Abstract
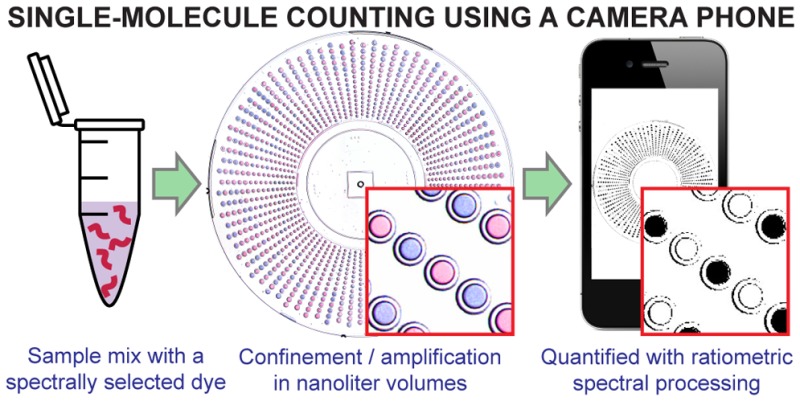
Digital single-molecule technologies are expanding diagnostic capabilities, enabling the ultrasensitive quantification of targets, such as viral load in HIV and hepatitis C infections, by directly counting single molecules. Replacing fluorescent readout with a robust visual readout that can be captured by any unmodified cell phone camera will facilitate the global distribution of diagnostic tests, including in limited-resource settings where the need is greatest. This paper describes a methodology for developing a visual readout system for digital single-molecule amplification of RNA and DNA by (i) selecting colorimetric amplification-indicator dyes that are compatible with the spectral sensitivity of standard mobile phones, and (ii) identifying an optimal ratiometric image-process for a selected dye to achieve a readout that is robust to lighting conditions and camera hardware and provides unambiguous quantitative results, even for colorblind users. We also include an analysis of the limitations of this methodology, and provide a microfluidic approach that can be applied to expand dynamic range and improve reaction performance, allowing ultrasensitive, quantitative measurements at volumes as low as 5 nL. We validate this methodology using SlipChip-based digital single-molecule isothermal amplification with λDNA as a model and hepatitis C viral RNA as a clinically relevant target. The innovative combination of isothermal amplification chemistry in the presence of a judiciously chosen indicator dye and ratiometric image processing with SlipChip technology allowed the sequence-specific visual readout of single nucleic acid molecules in nanoliter volumes with an unmodified cell phone camera. When paired with devices that integrate sample preparation and nucleic acid amplification, this hardware-agnostic approach will increase the affordability and the distribution of quantitative diagnostic and environmental tests.
Keywords: nanoliter reactions, single-molecule counting, cell phone camera, digital, isothermal, microfluidics, ratiometric visual readout
This paper shows that single nucleic acid molecules confined in nanoliter volumes in microfluidic devices can be detected and counted by an unmodified cell phone camera, in combination with isothermal amplification chemistry, a judiciously chosen indicator dye and ratiometric image processing. We describe a novel methodology that can be used to develop a visual readout for digital single-molecule amplification of sequence-specific RNA and DNA that can be used with any camera phone, without modifications or attachments. Single-molecule visual readout has never been achieved before with an unmodified cell phone camera. Diagnostic tests that incorporate such a visual readout will greatly expand the applicability of emerging digital single-molecule technologies, including in limited resource settings (LRS). Ultrasensitive and quantitative detection of nucleic acid molecules is of particular interest for infectious disease diagnosis in LRS, such as the quantification of viral load for human immunodeficiency virus (HIV) and hepatitis C virus (HCV),1−3 as many of these infections occur far from centralized laboratories where diagnostic tests are routine. Increasing diagnoses in these locations will lead to faster and more appropriate treatment and have a major impact on disease burden.4,5 Most point of care (POC) tests are not amenable to LRS because they do not meet the World Health Organization’s ASSURED criteria of being affordable, sensitive, specific, user-friendly, rapid, robust, equipment-free and deliverable.5 The tests that do meet the requirements for LRS (e.g., immunochromatography to detect antigens or antibodies in a dipstick or lateral-flow format; or the visualization of antigen–antibody lattice formation) have poor reported sensitivities and thus are unable to detect and quantify analytes at low concentrations.4,6 Nucleic acid amplification tests (NAATs), such as PCR, have the desired high sensitivity and target specificity, providing accurate quantification, but these technologies are costly, time-consuming, and require skilled technicians and laboratory settings.7
Of the NAATs, isothermal amplification methods (e.g., loop-mediated isothermal amplification, LAMP) are among the most attractive for LRS because they do not require thermocycling or capital equipment and can be run in water baths, using simple heaters or with exothermic chemical heating that does not require electricity.8−11 Still, acquiring quantitative and ultrasensitive measurements outside of the lab remains challenging because the methods are not robust to variability in reaction conditions and readouts rely on precise measures of fluorescence intensity. Running isothermal amplification chemistries in a digital, single-molecule format maintains the high sensitivity and quantification capabilities typically achieved only in lab settings.12−15 In digital single-molecule isothermal amplification, single, stochastically confined DNA or RNA molecules are randomly distributed among discrete nanoliter or picoliter volumes and amplified under controlled conditions.16−18 This creates relatively high local concentrations of target DNA or RNA, making digital amplification more efficient and robust compared to bulk reactions with the same number of starting target molecules. Nucleic acid amplification of even a single target molecule produces a clear fluorescent signal and the results of digital amplification can be read by a modified cell phone (e.g., a phone camera with an optical filter) under dim lighting.14
Microfluidic technology has been an instrumental tool in developing single nucleic acid molecule capabilities,19−27 and the integration of sample-preparation modules into portable microfluidic devices will further enable their use by untrained users in any setting.28−30 To bring these emerging technological capabilities to LRS, however, such devices capable of ultrasensitive, quantitative measurements should provide a rapid, visual readout that can be captured easily, e.g., by any mobile phone without modifications or attachments. Cell phone cameras provide a convenient, nearly universal tool to pair with emerging diagnostic technologies to transform global healthcare as ∼7 billion mobile cellular subscribers exist worldwide and 70% of users live in developing countries.31 Mobile devices are emerging as a powerful platform to create cost-effective alternatives for molecular diagnostics in LRS32−42 and colorimetric diagnostics based on unmodified cell phones have been used before,38,43−46 but not in a digital format, where the short path lengths and nanoliter volumes have constrained visual-based methods. Here, we describe an approach that enables visual readout of single nucleic acid molecule amplification by (i) selecting an appropriate colorimetric indicator dye based on spectral properties that align well with the RGB sensitivities of common cell phone camera sensors and (ii) identifying the optimal ratiometric image-processing for the selected dye to achieve a readout that is robust to lighting conditions and camera hardware. With this approach, after sequence-specific single-molecule isothermal amplification, a visual readout is captured by an unmodified camera phone and the resulting image is analyzed using a ratiometric approach, wherein the measured intensities of two of the three RGB color channels are divided to provide a binary result (a positive or negative reaction) for each well. The automation of this ratiometric analysis provides a clear, reliable digital readout without requiring the user to differentiate color change by eye or manipulate lighting (Figure 1a). We further show how limitations related to reaction inhibition by the readout dye can be solved with SlipChip microfluidics technology to decouple the amplification and readout steps. We validated our visual readout method with SlipChip-based digital single-molecule isothermal amplification reactions using phage lambda DNA (λDNA) as a model and HCV RNA as a clinically relevant target, in reaction volumes as low as 5 nL, using a variety of common cell phones and a range of illumination conditions.
Figure 1.
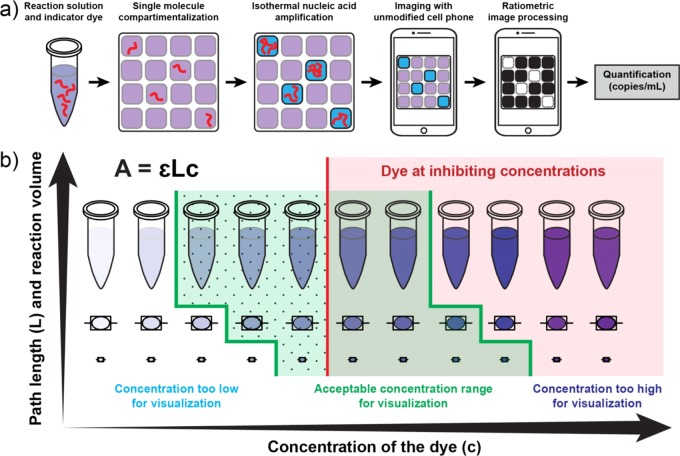
A visual readout approach for digital single-molecule isothermal amplification for use with an unmodified cell phone camera. (a) A workflow for visual readout of digital single-molecule amplification. Single nucleic acid molecules are compartmentalized on a microfluidic device with indicator dye and followed by isothermal nucleic acid amplification. Positive reaction solutions are blue; negative reactions are purple. After ratiometric image processing, positive reactions become white and negative reactions become black, an unambiguous binary result. The number of positive wells is then used to quantify the concentration of the input target. (b) A diagram for delineating the optimal range of dye concentrations as a factor of path length (reaction volume) and the threshold for reaction inhibition. The green-shaded region indicates the range of acceptable dye concentrations for visualization with an unmodified cell phone camera. Concentrations to the left of the green region are too low for visualization; concentrations to the right of the green region are too high. Within this green region, the dotted area indicates dye concentrations that both enable readout with an unmodified cell phone camera and do not inhibit the amplification reaction. The area to the right of the red line indicates dye concentrations that interfere with amplification making accurate quantification based on real-time data challenging.
