Abstract
We report programmable self-assembly of branched, 3D globular, monodisperse and nanoscale sized dendrimers using RNA as building blocks. The central core and repeating units of the RNA dendrimer are derivatives of the ultrastable three-way junction (3WJ) motif from the bacteriophage phi29 motor pRNA. RNA dendrimers were constructed by step-wise self assembly of modular 3WJ building blocks initiating with a single 3WJ core (Generation-0) with overhanging sticky-end and proceeding in a radial manner in layers up to Generation-4. The final constructs were generated under control without any structural defects in high yield and purity, as demonstrated by gel electrophoresis and AFM imaging. Upon incorporation of folate on the peripheral branches of the RNA dendrimers, the resulting constructs showed high binding and internalization into cancer cells. RNA dendrimers are envisioned to have a major impact in targeting, disease therapy, molecular diagnostics and bioelectronics in the near future.
Keywords: pRNA, Three-way junction, RNA nanotechnology, RNA nanoparticle
GRAPHICAL ABSTRACT
We report the construction of RNA dendrimers utilizing the highly stable pRNA-3WJ motif as a core scaffold. Each of the component strands were synthesized and then self-assembled sequentially to construct 3D globular Generation–0 to Generation–4 RNA dendrimers with high yield and purity. RNA dendrimers are envisioned to have a major impact in nanotechnology and nanomedicine, due to their structural diversity, thermodynamic stability, monodisperse nature, nanoscale size, polyvalent property, ease of synthesis, high loading capacity, and simple derivatization with desired functional modules prior to assembly.
INTRODUCTION
In recent years, extensively branched 3D structures called dendrimers1–3 have become an attractive platform for building multifunctional macromolecular nanomaterials as diagnostic and therapeutic agents4–8. This is primarily because of the polyvalent nature, nanoscale size, highly branched, and void-space containing framework of dendrimers, which offers high loading capacity of desired functionalities. The attractive and intriguing dendrimers were conceptualized over four decades ago1; however, improvements are desirable for realizing their commercial availability in clinical settings. For example, the multi-step synthetic reactions necessary to generate defect-free dendrimer that impact product yield and purity. Incorporation of different functional modules to generate a multifunctional framework often requires series of protection–deprotection steps that affects synthetic yield, water solubility and even functionality. Traditional small molecule building blocks, such as high molecular weight carrier poly(amidoamine) (PAMAM) and poly (propyleneimine) (PPI) dendrimers are mostly smaller than 10 nm with angstrom level size differences within successive generations, make it desirable to seek alternate building blocks to enhance their size in order to avoid rapid renal clearance for in vivo applications.
Larger building blocks such as nucleic acids can overcome the size limitation. The self-assembly properties of nucleic acids can prevail over the complex synthetic routes to generate precisely assembled dendrimers under control. It has been proposed that DNA can serve as building blocks3, and the construction of DNA dendrimers by enzymatic ligation9, and recently, by hybridization using sticky end segments6,10–13 have been reported. RNA dendrimers have not been realized to date due to challenges in predicting intra- and inter-molecular RNA folding involving canonical and non-canonical base pairing, base stacking, and tertiary interactions14,15, as well as concerns regarding the chemical stability of RNA16–18.
Recent progress in RNA nanotechnology has enabled construction of varieties of RNA nanostructures with precise control of size, shape and stoichiometry16–21. Several structural RNA motifs have been used as scaffold to construct varieties of 2D and 3D architectures including, branched RNA nanoparticles22–24, packaging RNA (pRNA) hexamers24,25, nanorings26,27, triangles28,29, square30–32, pentagons33; cube34, and origami structures35. Recently we discovered a robust 3WJ (three-way junction) motif derived from pRNA36 of the phi29 bacteriophage DNA packaging motor22 that can serve as a scaffold for fabricating RNA nanoparticles. The pRNA-3WJ assembles from three pieces of RNA oligonucleotides, is unusually thermodynamically stable (Tm of 59°C; ΔG°37°C of −28 kcal/mol), is resistant to denaturation even in presence of 8 M urea, and remains intact at ultra-low concentrations in vivo22,23,37. We have functionalized the pRNA-3WJ with targeting, imaging and therapeutic modules, and the resulting RNA nanoparticles have been used for cancer targeting22–24,38–40 and therapy41,42, as well as for resistive biomemory applications 43. More recently, we solved the crystal structure of the pRNA-3WJ scaffold44, which has facilitated the designs of multifunctional RNA nanoparticles. RNA has also been reported as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry28,45. Herein, we report for the first time the construction of RNA dendrimers utilizing the highly stable pRNA-3WJ motif as core scaffold. Each of the component strands were synthesized by transcription or chemically using phosphoramidite chemistry and then self-assembled sequentially to construct 3D globular Generation-0–4 (G-0–G-4) RNA dendrimers. The assembly of RNA dendrimers employs modular design principles and is highly controllable. Functional groups can be introduced with relative ease both internally as well as at the peripheral ends of the component strands prior to the assembly process, thus resulting in homogenous (monodisperse) RNA dendrimers with high yield and purity.
METHODS
RNA synthesis and purification
RNA oligonucleotides were prepared by chemical synthesis using a oligo synthesizer, custom ordered from Trilink Biotechnologies, Inc.(San Diego, CA, USA) or generated by in vitro transcription of respective PCR amplified dsDNA containing the T7 promoter46. RNA strands were purified by HPLC or by 8% Urea-PAGE. Single-stranded DNA templates and primers were purchased from Integrated DNA Technologies (Coralville, IA, USA).
