseases in poultry, they are very similar in genetically and phenotypically and it is very difficult to differentiate these two Salmonellae. Salmonella Gallinarum is responsible for fowl typhoid, and Salmonella Pullorum causes Pullorum disease, both are non-motile and contain the same somatic antigenic structure [31]. The differentiation between Salmonella Pullorum and Salmonella Gallinarum is very important from epidemiological and preventive perspectives and also for the development of vaccine. As these two organisms are very similar, and cannot be distinguished by conventional and serological methods [31]. The biovars Salmonella Gallinarum and Pullorum were differentiated in the study based on different sugar fermentation tests particularly dulcitol fermentation test. This biovar differentiation concurs with the report of Rahaman et al. [30] who differentiated biovar by the use of sugars such as maltose, dulcitol and glucose.
Antibimicrbial resistance in Salmonella has assumed alarming proportion worldwide [32]. It is associated with improper use of antimicrobial agent and has been reported to occur mostly in hosts that receive the antimicrobial drugs. In this study, multidrug resistance Salmonella were isolated might be due to wide use of antibiotics in commercial poultry farms. Antibiogram using six drugs revealed 54.54% isolates were sensitive to ciprofloxacin and 81.81% isolates were resistant to amoxycillin, doxycycline, kanamycin, gentamycin, and tetracycline. Ramya et al. [33] described that the sensitivity of Salmonella spp. was 100% for ciprofloxacin followed by amoxycillin (82%). Hyeon et al. [34] and Roy et al. [23] stated that in Salmonella the highest antibiotic resistance observed was to erythromycin (100%) followed by streptomycin (22.2%) and tetracycline and chloramphenicol (16.7%).
The isolation of Salmonella Gallinarum and Salmonella Pullorum from cloacal swab samples indicates the existence of fowl typhoid and Pullorum disease in intensive poultry farms in Mymensingh. All of the isolates showed identical restriction cleavage patterns in PFGE. The PFGE pattern of all the strains matched with each other in the number and position of the DNA fragments, suggesting genetic homogeneity. Pulsed-field gel electrophoresis of the XbaI-digested genomic DNA exhibited banding patterns that were identical (the similarity coefficient was 100%) for all the tested isolates which formed a tight cluster when dendrogram was constructed using the PFGE patterns, suggesting a high level of clonal relatedness among them. The data in this study suggest the prevalence of Salmonella Enterica, which is multidrug resistant and highly clonal for commercial layers of Bangladesh.
Conclusion
Salmonella is regarded as the major bacterial foodborne pathogen causing human illnesses worldwide. It cannot be debated that poultry meat and eggs are major vehicles for Salmonella transmission to human. As such, researchers, veterinarians, processors and the government are working in tandem to search for a new method of testing and characterization for Salmonella. The present study was conducted to find out the prevalence of poultry Salmonella in layer chicken and to find out the clonal relationship among them. The data in this study suggest the prevalence of Salmonella Enterica, which is multidrug resistant and highly clonal for commercial layers of Bangladesh.
Authors’ Contributions
MSP, MJ and MFRK have participated in isolation of samples and conventional identification of Salmonella spp., and drafting the manuscript. KHMNHN and MR have conducted the molecular identification of the bacteria. MBR and KHMNHN have critically checked the manuscript for finalization.