Results and Discussion
Selecting an Indicator Dye
To eliminate the need for a fluorescent readout in single-molecule amplification and produce a readout that can be imaged by any cell phone camera under various illumination conditions, one can use a nucleic acid amplification-indicator dye that changes color in response to amplification. A robust colorimetric readout balances two opposing requirements: the indicator dye must be sufficiently concentrated (or present in a large enough volume) to provide readable absorbance (i.e., smaller volumes and shorter path lengths require greater concentrations of dye for sufficient absorbance to be detected) but not so concentrated that the dye interferes with the amplification reaction. To optimize a visual readout system for single-molecule counting with an unmodified cell phone camera, we first identified the factors that contribute to hypothetical limitations of a visual readout system, including the range of reaction volumes (or path lengths) at which a particular indicator could be used to monitor amplification and the range of indicator concentrations that would not interfere with the amplification reaction. Where these ranges overlap are the optimal volumes and dye concentrations at which a reaction is not inhibited and can provide a change in absorbance that is sufficient for readout with an unmodified camera phone (dotted green region of Figure 1b).
We validated this visual readout approach using loop-mediated isothermal amplification (LAMP)47,48 (Supporting Information Tables S1 and S2) because this method has been well characterized and validated previously for single-molecule analyses.12,14−17,49 LAMP chemistry is based on an autocycling strand displacement reaction performed at a constant temperature to synthesize large amounts of amplified product; a LAMP reaction generates more than 109 copies of template within 1 h of incubation at 60–65 °C.48 We used a cubic reaction volume of 8 nL (200 × 200 × 200 μm3), which is in the range of volumes used in digital experiments.12,14,15,17,49 We assume that an appropriate indicator of an amplification reaction will have a change in absorbance that equates to a change of extinction coefficient of ∼25 000 L mol–1 cm–1 upon reaction (this number approaches the maximum achievable change in absorbance for small-molecule dyes). We use the Beer–Lambert law (A = εLc), which describes the relevant parameters to consider for visualization, wherein A = absorbance; ε = extinction coefficient (L mol–1 cm–1); L = length of the light’s path through the solution (cm); c = concentration of absorbing species (mol/L). At a path length of 0.2 mm, an estimated ∼2 mM concentration of the dye is required to reach a change of absorbance of 1 unit. Given these parameters, to obtain a readout that can be captured by an unmodified mobile phone, we predicted that an appropriate indicator dye would be one that responds to the incorporation of each nucleotide (present in mM concentrations), as opposed to responding only to the number of produced molecules (amplicons), which would not exceed primer concentration (present in the μM range).
Colorimetric approaches to visual detection of nucleic acid amplification typically measure absolute changes in color intensity;50−54 however, distinguishing color change, e.g., purple vs blue, is difficult and therefore not an appropriate way to quantify readout under variable conditions, such as in LRS. Ratiometric measurements, which take the ratio of two independent measurements under the same conditions, improve the robustness of a colorimetric approach, converting results to a yes/no binary outcome, eliminating the need for the user to differentiate colors. We hypothesized that a cell phone camera’s sensor, which reads in three color channels (red, green, and blue, RGB) could provide suitable information for using a ratiometric approach to read amplification reactions at the single molecule level. The example we considered here is the back-illuminated Exmor R CMOS image sensor55 used on popular cell phones such as the Samsung Galaxy 4, iPhone 4S, and iPhone 5, which has a sensitivity maxima of ∼520 nm (green), ∼459 nm (blue), and ∼597 nm (red) (Figure 2a).
Figure 2.
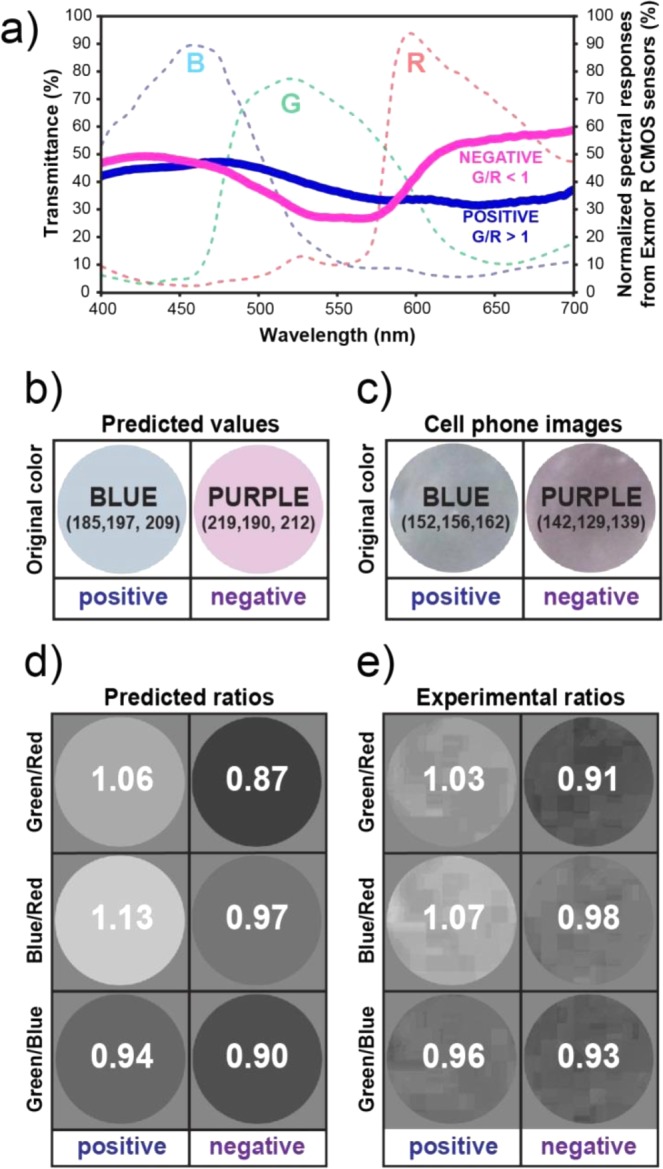
Predicted values and experimental validation of the first step of the ratiometric approach. (a) Measured spectral transmittance (%) in the range of visible light (400–700 nm) for positive (solid blue line) and negative (solid purple line) RT-LAMP reaction solutions, each containing 0.7 mM of eriochrome black T (EBT) as the amplification indicator dye. Dashed lines correspond to normalized spectral responses for red (R), green (G), and blue (B) channels of an Exmor R CMOS sensor, a common sensor in cell phone cameras. (b–e) Analysis of the three possible RGB ratiometric combinations for positive and negative RT-LAMP reaction solutions. (b) The predicted RGB values and corresponding colors for positive and negative LAMP amplification reactions obtained by convoluting the transmittance spectrum and Exmor R spectral responses described in panel a. (c) The cropped and enlarged color images collected with an Apple iPhone 4S for positive and negative RT-LAMP reaction solutions containing 90 μM of EBT dye. (d) Predicted images and ratiometric values for positive and negative amplification reactions processed for each ratiometric combination, G/R, B/R, and G/B. (e) Experimental images and ratiometric values for positive and negative amplification reactions for each combination: G/R, B/R, and G/B. All experiments were performed with HCV RNA as template.
To illustrate our methodology for a hardware-agnostic visual readout with a ratiometric approach, we selected eriochrome black T (EBT), a magnesium ion indicator that meets the aforementioned dye specifications and has been used previously for visualization of LAMP products.56,57 During an isothermal amplification reaction, as nucleotides are incorporated, protons and byproduct pyrophosphate ions (P2O74–) are produced, and these ions can strongly bind metal ions (e.g., Mg2+ ions) and form insoluble salts, decreasing the concentration of metal ions in the reaction solution. Before the amplification reaction, EBT is bound to magnesium ions and the reaction solution is purple. As a LAMP reaction proceeds in the presence of target nucleic acid, it is suggested that EBT is deprived of Mg2+ by newly generated pyrophosphate ions, and the reaction solution turns blue.
We hypothesized that EBT would be amenable to colorimetric analysis with a cell phone camera because, in RGB terms, in a positive LAMP reaction containing EBT dye, there is a peak in the spectral transmittance in the blue channel (blue LAMP reaction solution), while in a negative LAMP reaction, transmittance remains high in the blue and red channels (purple LAMP reaction solution) (Figure 2a). These observed changes in transmittance between positive and negative reactions can be captured by the Exmor R optical sensor (Figure 2a), which matches well with the observed differences between positive and negative transmittance profiles of LAMP reactions containing EBT (Figure 2a).
Selecting the Optimal Ratiometric Approach
We tested whether the suitability of an indicator dye can be evaluated for a ratiometric approach prior to experimental validation by predicting the RGB values read by a cell phone camera for a positive and a negative reaction. First, we took the transmittance spectra for positive and negative amplification reactions containing EBT and convoluted them with the normalized spectral responses for each of the RGB channels in an Exmor R CMOS sensor58 providing six curves (a positive and negative for each of the three color channels). Next, we calculated the area under each curve and took its square root (to account for the standard square-root scaling that occurs with nonscientific devices used for imaging), providing the predicted RGB values (Figure 2b) for positive (R = 185, G = 197, and B = 209) and negative (R = 219, G = 190, and B = 212) RT-LAMP reaction solutions in the presence of EBT at this particular concentration. These values can then be evaluated to select the optimal ratiometric approach for this particular indicator dye. In an RGB color scheme, there are three possible combinations for ratiometric analysis: G/R, B/R, or G/B. The predicted RGB values for a positive and a negative reaction are used to calculate the ratios for each channel combination (Figure 2d); the ratio with the greatest difference between positive and negative outcomes (G/R in this example) is predicted to be the most robust ratiometric analysis.