Construction of G-0 to G-4 dendrimers
Dendrimers structure G-0, G-1, G-2 and G-3 were constructed using one-pot self-assembly of component strands (see sequences below). For G-0, the three component strands (3WJ-a):(3WJ-b):(3WJ-c) = 1:1:1 molar ratio; For G-1, the five component strands (SquareA): (SquareB):(SquareC):(SquareD):(SquareE) = 1:1:1:1:1 molar ratio. For G-2, the seven component strands (b-SquareA):(b-SquareB):(b-SquareC):(b-SquareD):(SquareE): (3WJ-a):(3WJ-c) = 1:1:1:1:1:4:4 molar ratio. For G-3, the nine component strands (b-SquareA):(b-SquareB):(b-SquareC):(b-SquareD):(SquareE):(3WJ-c-b_rev):(3WJ-a-b_rev):(3WJ-a_rev): (3WJ-c_rev) = 1:1:1:1:1:4:4:8:8 molar ratio. The required dendrimer RNA strands were mixed together in stoichiometric ratio in TMS buffer (25 mM Tris, 50 mM NaCl, 5 mM MgCl2) and the mixture was heated to 95°C for 5 min and then slowly cooled to 4°C at a rate of 2°C/min on an Eppendorf thermocycler. The assembly efficiency was verified using 8% native PAGE or 2% agarose gel.
For the construction of G-4 dendrimer, a two-step approach was used. First, the dendrimer nanostructure G-2 with sticky ends was constructed by mixing together required RNA strands with appropriate stoichiometry in TMS buffer. The mixture was heated to 95°C for 5 min and then slowly cooled as described above for the construction of G-1, G-2 and G-3 dendrimer generations. The free strands were removed by passing the mixture through Amicon Ultracentrifugal spin filters with 100 KDa cut off following manufacturer protocol and washed twice with TMS buffer. The pure G-2 with sticky ends was collected from column. In second step, the G-2 with sticky ends was mixed in stoichiometric ratio with other RNA strands needed to form G-4. The final molar ratio of the 11 G-4 component strands are (b-SquareA):(b-SquareB):(b-SquareC):(b-SquareD):(SquareE):(3WJ-c-b_rev):(3WJ-a-b_rev):(3WJ-b-a_rev): (3WJ-b-c_rev):(3WJ-a):(3WJ-c) = 1:1:1:1:1:4:4:8:8:16:16. The mixture was incubated at room temperature for 1 hour. Then, an agarose gel was run to check assembly efficiency. G-4 was constructed with a maximum final concentration of 1 μM in TMS buffer.
The sequences of all the strands of the RNA dendrimers are as follows (Sequence 5′→ 3′):
3WJ-a: UUG CCA UGU GUA UGU GGG
3WJ-b: CCC ACA UAC UUU GUU GAU CC
3WJ-c: GGA UCA AUC AUG GCA A
3WJ-a_rev: GGG UGU AUG UGU ACC GUU
3WJ-b_rev: CCU AGU UGU UUC AUA CAC CC
3WJ-c_rev: AAC GGU ACU AAC UAG G
3WJ-c-folate: GGA UCA AUC AUG GCA A–folate
SquareA: GGG AGC CGU CAA UCA UGG CAA GUG UCC GCC AUA CUU UGU UGC ACG CAC
SquareB: GGG AGC GUG CAA UCA UGG CAA GCG CAU CGC AUA CUU UGU UGC GAC CUA
SquareC: GGG AGG UCG CAA UCA UGG CAA CGA UAG AGC AUA CUU UGU UGG CUG GAG
SquareD: GGG ACC AGC CAA UCA UGG CAA UAU ACA CGC AUA CUU UGU UGA CGG CGG
SquareE: GGA CAC UUG UCA UGU GUA UGC GUG UAU AUU GUC AUG UGU AUG CUC UAU CGU UGU CAU GUG UAU GCG AUG CGC UUG UCA UGU GUA UGG C
b-SquareA: GGC CCA CAU ACU UUG UUG AUC CAU GGU GCG UAG GGU CGU CAA UCA UGG CAA GUG UCC GCC AUA CUU UGU UGC ACU CCC UUG CUC AUC A
b-SquareB: GGC CCA CAU ACU UUG UUG AUC CUG AUG AGC AAG GGA GUG CAA UCA UGG CAA GCG UAU CGC AUA CUU UGU UGA GAA CCC UAU GUG ACU U
b-SquareC: GGC CCA CAU ACU UUG UUG AUC CAA GUC ACA UAG GGU UCG CAA UCA UGG CAA CGA UAG AGC AUA CUU UGU UGG AGU CCC UUA GAG UAG A
b-SquareD: GGC CCA CAU ACU UUG UUG AUC CUC UAC UCU AAG GGA CUC CAA UCA UGG CAA UAU ACA CGC AUA CUU UGU UGA CGA CCC UAC GCA CCA U
3WJ-a-b_rev: UUG CCA UGU GUA UGU GGG CCU AGU UGU UUC AUA CAC CC
3WJ-c-b_rev: GGA UCA AUC AUG GCA A CCU AGU UGU UUC AUA CAC CC
3WJ-b-a_rev: CCC ACA UAC UUU GUU GAU CC GGG UGU AUG UGU ACC GUU (20) 3WJ-b-c_rev: CCC ACA UAC UUU GUU GAU CC AAC GGU ACU AAC UAG G
Gel Analysis
RNA dendrimers were analyzed using 2% agarose gel and 6–8% PAGE. The gels were prepared and ran in TMS buffer at 90 V for 1 hr. The gels were stained with ethidium bromide and scanned using Typhoon FLA 7000 (GE Healthcare, Cincinnati, OH, USA).
AFM Imaging
Dendrimer structures were imaged using APS modified mica surfaces with a Multi-Mode AFM Nano-Scope IV system (Veeco/Bruker) Santa Barbara , CA, USA), operating in tapping mode, as described previously47.
Serum stability assay
The chemical stability of RNA dendrimers was studied by incubating the dendrimers with 10% Fetal Bovine Serum (FBS) at 37°C at final concentration of 1 μM. 10 μL of samples were collected at each time point (0, 0.16 hr, 1 hr, 3 hr, 6 hr, 18 hr, and 24 hr) and were subjected to a 2% agarose gel assay with TBM running buffer. The gel was run at 120 V for 120 minutes, imaged by Typhoon FLA 7000 (GE Healthcare, Cincinnati, OH, USA), and gel bands quantified by Image J.