Acknowledgements
The authors are grateful to all researchers of the Department of Microbiology and Hygiene, Bang- ladesh Agricultural University, Mymensingh-2202, Bangladesh. The financial support from the Bang ladesh Agricultural Research Council (BARC) is accordingly acknowledged.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Bennasar A.G, Luna D, Cabrer B, Lalucat J. Rapid identification of Salmonella Typhimurium S. Enteritidis and S. virchow isolates by polymerase chain reaction based fingerprinting methods. Int. Microbiol. Microbiol. 2000;3:31–38. [PubMed] [Google Scholar]
- 2.Grimont P.A, Grimont F, Bouvet P. In: Taxonomy of the genus Salmonella, Salmonella in Domestic Animals. Wra C, Wray A, editors. New York, NY: CAB International; 2000. pp. 1–17. [Google Scholar]
- 3.Baird-Parker A.C. Foodborne salmonellosis. Lancet. 1990;336:1231–1235. doi: 10.1016/0140-6736(90)92844-8. [DOI] [PubMed] [Google Scholar]
- 4.Jalil M.A, Islam M.T. Serological survey of Salmonella infection in non-vaccinated commercial layer birds in Khulna District of Bangladesh. Bangladesh J. Vet. Med. 2011;9:27–31. [Google Scholar]
- 5.Tassios P.T. Molecular epidemiology of antibiotic resistance of Salmonella enteritidis during a 7-Year period in Greece. J. Clin. Microbiol. 1997;35:1316–1321. doi: 10.1128/jcm.35.6.1316-1321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DLS. Livestock Population. Khamarbari, Farmgate, Dhaka: Directorate of Livestock Serviece; 2009. [Google Scholar]
- 7.Kabir S.S.K. Poultry in Bangladesh:Awaiting the awakening. Proceeding of 4thInternational Poultry Show and Semina., from February 28 to March 2, 2003, Held in Bangladesh China Friendship Conference Centre, Agargaon, Dhaka. 2005:8–15. [Google Scholar]
- 8.Haider M.G, Chowdhury E.H, Khan M.A.H.N, Hossain M.T, Rahman M.S, Song H.J, Hossain M.M. Experimental pathogenesis of Pullorum disease with local isolate of Salmonella Entericaserovar. Entericasubspecies Pullorum in pullets in Bangladesh. Korean J. Poult. Sci. 2008;35:341–350. [Google Scholar]
- 9.Jones F.T, Axtell R.C, Rives D.V, Scheideler S.E, Tarver J, Walker R.L, Wineland M.J. A survey of Salmonella contamination in modern broiler production. J. Food Prot. 1991;54:502–507. doi: 10.4315/0362-028X-54.7.502. [DOI] [PubMed] [Google Scholar]
- 10.Hoover N.J, Kenney P.B, Amick J.D, Hypes W.A. Preharvest sources of Salmonella colonization in turkey production. Poult. Sci. 1997;76:1232–1238. doi: 10.1093/ps/76.9.1232. [DOI] [PubMed] [Google Scholar]
- 11.Amick-Morris J. Insect’s contribution to Salmonella transmission in turkey flocks. Morgantown, WV, USA: MS Thesi., West Virginia University; 1998. [Google Scholar]
- 12.Khan A.A, Melvin C.D, Dagdag E.B. Identification and molecular characterization of Salmonella spp. from unpasteurized orange juices and identification of new serotype Salmonella strain S. Enterica serovar. Food Microbiol. 2007;24:39–43. doi: 10.1016/j.fm.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 13.White D.G, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, McDermott P.F, McDermott S, Wagner D.D, Meng J. The isolation of antibiotic-resistant Salmonella from retail ground meats. New Eng. J. Med. 2001;345:1147–1154. doi: 10.1056/NEJMoa010315. [DOI] [PubMed] [Google Scholar]
- 14.Padungtod P, Kaneene J.B. Salmonella in food animals and humans in northern Thailand. Int. J. Food Microbiol. 2006;108:346–354. doi: 10.1016/j.ijfoodmicro.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Waldroup A.L. Contamination of raw poultry with pathogens. World Poult. Sci. J. 1996;52:7–25. [Google Scholar]
- 16.Ahmed A.K.M, Islam M.T, Haider M.G, Hossain M.M. Seroprevalence and pathology of naturally infected Salmonellosis in poultry with isolation and identification of causal agents. J. Bangladesh Agric. Univ. 2008;6:327–334. [Google Scholar]