Using the approach described above, we predicted the RGB ratios for a positive and negative RT-LAMP reaction in the presence of two additional indicator dyes: hydroxynaphthol blue (HNB) and calmagite. HNB is being reported increasingly in the literature for LAMP visualization50,59−62 and calmagite is an analogue of EBT dye with the nitro group absent (more stable version).63 A side-by-side comparison showed that the greatest predicted difference between positive and negative RT-LAMP reaction, as captured by an unmodified cell phone camera, would be achieved using EBT as the indicator dye and G/R as the ratiometric combination (Figure S11). On the basis of these predicted ratios, we decided to validate our methodology using EBT as the indicator dye. We confirmed the storage stability of the EBT dye stock solution in the dried state (Figure S13), as this is a critical requirement for the use of a dye in real point-of-need diagnostic applications. EBT serves as our validation dye in this paper, however our methodology is designed to be applicable to alternative dyes.
To experimentally validate this approach of predicting an optimal ratiometric combination, we performed an RT-LAMP reaction for HCV RNA containing EBT as the indicator dye and captured an image of the readout with an unmodified camera phone (iPhone 4S) (Figure 2c). We processed the readout image; color channels of the original image were split and all three channel ratios (G/R, B/R, G/B) were calculated to derive a ratiometric image for each ratiometric combination. These experimental ratios obtained with an unmodified cell phone camera (Figure 2e) matched well with the predicted values (Figure 2d) for each of the three ratiometric combinations, confirming the predictive power of this approach. The G/B ratio was identified as less appropriate for distinguishing positive and negative reactions because the values for positive and negative reactions were similar; G/R and B/R ratios were identified as suitable because there was sufficient contrast between the values for positive and negative reactions. For the G/R combination, the ratio obtained after a negative reaction was 0.91 and the ratio from a positive reaction was 1.03, a difference of 0.12 (Figure 2e). For the B/R combination, the ratios for negative and positive reactions were 0.98 and 1.07, a difference of 0.09 (Figure 2e). Therefore, we selected the G/R combination for our subsequent validation experiments. Counting positives is a more intuitive approach, so the B/R ratio (where the positive ratio had the greatest difference from the background) can be a useful and attractive method. However, it is generally more desirable to select a ratio that includes the green channel because most single-chip digital image sensors used in digital cameras, including cell phones, utilize a Bayer filter mosaic pattern that is composed of 50% green, 25% red, and 25% blue pixels.64
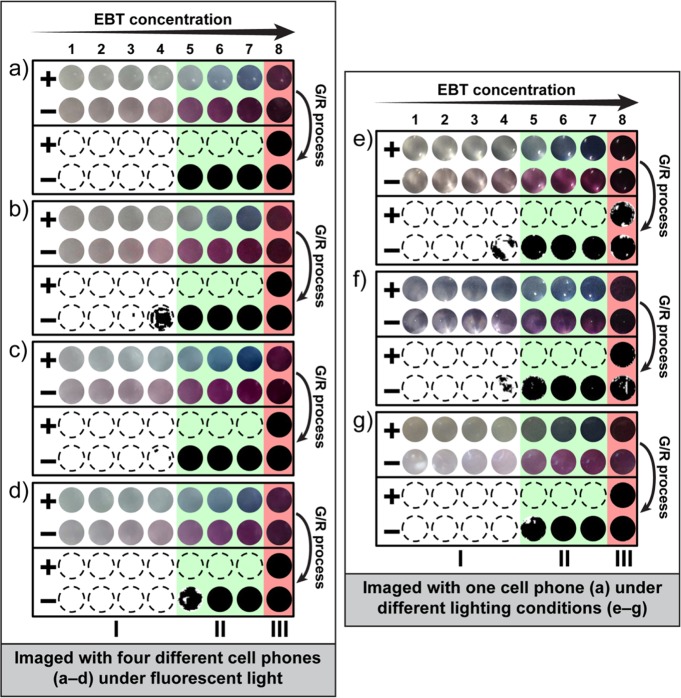
To test the robustness of our approach to different hardware and illumination conditions, we used HCV RNA amplified by RT-LAMP at 2-fold increasing concentrations of indicator dye ranging from 10.9 μM to 1.4 mM (for a total of eight dye concentrations). After RT-LAMP amplification, 50 μL of each reaction solution was transferred to 96-well plates (path length of ∼1.5 mm) and the readout was imaged with cameras from four common cell phone models: Apple iPhone 4S (Figure 3a), HTC inspire 4G (Figure 3b), Motorola Moto G (Figure 3c), and Nokia 808 PureView (Figure 3d). Under fluorescent light and using the G/R ratiometric process (green channel divided by red channel followed by a threshold adjustment to generate a binarized black and white image), we determined that EBT concentrations lower than 0.175 mM provided an insufficient color change for detection with a cell phone camera (Figure 3, region I, white background), while concentrations of 1.4 mM inhibited the amplification reaction (Figure 3, region III, red background). For this particular indicator dye, the range of concentrations at which color change could be detected by an unmodified cell phone camera and no inhibition was observed at the end point of the reaction (Supporting Information Figure S1) was identified as 0.175–0.7 mM (Figure 3, region II, green background). Some cell phone cameras were more sensitive (e.g., HTC inspire 4G was able to distinguish a positive result at EBT concentrations as low as 0.0875 mM) (Figure 3b), but all four cell phone models distinguished a positive reaction at concentrations between 0.175 and 0.7 mM (Figure 3, region II, green background). We then chose one cell phone with the most representative performance (Apple iPhone 4S) to test the robustness of the G/R approach to different lighting conditions. Under all conditions tested: incandescent light (Figure 3e), direct sunlight (Figure 3f) and indirect sunlight (Figure 3g), the optimal EBT concentration range that we identified under fluorescent light (0.175–0.7 mM) could be read clearly, confirming the robustness of the ratiometric approach to variations in illumination.
Figure 3.
Validation of the robustness of the G/R ratiometric approach to different hardware (cell phone cameras) and lighting conditions. (a–g) Enlarged and cropped color images (top two rows of each individual panel) captured by an unmodified cell phone camera from positive (+) and negative (−) RT-LAMP reactions at 2-fold increases in EBT concentration from 10.9 μM to 1.4 mM (1 = 0.011 mM; 2 = 0.022 mM; 3 = 0.044 mM, 4 = 0.088 mM, 5 = 0.175 mM; 6 = 0.35 mM; 7 = 0.7 mM; 8 = 1.4 mM). Positive wells are blue and negative wells are purple. After G/R ratiometric processing (bottom two rows of each individual panel), negative wells are black. Regions I, II, III in each panel indicate the effect of dye concentration: (II) acceptable concentration range for visualization (green regions); (I) concentrations too low for visualization (white regions); and (III) concentrations too high for visualization (red regions). (a–d) Images captured by four common cell phones under fluorescent light: (a) Apple iPhone 4S, (b) HTC inspire 4G, (c) Motorola Moto G, and (d) Nokia 808 PureView. (e–g) Images captured by an Apple iPhone 4S under three additional light conditions: (e) incandescent light, (f) direct sunlight, and (g) indirect sunlight. All experiments were performed with HCV RNA as a clinically relevant target. All images were acquired with unmodified cell phone cameras. Detailed information for the G/R ratiometric process (Figure S2) and additional cell phone camera images (Figure S3) are provided in the Supporting Information.
One-Step Method for Digital Visual Readout
Microfluidic devices enable ultrasensitive digital quantification. Small well volumes are valuable because they enable faster reactions (because concentrations are high in single wells), minimize the effects of inhibitory materials (due to their isolation into wells) and expand the upper limit of the dynamic range (because single molecules can be confined from samples containing high template concentrations).18,65,66 However, as well volumes (and path lengths) decrease, color visualization becomes challenging for a mobile phone. To compensate, the concentration of the indicator dye can be increased; however, high concentrations of some dyes inhibit amplification reactions. Thus, there are inherent physical limits to a colorimetric approach. To validate that this visual readout approach could be applied to single-molecule amplification at nanoliter volumes, we used digital LAMP (dLAMP) and phage λDNA as a target. We specifically aimed to resolve three questions: (i) Can we obtain a visual readout for amplified single molecules that can be captured by an unmodified cell phone camera? (ii) Is volume a factor in achieving a digital visual readout? (iii) Does ratiometric processing work for small volumes?
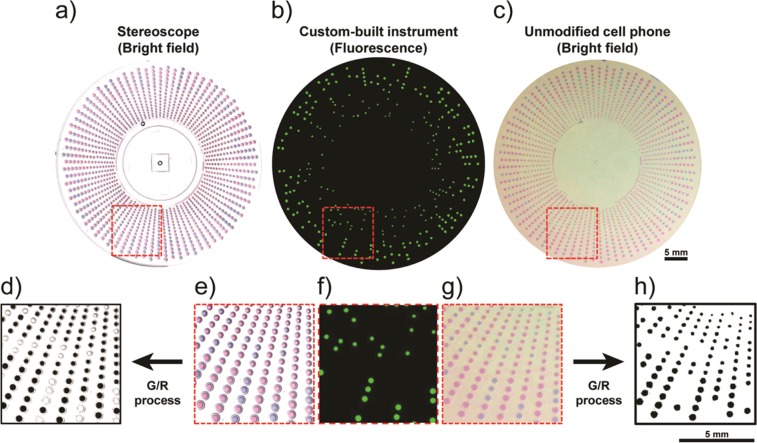
To answer these questions, we designed a multivolume rotational SlipChip device containing 1240 wells of eight volumes ranging from 15 to 50 nL (Figures S4 and S5). We loaded these devices with LAMP reaction solution containing an appropriate target concentration in the middle of the device’s dynamic range, a fluorescent DNA-detecting intercalation dye (Syto 9), and EBT dye at 0.7 mM (the highest noninhibiting concentration identified in Figure 3). We imaged this device with a house-built real-time fluorescence imager, with a Leica stereoscope (optimal imaging conditions) and with an Apple iPhone 4S. The number of positive counts based on fluorescence was 261, while 260 positives were counted using the indicator dye and G/R process both with the stereoscope and the cell phone (Figure 4). This experiment showed that the G/R method could be used in place of fluorescence readout to count amplified single molecules and that the readout capture and G/R processing performed on an unmodified cell phone matched the results obtained under optimal lighting conditions (stereoscope). Additionally, using a device containing 800 wells of 27 nL, we observed excellent correlation among positive counts obtained from the stereoscope, fluorescence imager and cell phone camera (Figure S6).