Flow cytometry analysis
Folate receptor (+) KB Cells were cultured in T75 flask and harvested at a density of 1×106 cells/ml with 0.25% trypsin. Cells were washed with Opti-MEM medium once and then incubated with different concentration of Cy5 labeled 2′-F RNA dendrimers in Opti-MEM medium for 1 hr at 37°C, protected from light. Cells were then washed three times and suspended in PBS (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) for analysis.
Confocal Imaging
KB cells were grown in cover glass (Thermo Fisher Scientific, Waltham, MA, USA) in 24 well bottom culture dishes in its complete medium overnight. Cells were seeded at a density of 2×105 cells/ml in a volume of 500 μl. On the day of testing, cells were washed with Opti-MEM medium twice, and RNA dendrimer (with whole chain Cy5 labeled SquareE-strand) were suspended in 200 μl of Opti-MEM medium and incubated with cells at 37°C for 1 hr. Cells were washed with PBS twice, fixed with 4 % paraformaldehyde solution (Microscopy Sciences, Hatfield, PA, USA) in PBS at room temperature for 20 min, then washed with PBS and permeabilized with 0.05% Triton-X 100 in PBS at room temperature for 3 min. The cells were then stained with Alexa488 phallodin (Thermo Fisher Scientific, Waltham, MA, USA) at room temperature for 20 min, the wells washed with PBS and air dried. The cells were stained with gold antifade (Thermo Fisher Scientific, Waltham, MA, USA) for DAPI staining for confocal microscopy imaging with Zeiss LSM 510 laser scanning system.
RESULTS
Design and assembly of parts and intermediates for RNA dendrimers
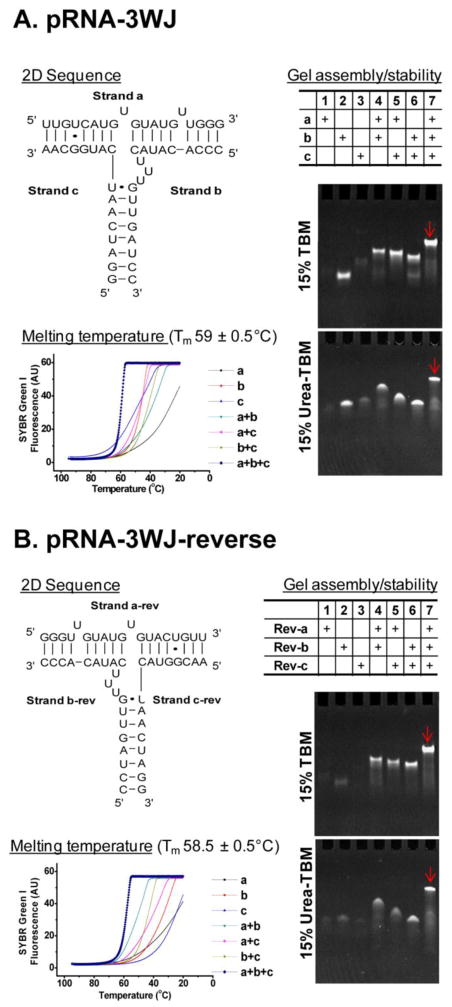
We developed three modules as building blocks for construction of RNA dendrimers. Module-1 is the 3WJ motif composed of three individual strands (3WJ-a, 3WJ-b and 3WJ-c) with three terminal ends (Figure 1A). In order to build higher order structures, we need to interconnect the 3WJ motifs in a radial manner, and this requires using two different 3WJs with near identical folding properties to avoid cross-talk between layers. Otherwise, misfolding and aggregation can occur. We generated a reverse pRNA-3WJ, denoted pRNA-3WJ-rev, which is a mirror image of the pRNA-3WJ with the 5′- and 3′-ends switched (Figure 1B). The pRNA-3WJ-rev has near identical properties as the conventional pRNA-3WJ with regards to thermodynamic stability, as well as assembly in native and 8 M denaturing PAGE.
Figure 1.
(A–B) 2D sequence; assembly in 15% native PAGE and stability in 8 M urea 15% PAGE; and thermodynamic properties of pRNA-3WJ (A) and pRNA-3WJ-rev (B) scaffold.
Module-2 consists of three sets of pRNA-3WJ building blocks, each harboring one of the component strands of the pRNA-3WJ-rev serving as sticky ends (3WJ-a_rev, 3WJ-b_rev and 3WJ-c_rev). The use of 3WJ core strands as sticky ends ensures near spontaneous 3WJ assembly with minimal non-specific hybridization, which can otherwise lead to misfolding, and also eliminates the need for additional linking sequences between 3WJs. Upon mixing the three building blocks in stoichiometric ratio, the pRNA-3WJ-rev assembles from the three sticky-ends thereby generating module-2 with high efficiency (Figure 2). Module-2 is composed of 5 component strands and has 6 terminal ends.
Figure 2.