- 17.Rahman M.R, Shahinuzzaman A.B.M, Saha A.K, Sufian M.A, Rahman M.H, Hossain M.M. Prevalence of Salmonella infection in naturally infected layer birds in Bangladesh. Bangladesh Vet. 2011;28:8–18. [Google Scholar]
- 18.Saha A.K, Sufian M.A, Hossain M.I, Hossain M.M. Salmonellosis in layer chickens:Pathological features and isolation of bacteria from ovaries and inner content of laid eggs. J. Bangladesh Agric. Univ. 2012;10:61–67. [Google Scholar]
- 19.Akond M.A, Shirin M, Alam S, Hassan S.M.R, Rahman M.M, Hoq M. Frequency of drug resistant Salmonella spp. isolated from poultry samples in Bangladesh. Sci. J. Microbiol. 2012;2:15–19. [Google Scholar]
- 20.Rybolt M.L, Wills R.W, Bailey R.H. Use of secondary enrichment for isolation of Salmonella from naturally contaminated environmental samples. Poult. Sci. 2005;84:992–997. doi: 10.1093/ps/84.7.992. [DOI] [PubMed] [Google Scholar]
- 21.Husna A.A, Islam M.A, Rahman M.T, Khatun M.M. Efficacy of vinegar, sorbitol and sodium benzoate in mitigation of Salmonella contamination in betel leaf. J. Adv. Vet. Anim. Res. 2015;2:190–194. [Google Scholar]
- 22.Haider M.G, Chowdhury E.H, Ahmed A.K.M, Hossain M.M. Experimental pathogenesis of Pullorum disease in chicks by local isolate of Salmonella Pullorum in Bangladesh. J. Bangladesh Agric. Univ. 2012;10:87–94. [Google Scholar]
- 23.Roy S.R, Rahman M.B, Hassan J, Nazir K.H.M.N.H. Isolation and identification of bacterial flora from internal organs of broiler and their antibiogram studies. Microbes Health. 2012;1:72–75. [Google Scholar]
- 24.Hasan M.A, Mamun A.A, Uddin A.S.M.A, Hassan M.Z, Hasan H, Rahman M.H. Investigation into gyneco-pathological disorders and identification of associated bacteria from the genital organs of cows in Dinajpur, Bangladesh. J. Adv. Vet. Anim. Res. 2015;2:165–169. [Google Scholar]
- 25.Ribot E.M, Fair M.A, Gautom R. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7 Salmonella and Shigella for pulse net. Foodborne Pathog. Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 3. Vol. 32. Wayne, PA: CLS; 2007. pp. 32–35. [Google Scholar]
- 27.Shivaprasad H.L. Fowl typhoid and Pullorum disease. Rev. Sci. Tech. 2000;19:405–424. doi: 10.20506/rst.19.2.1222. [DOI] [PubMed] [Google Scholar]
- 28.Pomeroy B.S, Hofsta M.S, Barnes H.J, Calnek B.W, Reid W.M, Yoder H.W. Diseases of Poultry. 8th ed. Ames: Iowa State University Press; 1984. Fowl typhoid; pp. 79–91. [Google Scholar]
- 29.Al-Abadi I.K.M, AL-Mayah A.A.S. Isolation and identification of Salmonella spp. from chicken and chicken environment in Basrah province. Afr. J. Biol. Sci. 2011;7:33–43. [Google Scholar]
- 30.Rahaman M.T, Rahman M, Rahman M.B, Khan M.F.R, Hossen M.L, Parvej M.S, Ahmed S. Poultry Salmonella specific bacteriophage isolation and characterization. Bangladesh J. Vet. Med. 2014;12:107–114. [Google Scholar]
- 31.Paiva J.B, Cavallini J.S, Silva M.D, Almeida M.A, Angela H.L, Berchieri Junior A. Molecular differentiation of Salmonella Gallinarum and Salmonella Pullorum by RFLP of flic gene from Brazilian isolates. Braz. J. Poultr. Sci. 2009;11:271–276. [Google Scholar]
- 32.Aggarwal P, Sarkar R, Singh M, Grover B.D, Anand B.R, Raichowdhuri A.N. Salmonella bareilly infection in a paediatric hospital of New Delhi. Indian J. Med. Res. 1983;78:22–25. [PubMed] [Google Scholar]
- 33.Ramya P, Thirtham M, Eevuri T.R. Antimicrobial sensitivity and resistance of Salmonella enteritidis isolated from natural samples. Vet. World. 2013;6(4):185–188. [Google Scholar]
- 34.Hyeon J.Y, Chon J.W, Hwang I.G, Kwak H.S, Kim M.S, Kim S.K, Choi I.S, Song C.S, Park C, Seo K.H. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J. Food Prot. 2011;74:161–166. doi: 10.4315/0362-028X.JFP-10-327. [DOI] [PubMed] [Google Scholar]