Figure 4.
Readout from single-molecule digital LAMP reactions performed with λDNA on a multivolume rotational SlipChip device imaged by (a) a stereoscope, (b) a fluorescence microscope, and (c) an unmodified cell phone camera. (e–g) Callouts are magnified to show visual correlation among the three imaging methods. (d) The results of the ratiometric processing for the stereoscope G/R-processed image and (h) the cell phone G/R-processed image. Colors were enhanced in these figures for clarity of publication; raw images were used in all ratiometric analyses. These devices contained 1240 wells of eight volumes ranging from 15 to 50 nL.
While investigating the limits that reaction volume may impose on visual readout, we observed that the estimated template concentration determined from each of the eight well volumes produced similar Most Probable Numbers (MPN) of molecules (mean 8500 ± 1500 copies/mL) (Figure 5a) (estimated concentration from all volumes are within 95% confidence interval at each volume, detailed in Figure S7). In addition, all SlipChip devices, analyzed independently, gave similar target concentrations (8400 ± 500 copies/mL) (Figure 5b), suggesting that the selected indicator dye did not impair quantification of single molecules in well sizes 15–50 nL and that these well volumes can be imaged reliably with either a stereoscope or an unmodified cell phone camera. However, the cell phone camera images of well volumes of 15 nL were less clear than those obtained from the stereoscope, suggesting that volumes of ∼15 nL may approach the limit of colorimetric imaging with current camera phone sensors, although as higher quality sensors are integrated into commercial cell phones, this limit would change.
Figure 5.
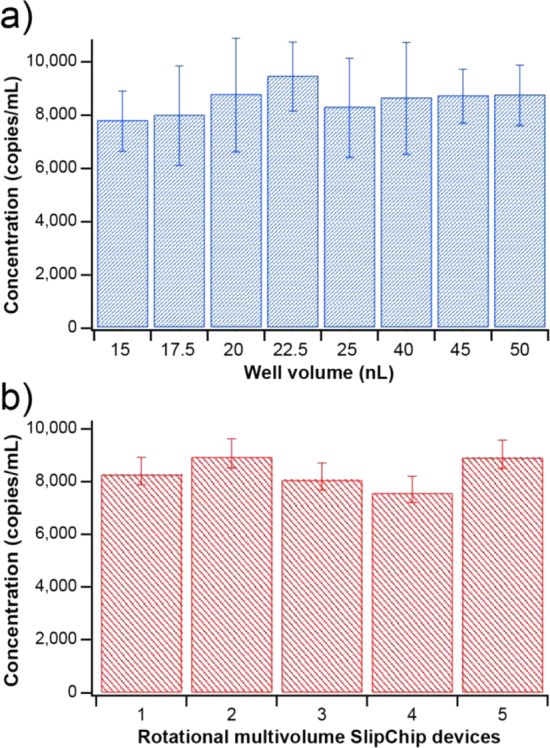
Robustness of digital visual readout at different well volumes. Concentration of λDNA was estimated by digital LAMP using five multivolume rotational SlipChip devices, each of which contained eight well volumes ranging from 15 to 50 nL. (a) Measured template concentration for each well volume averaged over five devices. (b) Mean template concentration for each of five rotational SlipChip devices. Concentrations were calculated using MPN theory65 and error bars represent standard deviation. Images were captured by a stereoscope and processed with the ratiometric approach (G/R process).
Two-Step Method for Digital Visual Readout
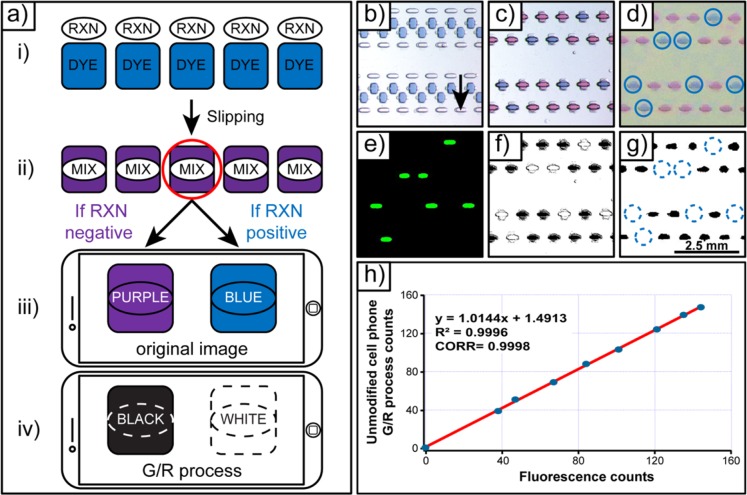
We next developed a method to apply the visual readout approach to digital devices that contain smaller well volumes. To be able to image at small volumes (e.g., 5 nL) on a microfluidic device, one must balance the need for greater indicator color intensity for visualization with the need to keep dye concentrations below the level of inhibition (Figure 3, region III) for an amplification reaction. High concentrations of indicator dye can completely halt an amplification reaction, and we knew from performing real-time bulk experiments that even when reactions are positive, an indicator dye can still interfere to some extent with isothermal nucleic acid amplification—for both RNA and DNA, we observed delays in the time-to-positive, and this delay increased at greater concentrations of the indicator dye, even though reactions were positive (Figure S8). We hypothesized that we could prevent inhibition completely by decoupling the amplification step from the readout step. To do this, we designed a two-step SlipChip device (based on previous SlipChip designs)13 (Figures S9 and S10) in which the amplification solution and the detection solution are loaded into separate wells (Figure 6a). We validated this two-step protocol with a clinically relevant target, purified HCV RNA, using digital reverse transcription-LAMP (dRT-LAMP). First, we performed digital isothermal amplification in the set of small (5 nL) amplification wells (in the absence of the indicator dye) (Figure 6a(i)). After amplification, a “slip” was performed and the amplification wells came into contact with a second set of larger (9.5 nL) wells, which contained the indicator dye, for a total well volume of 14.5 nL (Figure 6a(ii)). After mixing, negative wells lacking target molecules are purple and wells containing positive reactions are blue (Figure 6a(iii)). Counts obtained by a house-built real-time imaging instrument (to read fluorescence), and counts obtained by G/R processing from an image captured by an unmodified cell phone camera were significantly correlated (Pearson’s Corr = 0.9998; R2 = 0.9996) (Figure 6h), showing that this two-step SlipChip-based protocol provides a suitable visual readout for digital single-molecule amplification for devices containing wells of small volumes.
Figure 6.
Experimental validation of two-step SlipChip devices for single molecule counting with an unmodified cell phone camera. (a) A flow-chart of detection of single molecules in two-step SlipChip: (i) 5 nL amplification wells are loaded with amplification reaction solution (RXN) and 9.5 nL detection wells are loaded with indicator dye (DYE). (ii) After amplification, a slip is performed and the RXN and DYE wells are combined. (iii) Immediately after mixing, positive reaction solutions become blue, while negative reactions remain purple. The readout is imaged by an unmodified cell phone camera. (iv) Ratiometric image processing (G/R process) provides a single binary result (positive or negative). (b) Stereoscope image of the device before the amplification and readout wells are merged (arrow designates direction of slip). (c) Stereoscope, (d) cell phone camera and (e) fluorescent images after the device is slipped and the wells are merged. (f) Stereoscope and (g) cell phone camera images after G/R image processing. (h) Correlation between fluorescence counts and cell phone (G/R processed) counts. Colors were enhanced in figure panels b–d, and f for clarity of publication; raw images were used in all ratiometric analyses. In these experiments, HCV RNA was amplified by dRT-LAMP.