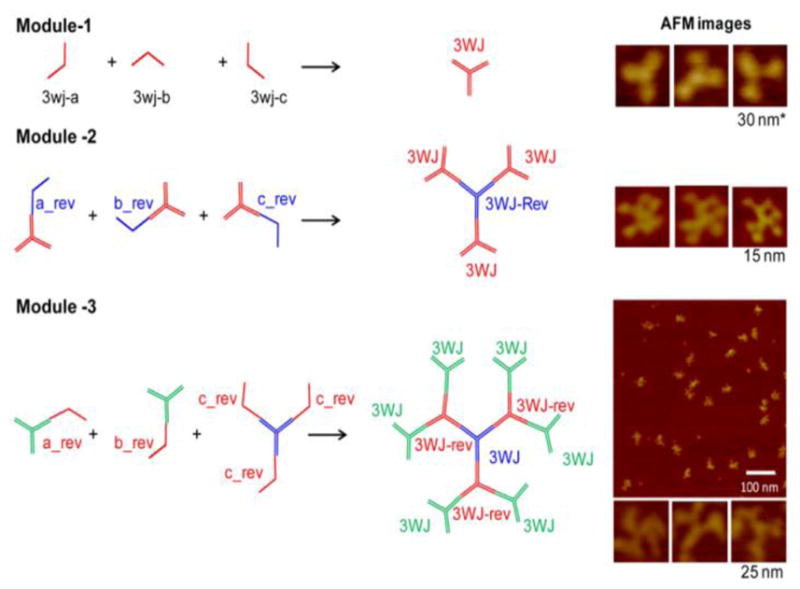
Step-wise assembly of three modules used as building blocks for RNA dendrimer construction. Module-1 assembles for three strands: 3WJ-a, 3WJ-b, and 3WJ-c. Module-2 assembles from three units: pRNA-3WJ harboring a_rev, pRNA-3WJ harboring b_rev, and pRNA-3WJ harboring c_rev sticky-ends. Module-3 assembles from three units: pRNA-3WJ harboring a_rev, pRNA-3WJ harboring b_rev, and pRNA-3WJ harboring three c_rev sticky ends. Right panel: AFM images of the modular building blocks. For module-1, the AFM image is for 3WJ with three pRNA subunits at the three branches.
Similarly, module-3 was constructed from iterative step-wise assembly on top of module-2 complexes by branch extension using one of the 3WJ component strands as sticky ends. The final complex is composed of 7 strands and 12 terminal ends with alternating pRNA-3WJ core and pRNA-3WJ-rev cores (Figure 2).
All three modules can be assembled either in one step or step-wise by simply mixing the strands at room temperature in TMS buffer (40 mM Tris, 10 mM MgCl2, 100 mM NaCl) or by annealing (heating to 95°C and cooling to room temperature over the course of 1 hour). The use of these modules will enable construction of RNA dendrimers with desired functional moieties, such as RNAi reagents siRNA, miRNA, anti-miRNA; chemotherapeutic drug; receptor targeting RNA aptamer or chemical ligand; riboswitch; ribozyme; endosome disrupting agents; and imaging fluorophores or radiolabels (Figure 3).
Figure 3.
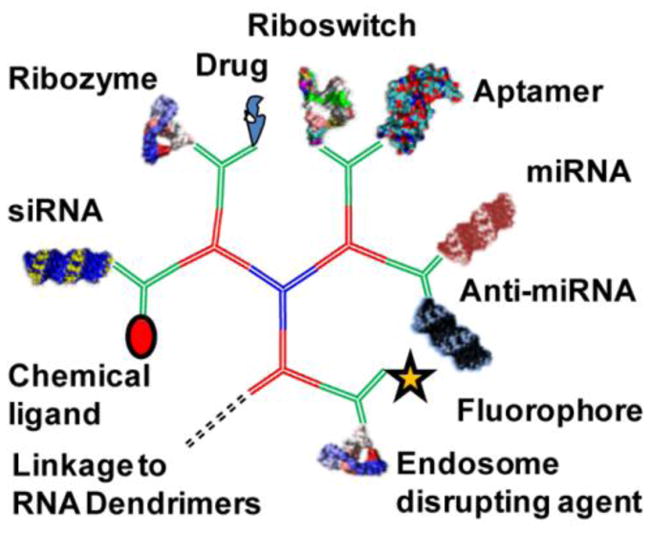
Construction of multifunctional RNA dendrimers. Functionalization of module-3 with varieties of moieties for targeting (chemical ligand or RNA aptamer), imaging (fluorophore or radiolabel), and therapeutic (siRNA, miRNA, anti-miRNA, chemotherapuetic drugs, ribozyme, riboswitch, endosome disrupting agents) units.
Design and assembly of RNA dendrimers G-0 to G-4
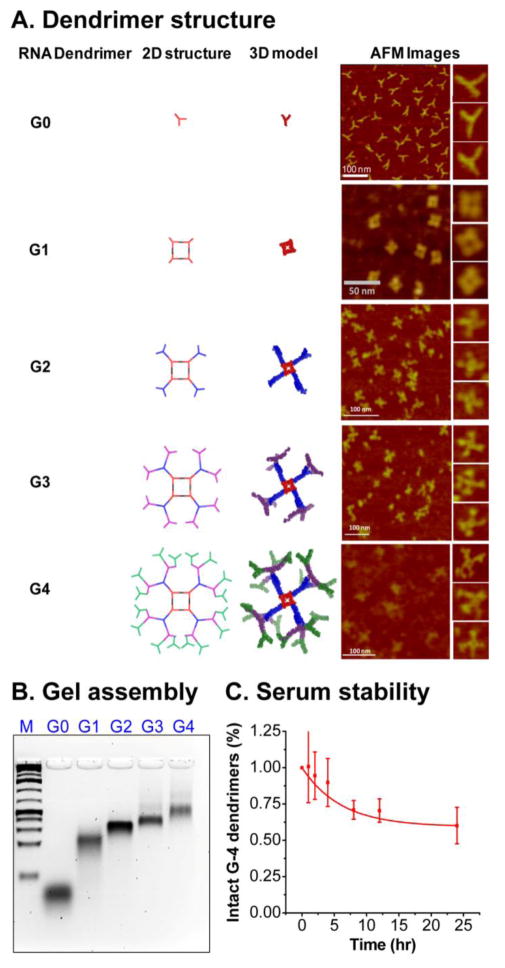
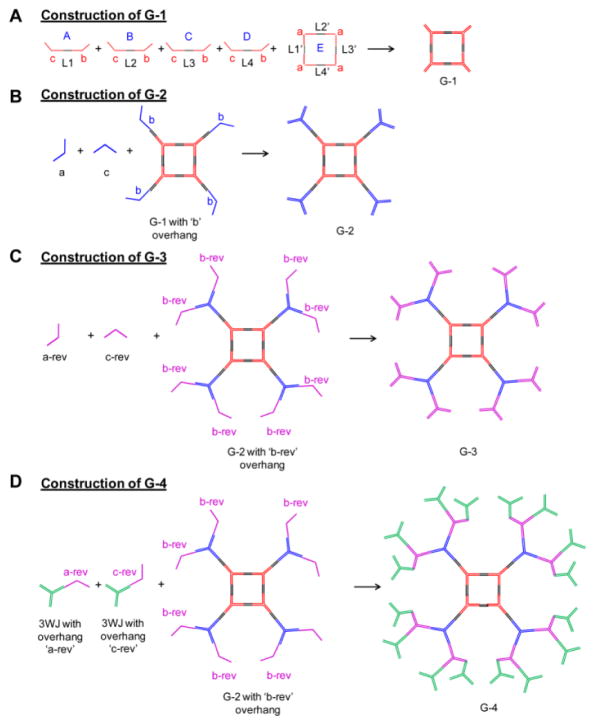
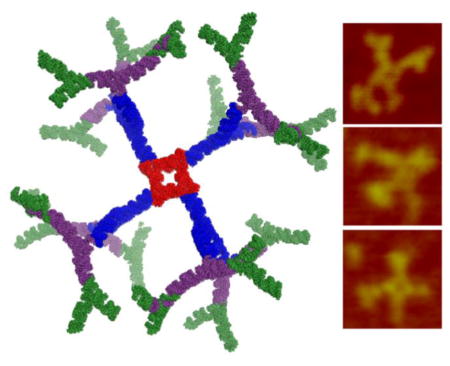
RNA dendrimers have similar architectures as small molecule based dendrimers consisting of an initiating core, interior layer of repeating units, and terminal units at the peripheral ends of the outermost generation. In the traditional sense, we adopted a ‘divergent growth’ method, whereby the dendrimer initiates from a core site and grows in a radial manner layer by layer. We used the pRNA-3WJ as G-0 initiating core (Figure 2A, 5A–B) and then constructed a planar square nanoparticle to serve as G-1 (Figure 4A, 5A–B). The pRNA-3WJ scaffold is highly tunable and the internal angle can be stretched from native 60° to 90° in the square configuration, as demonstrated previously30. The pRNA-3WJ cores at each vertex is linked by double stranded RNA sequences, with the entire square construct (G-1) composed of 5 strands and 4 terminal ends (MW ~83 KDa) (Figure 4A). The square pattern ensures equal spacing between the protruding corner branches and minimizes steric hindrances for addition of building blocks. We then adopted a similar strategy as aforementioned using one of the 3WJ component strands as sticky ends to sequentially construct G-2 (7 unique strands; 8 terminal ends; MW ~187 kDa) (Figure 4B, 5A–B), G-3 (9 unique strands; 16 terminal ends; MW ~ 328 kDa) (Figure 4C, 5A–B) and finally G-4 (11 unique strands; 32 terminal ends; MW ~ 610 kDa) (Figure 4D, 5A–B) (Table 1). G-0 through G-3 dendrimers can be assembled with high yield by mixing the component strands in equimolar ratio followed by annealing, except for G-4, which requires an additional incubation step with module-2 (Figure 2, 4D). Our design does not require any enzymatic ligation step and simply relies on self-assembly of individual blocks for the construction of higher order dendrimers. All the sequences for dendrimer generations were optimized to avoid any nonspecific interactions using the Mfold algorithm 48.
Figure 5.
(A) 2D structure (left), 3D model (middle) and AFM images (right) (using Pymol and Swiss PDB viewer) of G-0 to G-4 RNA dendrimers. (B) 2% agarose gel showing assembly of G-0 to G-4 RNA dendrimers. (C) Serum stability assay of G-4 RNA dendrimers assayed in 2% agarose gel and quantified by ImageJ.
Figure 4.
Assembly of G-1 to G-4 RNA dendrimers from component strands. G-0 assembles for three strands: 3WJ-a, 3WJ-b, and 3WJ-c, as shown in Figure 2A module 1. (A) G-1 assembles from five strands: A, B, C, D and E with complementary linkers (black) and 3WJ strands (red). (B) G-2 assembles from G-1 harboring four 3WJ-b sticky ends, and 3WJ-a and 3WJ-c strands. (C) G-3 assembles from G-2 harboring eight 3WJ-b_rev sticky ends, and 3WJ-a_rev and 3WJ-c_rev strands. (D) G-4 assembles from G-2 harboring eight 3WJ-b_rev sticky ends, and a dimer module harboring 3WJ-a_rev and 3WJ-c_rev strands. Please refer to Table 1 for defining characteristics.
Table 1.
| RNA Dendrimer | Predicted Size (nm) | Molecular Weight (kDa) | # of strands | # of nucleotides | Terminal units |
|---|---|---|---|---|---|
| G-0 | 6 | 17784 | 3 unique 3 total |
54 | 3 |
| G-1 | 12 | 82843 | 5 unique 5 total |
440 | 4 |
| G-2 | 37 | 186675 | 7 unique 13 total |
576 | 8 |
| G-3 | 50 | 327675 | 9 unique 29 total |
1008 | 16 |
| G-4 | 65 | 609675 | 11 unique 61 total |
1872 | 32 |
Characterization of RNA dendrimers for thermodynamic stability and serum stability
The thermodynamic stability of individual building blocks is of paramount importance to ensure that the RNA dendrimers remain intact without dissociating in vivo. Both the pRNA-3WJ and pRNA-3WJ-rev scaffold assemble with high efficiency, as shown in native PAGE (Figure 1A–B; top gel). Both 3WJs remain stable in presence of strongly denaturing 8 M urea, thereby demonstrating the robust attributes (Figure 1A–B; bottom gel). Melting experiments indicated that the three component strands have a much higher affinity to interact with one another than any of the one or two component strands. The slope of the melting curve is very steep indicating cooperative assembly of the three strands with very low free energy (ΔG°37°C of −28 kcal/mol), as calculated previously37. The Tm values are near identical for pRNA-3WJ and pRNA-3WJ-rev scaffolds with 59±0.5°C and 58.5±0.5°C, respectively (Figure 1A–B). Tm analysis of RNA dendrimers is challenging involving multiple strands and multi-step folding intermediates.