Devices shown in this manuscript were not designed to achieve clinically relevant concentrations in the lower detection limit of quantification (LDL) because larger well volumes do not represent a challenge when imaging with a mobile phone. Instead, we studied the performance of our approach with wells of small volumes to ensure that this method meets the upper limit of quantification (ULQ) required for clinical relevance. The ULQ is determined by the smallest well volume and the total number of wells at that volume. As an example, for SlipChip devices with 800 wells of 5 nL, the ULQ is 1 162 413 copies/mL, while a SlipChip device with 10 000 wells of 5 nL, the ULQ is 1 622 660 (calculations performed according to Kreutz et al. 2011).65
Conclusions
Here we show that single nucleic acid molecules can be detected and counted with an unmodified cell phone camera by employing microfluidic technology, sequence-specific isothermal amplification, and a judiciously chosen amplification-indicator dye. We further show that ratiometric processing of the cell phone image enables robust quantification without the need for a user to differentiate colors. The general methodology we developed can be used as a guideline to enable others to develop their own cell phone based single-molecule counting approach. The methodology includes the following steps: First, an appropriate amplification indicator should be selected. Indicators should respond optically to each nucleotide incorporation event (as opposed to responding to number of produced molecules) resulting in a change in the transmittance profile in the wavelength range of visible light (400–700 nm). The indicator dye should have a change in absorbance matched to the spectral sensitivity of the image sensor in an unmodified cell phone; for ratiometric processing, the solution should have a large relative change in transmittance in color channels for which the camera’s image sensor is most sensitive. Second, the color ratio used in the ratiometric approach is chosen based on the spectral sensitivity of the image sensor in an unmodified cell phone. This step can be done in silico to identify the dye with the ratio that provides an unambiguous binary readout of positive and negative reactions that is robust to illumination and hardware conditions. We hope others will use this algorithm to identify even better dyes that will move this field forward. Third, the selected dye and ratiometric approach should be validated using the desired amplification chemistry. Experiments should be performed to establish the range of dye concentrations and well volumes at which an amplification reaction is not inhibited and at which imaging can be done with an unmodified cell phone. For some indicator dyes, the range of suitable well volumes and concentrations will be too narrow. In such situations, an alternative approach is to use a two-step device that separates the amplification and readout steps. Processing can be done directly on a cell phone or uploaded wirelessly to a cloud server to swiftly communicate results, as we have shown previously.14 We anticipate that the capabilities of visual readout for counting single molecules will extend further as cell phone camera technology advances, as additional indicators are available (e.g., metal ions, pH indicators) and as additional types of amplification reactions are developed. Devices that integrate sample preparation, nucleic acid amplification, and a visual digital readout that can be captured easily will be a critical breakthrough toward bringing quantitative, ultrasensitive measurements outside of central laboratories, a key step for in vitro diagnostics, pandemic surveillance, and environmental monitoring. We hope this work will stimulate regulatory agencies such as the FDA to consider the use of cell phones as valuable diagnostic components.
Methods
Chemicals and Materials
All chemicals were purchased from commercial sources. The LoopAmp RNA amplification kit (Eiken Chemical Co., Ltd., Japan) was purchased from SA Scientific (San Antonio, TX, USA). The LoopAmp RNA amplification kit contains 2× Reaction Mix (RM) (40 mM Tris-HCl, pH 8.8, 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6 M betaine, and dNTPs 2.8 mM each), Enzyme Mix (EM) (mixture of Bst DNA polymerase and AMV reverse transcriptase), and distilled water (DW). Bovine serum albumin (BSA) was purchased from Roche Diagnostics (Indianapolis, IN, USA). Phage λDNA (500 μg), SUPERase In RNase Inhibitor (20 U/μL), Eriochrome Black T (EBT) dye, mineral oil (DNase, RNase, and Protease free), tetradecane, Costar Clear Polystyrene 96-Well Plates, Corning Universal Optical Microplate Sealing Tape, and DEPC-treated nuclease-free water were purchased from Thermo Fisher Scientific (Hanover Park, IL, USA). Chelex 100 resin was purchased from Bio-Rad (Hercules, CA, USA). Trehalose solution (1 M) was purchased from Amersham Life Science (Cleveland, OH, USA). Tris-HCl buffer stock solution (1 M, pH 8.0) was purchased from Affymetrix (Santa Clara, CA, USA). All primers were produced by Integrated DNA Technologies (Coralville, IA, USA). Dichlorodimethylsilane was purchased from Sigma-Aldrich (St. Louis, MO, USA). SYTO 9 Stain and AcroMetrix HCV High Control were purchased from Life Technologies (Grand Island, NY, USA). Nucleic acid extraction kit QIAamp Viral RNA Mini kit was purchased from QIAGEN, Inc. (Valencia, CA, USA). Eppendorf Mastercycler Gradient PCR Themal Cycler was purchased from Eppendorf (Hamburg, Germany). POLARstar Omega microplate reader was purchased from BMG Labtech (Durham, NC, USA). Leica MZ Fl III stereoscope with PLAN 0.5× lens was purchased from Leica Microsystems (Bannockburn, IL, USA). Photomasks were designed in AutoCAD 2013 and ordered from CAD/Art Services, Inc. (Bandon, OR, USA). Soda-lime glass plates coated with layers of chromium and photoresist were ordered from the Telic Company (Valencia, CA, USA).
SlipChip Device Design
The multivolume rotational SlipChip device design was used to demonstrate the one-step method for digital visual readout; this device was composed of 1240 microfluidic wells, with the following volumes: 160 wells × 15 nL, 160 × 17.5 nL, 160 × 20 nL, 160 × 22.5 nL, 160 × 25 nL, 160 × 40 nL, 160 × 45 nL, 120 × 50 nL (Figure S4). The total combined volume of all wells was 35.6 μL. For loading, one inlet hole (in the middle ring structure) and four oil escape holes (in the outer ring structure) were drilled in the top plate. The two-step SlipChip device was used to demonstrate a two-step method for digital visual readout; this device was based on previously published SlipChip designs.13 For the two-step SlipChip design used in this study, the device was modified in the following ways: (i) the number of each type of well was reduced to 800; (ii) space was added between the arrays to allow for the incubation conformation; (iii) the sequence of well loading was reversed (the smaller 5 nL wells are loaded before the larger 9.5 nL wells). See Figure S9 for more details. Examples of SlipChip multivolume designs for HCV and HIV viral load quantification at clinically relevant dynamic ranges67−69 are provided in the Supporting Information (Table S3).
SlipChip Device Fabrication
The procedure for fabricating the multivolume rotational SlipChip and two-step SlipChip devices was based on previous work.70 The device features were etched to a depth of ∼100 μm for the multivolume rotational SlipChip devices and ∼67 μm for the two-step SlipChip devices. After etching and drilling through-holes, both devices were subjected to the same glass silanization process, previously described,66 where the glass plates were first thoroughly cleaned with piranha solution and dried sequentially with 200 proof ethanol and nitrogen gas, and then oxidized in a plasma cleaner for 2 min and immediately transferred into a vacuum desiccator for 1.5 h for silanization with dimethyldichlorosilane. After silanization, the devices were rinsed thoroughly with chloroform, acetone, and ethanol, and dried with nitrogen gas before use. When a glass SlipChip device needed to be reused, it was first cleaned with acid piranha solution and then subjected to the same silanization and rinsing procedure described above.
Assembling and Loading SlipChips
The SlipChips used for both the dLAMP and the dRT-LAMP reactions were assembled under degassed oil (mineral oil/tetradecane 1:4 v/v). Both top and bottom plates were immersed in the oil phase and placed face to face. The two plates were aligned under a stereoscope (Leica, Germany) and stabilized using binder clips. Through-holes were drilled into the top plate to serve as fluid inlets and oil outlets in dead-end filling. The reagent solutions were loaded through the inlets by pipetting.
HCV Viral RNA Purification from AcroMetrix HCV High Control
A total of 200 μL of plasma containing HCV RNA (viral load estimate provided by AcroMetrix: 1.1–3.5 IU/mL) was extracted using the QIAamp Viral RNA Mini Kit (QIAGEN, Inc., Valencia, CA, USA) according to the manufacturer’s instructions. The elution volume was 60 μL. The purified HCV viral RNA was analyzed immediately or stored at −80 °C until further analysis.
Preparation of EBT Solution
The EBT stock solutions were prepared by dissolving EBT dye in deionized water. The aqueous solution was sonicated for 10–20 min and the free volume was filled with argon gas and mixed on a rotator at 65 °C for 1 h. To remove any potential impurities from the EBT dye, Chelex 100 ion-exchange resin was added to the resulting solution (5% w/v) and placed on rotator for 1 h. Resin was centrifuged at 3000 rpm for 5 min and the top fraction was collected in a Falcon tube, flushed with argon, and stored at room temperature for no more than 2 days. A comparison of EBT, HNB, and calmagite indicator dye stock solutions before and after treatment with Chelex 100 is provided in the Supporting Information (Figure S12).
Storage Stability of Amplification Indicator Dyes by Drying in the Presence of Stabilizer Trehalose
EBT, HNB, and calmagite stock solutions at 0.7 mM were prepared by dissolving the dyes in 20 mM Tris-HCl buffer (pH 8.8) and adding 30 mM of trehalose. The solutions were sonicated for 10 min and mixed on a rotator at room temperature for 1 h. Chelex 100 ion-exchange resin was added (5% w/v) and placed on rotator for 1 h. Resin was centrifuged at 3000 rpm for 5 min and the top fraction was collected in a Falcon tube. The resulting stock solutions were transferred to a Costar Clear Polystyrene 96-Well Plate (40 μL per well) and sealed with Corning Universal Optical Microplate Sealing Tape before spectrophotometric analysis (time 0 h). Immediately after analysis, the sealing cover was removed and the plate was placed in a desiccator under vacuum overnight until the dye stock solutions were completely dry. Then, at 24-h time points over the next 120 h (for a total of 5 time points), three wells of each dried amplification indicator solution were resuspended with 40 μL of deionized water and spectrophotometric analyses were performed. After each measurement, the plate was sealed again (to prevent hydration of the dried solutions in the other wells) and kept in the dark at room temperature. The absorption spectra analyses were performed using the POLARstar Omega microplate reader with Omega Data analysis software. Absorbance in the range of 400–700 nm was recorded at 2 nm intervals. Blank solutions (20 mM Tris-HCl buffer with 30 mM trehalose) were also loaded at time 0 h, desiccated after the first measurement, and treated as the rest of the solutions. The measured spectral absorbance from these control solutions was subtracted at each time point from the plotted data (Figure S13).
RT-LAMP Amplification of HCV RNA in-Tube
The purified HCV RNA described above was used for in-tube RT-LAMP amplification. The RT-LAMP mix contained the following: 20 μL of RM, 2 μL of EM, 2 μL of SYTO 9 Stain from a 40 μM stock, 4 μL of LAMP primer mixture (20 μM BIP/FIP, 10 μM LB/LF, and 2.5 μM B3/F3), 1 μL of SUPERase In RNase Inhibitor (20 U/μL), EBT solutions of various concentrations and with various amounts of RNA template solution, and enough nuclease-free water to bring the volume to 40 μL. The solution was loaded into 0.2 mL PCR tubes and heated at 63 °C for 50 min and 85 °C for 5 min (heat inactivation) on an Eppendorf Mastercycler Gradient PCR Themal Cycler.