For successful clinical application of RNA dendrimers, a major hurdle is their stability in biological media, such as serum. Unmodified RNAs are intrinsically sensitive to degradation because of presence of nucleases in serum or in cells with a half-life varying from a few minutes to several hours40,49,50. Chemical modifications, such as 2′-Fluoro (2′-F) modification on the ribose sugar of the RNA, have been shown to be nuclease resistant 22,51,52 and also enhance the thermodynamic stability of RNA nanoparticles37. Within the dendrimer RNA sequences, the U and C nucleotides were replaced by 2′-F modified U and C nucleotides during solid phase synthesis or in vitro transcription. The 2′-F modified G-4 RNA dendrimers were assembled and showed similar assembly efficiency as unmodified G-4 RNA dendrimers in native gel assays. To test the serum stability, 2′-F modified G-4 dendrimers were incubated with 10% Fetal Bovine Serum (FBS) at 37°C. At specific time points, aliquots were extracted and snap frozen. Gel assays demonstrated that the 2′-F G-4 RNA dendrimers are resistant to nuclease degradation for more than 24 hours (Figure 5C), while unmodified constructs are degraded within 10–15 minutes.
Characterization of RNA dendrimer assembly by Atomic Force Microscopy (AFM) and gel electrophoresis
The assembly of dendrimers was tested by native gel electrophoresis (Figure 5B). G-0, G-1, G-2, G-3 and G-4 dendrimers all display a single band in the gels without any smears indicating high efficiency of the adopted assembly strategy. No significant change in assembly efficiency was observed by going from lower to higher generations. The free RNA strands were removed after dendrimer assembly using centrifugal membrane filter columns. The slow migration of the gel band from G-0 to G-4 clearly indicates the increasing size of the constructs, as expected.
To further confirm the results, all constructed dendrimer generations from G-0 to G-4 were examined by AFM imaging, which strongly support the formation of all dendrimers with branched conformations (Figure 5A). For G-4 dendrimers, the bifurcation in the arms of outer layer can be easily observed and the central square is visible in most of the structures from G-1 to G-4. The dendrimer structures are indeed three dimensional as designed in silico utilizing the crystal structure of the planer 3WJ structural motif44. Swiss PDB Viewer (www.spdbv.vital-it.ch) and Pymol (https://www.pymol.org/) were used to align 3WJ building blocks into three dimensional structure models (Figure 5A), based on the available crystal structure of pRNA-3WJ (PDB ID: 4KZ2)44. The three dimensional nature of dendrimer make the structures to appear in different orientations on the surface for AFM imaging thus bringing dendrimer arms appear close or farther away from each other in different images. RNA dendrimers are highly negatively charged and do not aggregate in solution. This anionic nature and aggregation-free physical property will minimize non-specific cell entry, and entrapment by lung and spleen macrophages and liver Kupffer cells53.
Characterization of RNA dendrimers for cell receptor targeting
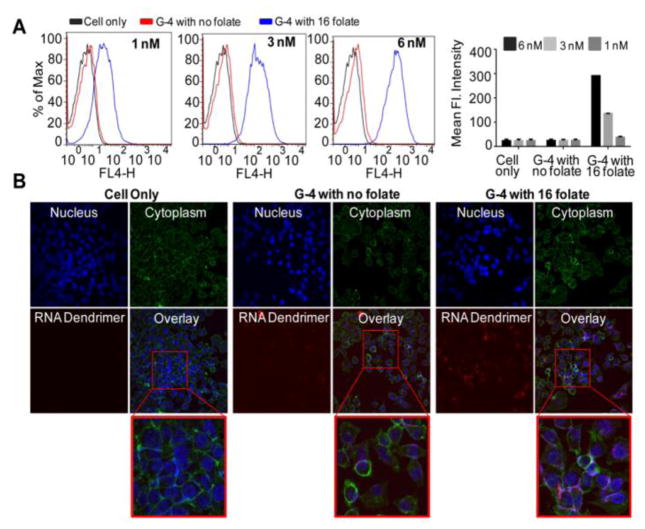
To study the potential applications of RNA dendrimers in targeting specific cells, 2′-F modified RNA dendrimers were assembled with targeting ligands at peripheral ends. Increased expression of folate receptors are frequently observed in cancer cells of epithelial origin54. Herein, we used folate as a targeting ligand. Within G-4, the core SquareE strand of G-1 was fluorescently tagged with Cy5 for detection. One of the terminal end sequences (3WJ-c) was labeled with folate at 3′-end (custom ordered from Trilink) and then used to assemble G-4 dendrimers resulting in 16 folates distributed evenly across the G-4 dendrimer surface. Folate receptor positive KB cells were incubated with G-4 dendrimers at different concentrations and the incubation efficiencies were analyzed using flow cytometry (Figure 6A). G-4 dendrimers with folate displayed increase in intracellular uptake with increasing RNA concentrations from 1 nM to 3 nM to 6 nM, compared to G-4 dendrimers without any folate. This is attributed to folate receptor mediated endocytosis. Confocal fluorescence microscopy (Figure 6B) images further confirmed the internalization of G-4 dendrimers with folates as indicated by strong overlap of cytoplasma (green) and dendrimers (red). The results demonstrate that RNA dendrimers would be useful to deliver various payloads including radioligand, drug molecules, and RNAi modules into the cells.
Figure 6.
(A) Flow cytometry data of G-4 RNA dendrimers at different concentrations (1 nM, 3 nM and 6 nM) with KB cells. Cell only (black), G-4 dendrimer with no folate (red); and G-4 with 16 folates (blue). Right: Summary of binding results. (B) Confocal microscope images of G-4 RNA dendrimers incubated with KB cells, nuclear staining by DAPI (blue), cell membrane staining by phalloidin Alexa488 (green), Cy5-labeled folate-G4 dendrimer (red) and a overlay of three panels. Bottom: Magnified view of overlayed image section boxed in red.
DISCUSSION
We demonstrated that RNA can serve as a new generation of building blocks to form homogenous supramolecular 3D dendrimers with defined size and shape. Our step-wise self assembly strategy utilizing a robust pRNA-3WJ motif is highly efficient and can generate homogeneous dendrimers under control with high yield and purity. Introduction of 2′-F RNA makes the dendrimers serum resistant and decoration of the dendrimers with targeting ligands results in high intracellular delivery to specific target cells.
Compared to small molecule based chemical polymers, RNA dendrimers can be made large enough (>10 nm) to avoid rapid renal excretion, yet small enough (65 nm for G-4 dendrimers) to enter cells via receptor-mediated endocytosis. The degradation of RNA can be timely controlled by adjusting the percent and location of chemically modified 2′-F nucleotides. Moreover, individual nucleotides within the RNA dendrimer can be labeled during chemical synthesis prior to dendrimer self-assembly, thus ensuring full utilization of void-spaces, which is an arduous challenge for small molecule based dendrimers. Compared to DNA counterparts, in addition to enhanced chemical stability after 2′-F modifications, RNA dendrimers are thermodynamically more stable15,16,22,37,55,56 and will therefore not dissociate at ultra-low concentrations in vivo, and therapeutic RNA interference modules, such as siRNA and miRNA can be seamlessly integrated into the sequence design.
The ease of chemical synthesis of RNA strands, the simplicity of self-assembly without synthetic reactions, the ease of functionalization of modular units prior to assembly, and the multivalent nature of RNA dendrimers will help in high loading of desired functional modules, such as drugs, targeting, therapeutic, and imaging agents for myriad of applications in bioelectronics, biomimetic membranes, imaging, diagnostics and therapeutics.
Acknowledgments
Funding source:
The research was supported by NIH grants U01 CA151648 (P.G.) and R01 EB019036 (P.G.). Service of Shared Resource Facilities was provided by University of Kentucky Markey Cancer Center P30 CA177558 (B.M.E). Funding to P.G.’s Endowed Chair in Nanobiotechnology position is by the William Fairish Endowment Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
The AFM work was supported by NIH grant P01GM091743 to Yuri Lyubchenko at University of Nebraska Medical Center.
Footnotes
Conflict of interest:
P.G. is a co-founder of Biomotor and RNA Nanotechnology Development Corp. Ltd.
Author contributions:
F.H. and P.G. conceived and designed the project as well as coordinated the projects and analyzed the data. A.S., F.H, and F.P. participated in experimental design and performed the experiments. L.S. performed the AFM imaging. P.G., F.H., B.M.E., A.S., F.P., contributed to manuscript preparation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buhleier E, Wehner W, Vogtle F. Cascade-Chain-Like and Nonskid-Chain-Like Syntheses of Molecular Cavity Topologies. Synthesis-Stuttgart. 1978;(2):155–158. [Google Scholar]
- 2.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR., Jr Efficient transfer of genetic material into mammalian cells using Starburst polyamidoamine dendrimers. Proc Natl Acad Sci U S A. 1996;93(10):4897–4902. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsen TW, Grayzel J, Prensky W. Dendritic nucleic acid structures. J Theor Biol. 1997;187(2):273–284. doi: 10.1006/jtbi.1997.0446. [DOI] [PubMed] [Google Scholar]
- 4.Astruc D. Electron-transfer processes in dendrimers and their implication in biology, catalysis, sensing and nanotechnology. Nat Chem. 2012;4(4):255–267. doi: 10.1038/nchem.1304. [DOI] [PubMed] [Google Scholar]
- 5.Nanjwade BK, Bechra HM, Derkar GK, Manvi FV, Nanjwade VK. Dendrimers: emerging polymers for drug-delivery systems. Eur J Pharm Sci. 2009;38(3):185–196. doi: 10.1016/j.ejps.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Hong CA, Eltoukhy AA, Lee H, Langer R, Anderson DG, Nam YS. Dendrimeric siRNA for Efficient Gene Silencing. Angew Chem Int Ed Engl. 2015;54(23):6740–6744. doi: 10.1002/anie.201412493. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi H, Brechbiel MW. Nano-sized MRI contrast agents with dendrimer cores. Adv Drug Deliv Rev. 2005;57(15):2271–2286. doi: 10.1016/j.addr.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Kaminskas LM, Boyd BJ, Porter CJ. Dendrimer pharmacokinetics: the effect of size, structure and surface characteristics on ADME properties. Nanomedicine (Lond) 2011;6(6):1063–1084. doi: 10.2217/nnm.11.67. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Tseng YD, Kwon SY, D’Espaux L, Bunch JS, McEuen PL, Luo D. Controlled assembly of dendrimer-like DNA. Nat Mater. 2004;3(1):38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 10.Meng HM, Zhang X, Lv Y, Zhao Z, Wang NN, Fu T, Fan H, Liang H, Qiu L, Zhu G, Tan W. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8(6):6171–6181. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi Y, Baker JR., Jr Targeting cancer cells with DNA-assembled dendrimers: a mix and match strategy for cancer. Cell Cycle. 2005;4(5):669–671. doi: 10.4161/cc.4.5.1684. [DOI] [PubMed] [Google Scholar]
- 12.Zhou T, Wang Y, Dong Y, Chen C, Liu D, Yang Z. Tetrahedron DNA dendrimers and their encapsulation of gold nanoparticles. Bioorg Med Chem. 2014;22(16):4391–4394. doi: 10.1016/j.bmc.2014.05.062. [DOI] [PubMed] [Google Scholar]
- 13.Zhou T, Chen P, Niu L, Jin J, Liang D, Li Z, Yang Z, Liu D. pH-responsive size-tunable self-assembled DNA dendrimers. Angew Chem Int Ed Engl. 2012;51(45):11271–11274. doi: 10.1002/anie.201205862. [DOI] [PubMed] [Google Scholar]