Spectrophotometric Analysis for Positive and Negative RT-LAMP Reactions
Fifty microliters of positive and negative RT-LAMP reaction solutions containing 0.7 mM of EBT, HNB, and calmagite dyes was transferred to a Costar Clear Polystyrene 96-Well Plates, the plate was sealed with a Corning Universal Optical Microplate Sealing Tape and then used for spectrophotometric analysis. An absorption spectra analysis was performed using the POLARstar Omega microplate reader with Omega Data analysis software. The instrument was first set to zero at 700 nm for distilled water, and absorbance in the range of 400–700 nm was recorded at 2 nm intervals. Transmittance was calculated from absorbance values using the following equation: T = 10(2–A).
Prediction of RGB Values
Predicted RGB values for a positive and negative LAMP amplification reaction containing EBT were calculated as follows: (i) The spectral response curves for a Exmor R CMOS image sensor were available only in a graphical format, so data was extracted using Plot Digitizer (ver. 2.6.6) and new plots were generated. (ii) The area under the curve for each of the three color channel spectra was normalized (selecting 1000 arbitrary values under each curve). Uniform white-balanced light source was assumed. (iii) Convolution of the spectral transmittance spectral profiles of the indicator dye for a positive and a negative LAMP reaction solution (experimentally obtained) with the normalized spectral responses from the Exmor R CMOS image sensor was performed. We ignored the light scattering caused by pyrophosphate release during the amplification reaction. As a result, six curves were generated (a positive and negative for each of the three color channels). (iv) The area under each curve was calculated and its square root taken, providing the predicted RGB values for positive and negative RT-LAMP reaction solutions in the presence of EBT at this particular concentration.
dLAMP Amplification of Phage λDNA on Multivolume Rotational SlipChip Devices
To amplify λ phage DNA using dLAMP method, the LAMP mix contained the following: 20 μL of RM, 2 μL of EM, 2 μL of SYTO 9 Stain from 40 μM stock, 4 μL of primer mixture (20 μM BIP/FIP, 10 μM LB/LF, and 2.5 μM B3/F3), 2 μL of BSA (20 mg/mL), various amounts of DNA template solution, 4.7 μL of 6 mM EBT dye (0.7 mM final concentration), and enough nuclease-free water to bring the volume to 40 μL. The solution was loaded onto a multivolume rotational SlipChip device and heated at 63 °C for 50 min on flat block PCR machine (Eppendorf Mastercycler). Five minutes of heating at 85 °C was used to stop the reaction.
Real-Time dRT-LAMP of HCV RNA on Two-Step SlipChip Devices
To amplify HCV viral RNA using dRT-LAMP method on house-built real-time instrument, the RT-LAMP mix contained the following: 20 μL of RM, 2 μL of EM, 2 μL of SYTO 9 Stain from 40 μM stock, 4 μL of primer mixture (20 μM BIP/FIP, 10 μM LB/LF, and 2.5 μM B3/F3), 2 μL of BSA (20 mg/mL), 1 μL of SUPERase In RNAase inhibitor, various amounts of RNA template solution, and enough nuclease-free water to bring the volume to 40 μL. The solution was loaded into the 5 nL wells of two-step SlipChip devices. Other set of wells (9.5 nL) were loaded with 2.4 mM solution of EBT solution (1.57 mM final concentration). SlipChips were heated at 63 °C for 50 min on a house-built real-time instrument; reactions were stopped by heating to 85 °C for 5 min.
House-Built Real-Time Instrument Imaging
Experiments were performed on a Bio-Rad PTC-200 thermocycler with a custom machined block. The block contains a flat 3 × 3 in.2 portion onto which the devices are placed ensuring optimal thermal contact. The excitation light source used was a Philips Luxeon S (LXS8-PW30) 1315 lm LED module with a Semrock filter (FF02-475). Image acquisition was performed with a VX-29MG camera and a Zeiss Macro Planar T F2-100mm lens. A Semrock filter (FF01-540) was used as an emission filter. Images acquired were analyzed using LabVIEW software.
House-Built Real-Time Instrument Data Analysis
Fluorescent images were analyzed using self-developed Labview software. The data were analyzed by first creating a binary mask that defined the location of each reaction volume within the image. The masked spots were then overlaid on the stack of images collected over the course of the experiment and the average intensity of each individual masked spot was tracked over the course of the stack. Background subtraction of the real-time trace was performed by creating a least mean square fit of each individual trace. Threshold was then manually set at the half height of the averaged maximum intensity, and the time-to-positive of each reaction was then determined as the point at which the real-time curve crossed the defined threshold.
Bright-Field Image Acquisition
A mobile phone was used to capture the readout under standard fluorescent light, using the camera’s default autofocus and autoexposure settings. Photographs of the 96-well plate were also taken using alternate commercial cell phones and under different lighting conditions (Figure 3 and Figure S3). Stereoscope imaging was done using Leica MZ Fl III stereoscope with a PLAN 0.5× lens. The stereoscope was equipped with a Diagnostic Instruments color mosaic model 11.2 megapixel camera and images were acquired using Spot imaging software. An automatic white-balance adjustment was done for each image using Spot software. Multiple images were acquired to capture all wells in the device, and assembled to form a complete image of the device to compare with the image acquired from the cell phone camera by using the freeware Image Composite Editor (ver. 2.0).
Bright Field Image Processing and Data Analysis
Images acquired with cell phone and stereoscope were processed using open source ImageJ software (ver.1.49) according to the standard procedure. Briefly: (i) white balance was corrected as needed, (ii) color channels of the original image were split, (iii) one channel was divided by a second channel (e.g., green channel divided by the red channel in the G/R approach) to derive a ratiometric image, and (iv) automatic thresholding was applied to make a binary (black and white) image. Semiautomatic counting on the two-step Slipchip images was accomplished using a freeware Fiji image processing. Acquired bright field images for the multivolume rotational SlipChips were counted manually.
Acknowledgments
This research was supported by DARPA Cooperative Agreement HR0011-11-2-0006. The content of this article does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. Multivolume SlipChip design and analysis were based on work supported by NIH Grant R01EB012946 administered by the National Institute of Biomedical Imaging and Bioengineering. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. R.F.I. holds an Innovation in Regulatory Science Award from BWF. This material is also based upon work supported by National Science Foundation Graduate Research Fellowships DGE-1144469 (to D.V.Z. and E.J). We thank T. Schlappi for multivolume device design calculations and N. Shelby for contributions to writing and editing this manuscript. R.F.I. has a financial interest in SlipChip Corp.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.5b07338.
DNA gel electrophoresis for RT-LAMP product in the presence of indicator dye; each step of the G/R process algorithm; original and G/R-processed images acquired with unmodified cell phone cameras; schematic of the multivolume rotational SlipChip device used in the one-step experiments before being assembled; schematic of the multivolume rotational SlipChip device used for one-step experiments after being assembled; positive counts obtained from single-molecule dLAMP reactions performed with λDNA on a one-step SlipChip device imaged by a house-built real-time fluorescence microscope, a Leica MZ Fl III stereoscope and an unmodified cell phone camera; estimated concentrations and 95% confidence interval range for the set of experiments (one-step) at each volume; performance of bulk LAMP reactions at increasing concentrations of indicator dye; schematic of the two-step SlipChip device before assembly; schematic of the two-step SlipChip device after assembly and its operation; sequence of primers used in RT-LAMP experiments for detection of hepatitis C RNA; and the sequence of primers used in LAMP experiments for detection of phage λDNA; measured spectral transmittance and predicted ratiometric values for positive and negative RT-LAMP reactions containing EBT, HNB, and calmagite; comparison of spectral absorbance of EBT, HNB, and calmagite stock solutions with and without treatment with Chelex 100 ion-exchange resin; stability experiments with the dye stock solutions in their dried states; a table of multivolume designs for clinically relevant dynamic ranges (PDF)
Author Contributions
† J.R.-M. and M.A.K. contributed equally to this work.
The authors declare the following competing financial interest(s): Rustem F. Ismagilov has a financial interest in SlipChip Corp.
Supplementary Material
References
- Calmy A.; Ford N.; Hirschel B.; Reynolds S. J.; Lynen L.; Goemaere E.; de la Vega F. G.; Perrin L.; Rodriguez W. HIV Viral Load Monitoring in Resource-Limited Regions: Optional or Necessary?. Clin. Infect. Dis. 2007, 44, 128–134. 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- Johannessen A. Where We Are with Point-of-Care Testing. J. Viral Hepatitis 2015, 22, 362–365. 10.1111/jvh.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Xu F.; Demirci U. Advances in Developing HIV-1 Viral Load Assays for Resource-Limited Settings. Biotechnol. Adv. 2010, 28, 770–781. 10.1016/j.biotechadv.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P.; Edwards T.; Fu E.; Helton K.; Nelson K.; Tam M. R.; Weigl B. H. Microfluidic Diagnostic Technologies for Global Public Health. Nature 2006, 442, 412–418. 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- Peeling R. W.; Holmes K. K.; Mabey D.; Ronald A. Rapid Tests for Sexually Transmitted Infections (STIs): The Way Forward. Sex. Transm. Infect. 2006, 82, 1–6. 10.1136/sti.2006.024265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P.; Domingo G. J.; Gerdes J. Point-of-Care Diagnostics for Global Health. Annu. Rev. Biomed. Eng. 2008, 10, 107–144. 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- Niemz A.; Ferguson T. M.; Boyle D. S. Point-of-Care Nucleic Acid Testing for Infectious Diseases. Trends Biotechnol. 2011, 29, 240–250. 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom N. K.; Yeung K.; Pillion M. B.; Phillips S. T. ″Fluidic Batteries″ as Low-Cost Sources of Power in Paper-Based Microfluidic Devices. Lab Chip 2012, 12, 1768–1770. 10.1039/c2lc40126f. [DOI] [PubMed] [Google Scholar]
- Labarre P.; Gerlach J.; Wilmoth J.; Beddoe A.; Singleton J.; Weigl B. Non-Instrumented Nucleic Acid Amplification (NINA): Instrument-Free Molecular Malaria Diagnostics for Low-Resource Settings. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2010, 2010, 1097–1099. 10.1109/IEMBS.2010.5627346. [DOI] [PubMed] [Google Scholar]
- Curtis K. A.; Rudolph D. L.; Nejad I.; Singleton J.; Beddoe A.; Weigl B.; LaBarre P.; Owen S. M. Isothermal Amplification Using a Chemical Heating Device for Point-of-Care Detection of HIV-1. PLoS One 2012, 7, e31432. 10.1371/journal.pone.0031432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Mauk M. G.; Hart R.; Qiu X.; Bau H. H. A Self-Heating Cartridge for Molecular Diagnostics. Lab Chip 2011, 11, 2686–2692. 10.1039/c1lc20345b. [DOI] [PubMed] [Google Scholar]
- Sun B.; Shen F.; McCalla S. E.; Kreutz J. E.; Karymov M. A.; Ismagilov R. F. Mechanistic Evaluation of the Pros and Cons of Digital RT-LAMP for HIV-1 Viral Load Quantification on a Microfluidic Device and Improved Efficiency Via a Two-Step Digital Protocol. Anal. Chem. 2013, 85, 1540–1546. 10.1021/ac3037206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F.; Davydova E. K.; Du W. B.; Kreutz J. E.; Piepenburg O.; Ismagilov R. F. Digital Isothermal Quantification of Nucleic Acids Via Simultaneous Chemical Initiation of Recombinase Polymerase Amplification Reactions on SlipChip. Anal. Chem. 2011, 83, 3533–3540. 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selck D. A.; Karymov M. A.; Sun B.; Ismagilov R. F. Increased Robustness of Single-Molecule Counting with Microfluidics, Digital Isothermal Amplification, and a Mobile Phone Versus Real-Time Kinetic Measurements. Anal. Chem. 2013, 85, 11129–11136. 10.1021/ac4030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B.; Rodriguez-Manzano J.; Selck D. A.; Khorosheva E.; Karymov M. A.; Ismagilov R. F. Measuring Fate and Rate of Single-Molecule Competition of Amplification and Restriction Digestion, and Its Use for Rapid Genotyping Tested with Hepatitis C Viral RNA. Angew. Chem., Int. Ed. 2014, 53, 8088–8092. 10.1002/anie.201403035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. Y.; Gao Y. B.; Yu B. W.; Ren H.; Qiu L.; Han S. H.; Jin W.; Jin Q. H.; Mu Y. Self-Priming Compartmentalization Digital LAMP for Point-of-Care. Lab Chip 2012, 12, 4755–4763. 10.1039/c2lc40774d. [DOI] [PubMed] [Google Scholar]
- Gansen A.; Herrick A. M.; Dimov I. K.; Lee L. P.; Chiu D. T. Digital LAMP in a Sample Self-Digitization (Sd) Chip. Lab Chip 2012, 12, 2247–2254. 10.1039/c2lc21247a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witters D.; Sun B.; Begolo S.; Rodriguez-Manzano J.; Robles W.; Ismagilov R. F. Digital Biology and Chemistry. Lab Chip 2014, 14, 3225–3232. 10.1039/C4LC00248B. [DOI] [PubMed] [Google Scholar]
- Sidorova J. M.; Li N.; Schwartz D. C.; Folch A.; Monnat R. J. Jr. Microfluidic-Assisted Analysis of Replicating DNA Molecules. Nat. Protoc. 2009, 4, 849–861. 10.1038/nprot.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. P.; Spence C.; Scherer A.; Quake S. A Microfabricated Device for Sizing and Sorting DNA Molecules. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 11–13. 10.1073/pnas.96.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus J. S.; Anderson W. F.; Quake S. R. Microfluidic Single-Cell mRNA Isolation and Analysis. Anal. Chem. 2006, 78, 3084–3089. 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- Ottesen E. A.; Hong J. W.; Quake S. R.; Leadbetter J. R. Microfluidic Digital PCR Enables Multigene Analysis of Individual Environmental Bacteria. Science 2006, 314, 1464–1467. 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- Beer N. R.; Hindson B. J.; Wheeler E. K.; Hall S. B.; Rose K. A.; Kennedy I. M.; Colston B. W. On-Chip, Real-Time, Single-Copy Polymerase Chain Reaction in Picoliter Droplets. Anal. Chem. 2007, 79, 8471–8475. 10.1021/ac701809w. [DOI] [PubMed] [Google Scholar]
- Kiss M. M.; Ortoleva-Donnelly L.; Beer N. R.; Warner J.; Bailey C. G.; Colston B. W.; Rothberg J. M.; Link D. R.; Leamon J. H. High-Throughput Quantitative Polymerase Chain Reaction in Picoliter Droplets. Anal. Chem. 2008, 80, 8975–8981. 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. O.; Wittwer C. T.; Gao C.; Gale B. K. Spinning Disk Platform for Microfluidic Digital Polymerase Chain Reaction. Anal. Chem. 2010, 82, 1546–1550. 10.1021/ac902398c. [DOI] [PubMed] [Google Scholar]
- Lagally E. T.; Medintz I.; Mathies R. A. Single-Molecule DNA Amplification and Analysis in an Integrated Microfluidic Device. Anal. Chem. 2001, 73, 565–570. 10.1021/ac001026b. [DOI] [PubMed] [Google Scholar]
- Shen F.; Du W.; Kreutz J. E.; Fok A.; Ismagilov R. F. Digital PCR on a Slipchip. Lab Chip 2010, 10, 2666–2672. 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. D.; Linder V.; Sia S. K. Lab-on-a-Chip Devices for Global Health: Past Studies and Future Opportunities. Lab Chip 2007, 7, 41–57. 10.1039/B611455E. [DOI] [PubMed] [Google Scholar]
- Chin C. D.; Laksanasopin T.; Cheung Y. K.; Steinmiller D.; Linder V.; Parsa H.; Wang J.; Moore H.; Rouse R.; Umviligihozo G.; Karita E.; Mwambarangwe L.; Braunstein S. L.; van de Wijgert J.; Sahabo R.; Justman J. E.; El-Sadr W.; Sia S. K. Microfluidics-Based Diagnostics of Infectious Diseases in the Developing World. Nat. Med. 2011, 17, 1015–1019. 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Chin C. D.; Linder V.; Sia S. K. Commercialization of Microfluidic Point-of-Care Diagnostic Devices. Lab Chip 2012, 12, 2118–2134. 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- International Telecommunication Union. 2015 ICT Facts and Figures—the World in 2015. http://www.itu.int/en/ITU-D/Statistics/Pages/facts/default.aspx.