- 14.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Searle MS, Williams DH. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 1993;21(9):2051–2056. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo P. The emerging field of RNA nanotechnology. Nature Nanotechnology. 2010;5(12):833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo P, Haque F, Hallahan B, Reif R, Li H. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther. 2012;22(4):226–245. doi: 10.1089/nat.2012.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev. 2014;66C:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo P, Haque F. RNA Nanotechnology and Therapeutics; CRC Press; Boca Raton, FL: 2013. [Google Scholar]
- 20.Shukla GC, Haque F, Tor Y, Wilhelmsson LM, Toulme JJ, Isambert H, Guo P, Rossi JJ, Tenenbaum SA, Shapiro BA. A Boost for the Emerging Field of RNA Nanotechnology. ACS Nano. 2011;5(5):3405–3418. doi: 10.1021/nn200989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leontis N, Sweeney B, Haque F, Guo P. Conference Scene: Advances in RNA nanotechnology promise to transform medicine. Nanomedicine. 2013;8:1051–1054. doi: 10.2217/nnm.13.105. [DOI] [PubMed] [Google Scholar]
- 22.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nature Nanotechnology. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA. 2013;19:766–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 26.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett. 2011;11(2):878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA. Multifunctional RNA nanoparticles. Nano Lett. 2014;14(10):5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khisamutdinov EF, Jasinski DL, Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014;8:4771–4781. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, Takeyasu K, Inoue T, Saito H. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol. 2011;6(2):116–120. doi: 10.1038/nnano.2010.268. [DOI] [PubMed] [Google Scholar]
- 30.Jasinski D, Khisamutdinov EF, Lyubchenko YL, Guo P. Physicochemically Tunable Poly-Functionalized RNA Square Architecture with Fluorogenic and Ribozymatic Properties. ACS Nano. 2014;8:7620–7629. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. Proc Natl Acad Sci U S A. 2011;108(16):6405–6408. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Severcan I, Geary C, VE, CA, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khisamutdinov E, Li H, Jasinski D, Chen J, Fu J, Guo P. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square, and pentagon nanovehicles. Nucleic Acids Res. 2014;42:9996–10004. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol. 2010;5(9):676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geary C, Rothemund PW, Andersen ES. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014;345:799–804. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- 36.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 37.Binzel DW, Khisamutdinov EF, Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry. 2014;53(14):2221–2231. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee TJ, Haque F, Shu D, Yoo JY, Li H, Yokel RA, Horbinski C, Kim TH, Kim SH, Nakano I, Kaur B, Croce CM, Guo P. RNA nanoparticles as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015;6:14766–14776. doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, Mustain W, Valentino J, Guo P, Evers BM. Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano. 2015;9(2):1108–1116. doi: 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelmawla S, Guo S, Zhang L, Pulukuri S, Patankar P, Conley P, Trebley J, Guo P, Li QX. Pharmacological characterization of chemically synthesized monomeric pRNA nanoparticles for systemic delivery. Molecular Therapy. 2011;19:1312–1322. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui D, Zhang C, Liu B, Shu Y, Du T, Shu D, Wang K, Dai F, Liu Y, Li C, Pan F, Yang Y, Ni J, Li H, Brand-Saberi B, Guo P. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Scientific reports. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, Xu R, Guo P. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–40. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee T, Yagati AK, Pi F, Sharma A, Chio JW, Guo P. Construction of RNA-quantum dot chimera for nanoscale resistive biomemory application. ACS Nano. 2015;9:6675–82. doi: 10.1021/acsnano.5b03269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. Crystal Structure of 3WJ Core Revealing Divalent Ion-promoted Thermostability and Assembly of the Phi29 Hexameric Motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang X, Guo P. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today. 2015;10:631–655. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu Y, Shu D, Haque F, Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat Protoc. 2013;8(9):1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyubchenko YL, Shlyakhtenko LS, Ando T. Imaging of nucleic acids with atomic force microscopy. Methods. 2011;54:274–283. doi: 10.1016/j.ymeth.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18(4):305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 50.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Guo S, Cinier M, Shlyakhtenko LS, Shu Y, Chen C, Shen G, Guo P. Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano. 2011;5(1):237–246. doi: 10.1021/nn1024658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helmling S, Moyroud E, Schroeder W, Roehl I, Kleinjung F, Stark S, Bahrenberg G, Gillen C, Klussmann S, Vonhoff S. A new class of Spiegelmers containing 2′-fluoro-nucleotides. Nucleosides Nucleotides Nucleic Acids. 2003;22:1035–1038. doi: 10.1081/NCN-120022729. [DOI] [PubMed] [Google Scholar]
- 53.Grodzinski P, Torchilin V, editors. Advanced Drug Delivery Reviews: Cancer Nanotechnology; Vol. 66. Elsevier; 2014. [Google Scholar]
- 54.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal Biochem. 2005;338(2):284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 55.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34(35):11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 56.Rauzan B, McMichael E, Cave R, Sevcik LR, Ostrosky K, Whitman E, Stegemann R, Sinclair AL, Serra MJ, Deckert AA. Kinetics and Thermodynamics of DNA, RNA, and Hybrid Duplex Formation. Biochemistry. 2013;52(5):765–772. doi: 10.1021/bi3013005. [DOI] [PubMed] [Google Scholar]