- Whitesides G. M. A Glimpse into the Future of Diagnostics. Clin. Chem. 2013, 59, 589–591. 10.1373/clinchem.2013.204347. [DOI] [PubMed] [Google Scholar]
- Berg B.; Cortazar B.; Tseng D.; Ozkan H.; Feng S.; Wei Q. S.; Chan R. Y. L.; Burbano J.; Farooqui Q.; Lewinski M.; Di Carlo D.; Garner O. B.; Ozcan A. Cellphone-Based Hand-Held Microplate Reader for Point-of-Care Testing of Enzyme-Linked Immunosorbent Assays. ACS Nano 2015, 9, 7857–7866. 10.1021/acsnano.5b03203. [DOI] [PubMed] [Google Scholar]
- Wei Q.; Luo W.; Chiang S.; Kappel T.; Mejia C.; Tseng D.; Chan R. Y.; Yan E.; Qi H.; Shabbir F.; Ozkan H.; Feng S.; Ozcan A. Imaging and Sizing of Single DNA Molecules on a Mobile Phone. ACS Nano 2014, 8, 12725–12733. 10.1021/nn505821y. [DOI] [PubMed] [Google Scholar]
- Laksanasopin T.; Guo T. W.; Nayak S.; Sridhara A. A.; Xie S.; Olowookere O. O.; Cadinu P.; Meng F.; Chee N. H.; Kim J.; Chin C. D.; Munyazesa E.; Mugwaneza P.; Rai A. J.; Mugisha V.; Castro A. R.; Steinmiller D.; Linder V.; Justman J. E.; Nsanzimana S.; et al. A Smartphone Dongle for Diagnosis of Infectious Diseases at the Point of Care. Sci. Transl. Med. 2015, 7, 273re1. 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- Yetisen A. K.; Akram M. S.; Lowe C. R. Paper-Based Microfluidic Point-of-Care Diagnostic Devices. Lab Chip 2013, 13, 2210–2251. 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- Besant J. D.; Das J.; Burgess I. B.; Liu W.; Sargent E. H.; Kelley S. O. Ultrasensitive Visual Read-out of Nucleic Acids Using Electrocatalytic Fluid Displacement. Nat. Commun. 2015, 6, 6978. 10.1038/ncomms7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Carrilho E.; Thomas S. W.; Sindi H.; Whitesides G. M. Simple Telemedicine for Developing Regions: Camera Phones and Paper-Based Microfluidic Devices for Real-Time, Off-Site Diagnosis. Anal. Chem. 2008, 80, 3699–3707. 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Whitesides G. M.; Carrilho E. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal. Chem. 2010, 82, 3–10. 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- Park T. S.; Li W. Y.; McCracken K. E.; Yoon J. Y. Smartphone Quantifies Salmonella from Paper Microfluidics. Lab Chip 2013, 13, 4832–4840. 10.1039/c3lc50976a. [DOI] [PubMed] [Google Scholar]
- Vashist S. K.; Mudanyali O.; Schneider E. M.; Zengerle R.; Ozcan A. Cellphone-Based Devices for Bioanalytical Sciences. Anal. Bioanal. Chem. 2014, 406, 3263–3277. 10.1007/s00216-013-7473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D. J.; Park T. S.; Yoon J. Y. Cell-Phone-Based Measurement of TSH Using Mie Scatter Optimized Lateral Flow Assays. Biosens. Bioelectron. 2013, 40, 180–185. 10.1016/j.bios.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Shen L.; Hagen J. A.; Papautsky I. Point-of-Care Colorimetric Detection with a Smartphone. Lab Chip 2012, 12, 4240–4243. 10.1039/c2lc40741h. [DOI] [PubMed] [Google Scholar]
- Tyburski E. A.; Gillespie S. E.; Stoy W. A.; Mannino R. G.; Weiss A. J.; Siu A. F.; Bulloch R. H.; Thota K.; Cardenas A.; Session W.; Khoury H. J.; O’Connor S.; Bunting S. T.; Boudreaux J.; Forest C. R.; Gaddh M.; Leong T.; Lyon L. A.; Lam W. A. Disposable Platform Provides Visual and Color-Based Point-of-Care Anemia Self-Testing. J. Clin. Invest. 2014, 124, 4387–4394. 10.1172/JCI76666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A.; Erenas M. M.; Marinetto E. D.; Abad C. A.; de Orbe-Paya I.; Palma A. J.; Capitan-Vallvey L. F. Mobile Phone Platform as Portable Chemical Analyzer. Sens. Actuators, B 2011, 156, 350–359. 10.1016/j.snb.2011.04.045. [DOI] [Google Scholar]
- Wang S. Q.; Zhao X. H.; Khimji I.; Akbas R.; Qiu W. L.; Edwards D.; Cramer D. W.; Ye B.; Demirci U. Integration of Cell Phone Imaging with Microchip ELISA to Detect Ovarian Cancer HE4 Biomarker in Urine at the Point-of-Care. Lab Chip 2011, 11, 3411–3418. 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K.; Hase T.; Notomi T. Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Mol. Cell. Probes 2002, 16, 223–229. 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Notomi T.; Okayama H.; Masubuchi H.; Yonekawa T.; Watanabe K.; Amino N.; Hase T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63. 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon G.; Garson J. A.; Grant P.; Nastouli E.; Foy C. A.; Huggett J. F. Comparative Study of Sensitivity, Linearity, and Resistance to Inhibition of Digital and Nondigital Polymerase Chain Reaction and Loop Mediated Isothermal Amplification Assays for Quantification of Human Cytomegalovirus. Anal. Chem. 2014, 86, 4387–4394. 10.1021/ac500208w. [DOI] [PubMed] [Google Scholar]
- Goto M.; Honda E.; Ogura A.; Nomoto A.; Hanaki K. I. Colorimetric Detection of Loop-Mediated Isothermal Amplification Reaction by Using Hydroxy Naphthol Blue. BioTechniques 2009, 46, 167. 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- Tanner N. A.; Zhang Y.; Evans T. C. Jr. Visual Detection of Isothermal Nucleic Acid Amplification Using pH-Sensitive Dyes. BioTechniques 2015, 58, 59–68. [DOI] [PubMed] [Google Scholar]
- Wang D. G. Visual Detection of Mycobacterium Tuberculosis Complex with Loop-Mediated Isothermal Amplification and Eriochrome Black T. Appl. Mech. Mater. 2014, 618, 264–267. 10.4028/www.scientific.net/AMM.618.264. [DOI] [Google Scholar]
- Yetisen A. K.; Martinez-Hurtado J. L.; Garcia-Melendrez A.; Vasconcellos F. D.; Lowe C. R. A Smartphone Algorithm with Inter-Phone Repeatability for the Analysis of Colorimetric Tests. Sens. Actuators, B 2014, 196, 156–160. 10.1016/j.snb.2014.01.077. [DOI] [Google Scholar]
- Safavieh M.; Ahmed M. U.; Sokullu E.; Ng A.; Braescu L.; Zourob M. A Simple Cassette as Point-of-Care Diagnostic Device for Naked-Eye Colorimetric Bacteria Detection. Analyst 2014, 139, 482–487. 10.1039/C3AN01859H. [DOI] [PubMed] [Google Scholar]
- Sony Corp. 2010. Sony “Exmor R” Cmos Image Sensors Achieve a Dramatic Increase in Performance. CX-News Archives , Vol. 59, http://www.sony.net/Products/SC-HP/cx_news_archives/img/pdf/vol_59/featuring_Exmorr.pdf.
- Oh S. J.; Park B. H.; Jung J. H.; Choi G.; Lee D. C.; Kim do H.; Seo T. S. Centrifugal Loop-Mediated Isothermal Amplification Microdevice for Rapid, Multiplex and Colorimetric Foodborne Pathogen Detection. Biosens. Bioelectron. 2016, 75, 293–300. 10.1016/j.bios.2015.08.052. [DOI] [PubMed] [Google Scholar]
- Wang D. G. Visual Detection of Mycobacterium Tuberculosis Complex with Loop-Mediated Isothermal Amplification and Eriochrome Black T. Appl. Mech. Mater. 2014, 618, 264–267. 10.4028/www.scientific.net/AMM.618.264. [DOI] [Google Scholar]
- Sony Corp. 2011. High Picture Quality Cellular Phone CMOS Image Sensors Feature Full HD Video. CX-News Archives, Vol. 65, http://www.sony.net/Products/SC-HP/cx_news_archives/img/pdf/vol_65/imx081_091_111pq.pdf.
- Ma X. J.; Shu Y. L.; Nie K.; Qin M.; Wang D. Y.; Gao R. B.; Wang M.; Wen L. Y.; Han F.; Zhou S. M.; Zhao X.; Cheng Y. H.; Li D. X.; Dong X. P. Visual Detection of Pandemic Influenza a H1N1 Virus 2009 by Reverse-Transcription Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue Dye. J. Virol. Methods 2010, 167, 214–217. 10.1016/j.jviromet.2010.03.027. [DOI] [PubMed] [Google Scholar]
- Nie K.; Zhao X.; Ding X.; Li X. D.; Zou S. M.; Guo J. F.; Wang D. Y.; Gao R. B.; Li X. Y.; Huang W. J.; Shu Y. L.; Ma X. J. Visual Detection of Human Infection with Influenza a (H7n9) Virus by Subtype-Specific Reverse Transcription Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue Dye. Clin. Microbiol. Infect. 2013, 19, E372. 10.1111/1469-0691.12263. [DOI] [PubMed] [Google Scholar]
- Duan Y. B.; Ge C. Y.; Zhang X. K.; Wang J. X.; Zhou M. G. Development and Evaluation of a Novel and Rapid Detection Assay for Botrytis Cinerea Based on Loop-Mediated Isothermal Amplification. PLoS One 2014, 9, e111094. 10.1371/journal.pone.0111094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B. Y.; Liu X. L.; Wei Y. M.; Wang J. Q.; He X. Q.; Jin Y.; Wang Z. J. Rapid and Sensitive Detection of Human Astrovirus in Water Samples by Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue Dye. BMC Microbiol. 2014, 14, 38. 10.1186/1471-2180-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom F.; Diehl H. Indicator for the Titration of Calcium Plus Magnesium with (Ethylenedinitrilo)Tetraacetate. Anal. Chem. 1960, 32, 1123–1127. 10.1021/ac60165a022. [DOI] [Google Scholar]
- Bayer B. E.Color Imaging Array, 1976, US 05/555,477.
- Kreutz J. E.; Munson T.; Huynh T.; Shen F.; Du W. B.; Ismagilov R. F. Theoretical Design and Analysis of Multivolume Digital Assays with Wide Dynamic Range Validated Experimentally with Microfluidic Digital PCR. Anal. Chem. 2011, 83, 8158–8168. 10.1021/ac201658s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F.; Sun B.; Kreutz J. E.; Davydova E. K.; Du W. B.; Reddy P. L.; Joseph L. J.; Ismagilov R. F. Multiplexed Quantification of Nucleic Acids with Large Dynamic Range Using Multivolume Digital RT-PCR on a Rotational Slipchip Tested with HIV and Hepatitis C Viral Load. J. Am. Chem. Soc. 2011, 133, 17705–17712. 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnedo M.; Alonso E.; Eisenberg N.; Ibanez L.; Ferreyra C.; Jaen A.; Flevaud L.; Khamadi S.; Roddy P.; Gatell J. M.; Dalmau D. Monitoring HIV Viral Load in Resource Limited Settings: Still a Matter of Debate?. PLoS One 2012, 7, e47391. 10.1371/journal.pone.0047391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney E. R.; Chung R. T. Antiviral Treatment of Hepatitis C. BMJ 2014, 349, g3308. 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B.; Pockros P. J.; Vilchez R. A.; Vierling J. M. HCV RNA Viral Load Assessments in the Era of Direct-Acting Antivirals. Am. J. Gastroenterol. 2013, 108, 471–475. 10.1038/ajg.2012.248. [DOI] [PubMed] [Google Scholar]
- Du W. B.; Li L.; Nichols K. P.; Ismagilov R. F. Slipchip. Lab Chip 2009, 9, 2286–2292. 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.