Abstract
Metastatic cancer remains the leading cause of death for patients with breast cancer. To understand the mechanisms underlying the development of distant metastases to specific sites is therefore important and of potential clinical value. From 157 primary breast tumours of the patients with known metastatic disease, gene expression profiling data were generated and correlated to metastatic behaviour including site-specific metastasis, metastasis pattern and survival outcomes. We analysed gene expression signatures specifically associated with the development of bone metastases. As a validation cohort, we used a published dataset of 376 breast carcinomas for which gene expression data and site-specific metastasis information were available. 80.5 % of luminal-type tumours developed bone metastasis as opposed to 41.7 % of basal and 55.6 % of HER2-like tumours. A novel 15-gene signature identified 82.4 % of the tumours with bone metastasis, 85.2 % of the tumours which had bone metastasis as first site of metastasis and 100 % of the ones with bone metastasis only (p 9.99e−09), in the training set. In the independent dataset, 81.2 % of the positive tested tumours had known metastatic disease to the bone (p 4.28e−10). This 15-gene signature showed much better correlation with the development of bone metastases than previously identified signatures and was predictive in both ER-positive as well as in ER-negative tumours. Multivariate analyses revealed that together with the molecular subtype, our 15-gene expression signature was significantly correlated to bone metastasis status (p <0.001, 95 % CI 3.86–48.02 in the training set; p 0.001, 95 % CI 1.54–5.00 in the independent set). The 15 genes, APOPEC3B, ATL2, BBS1, C6orf61, C6orf167, MMS22L, KCNS1, MFAP3L, NIP7, NUP155, PALM2, PH-4, PGD5, SFT2D2 and STEAP3, encoded mainly membrane-bound molecules with molecular function of protein binding. The expression levels of the up-regulated genes (NAT1, BBS1 and PH-4) were also found to be correlated to epithelial to mesenchymal transition status of the tumour. We have identified a novel 15-gene expression signature associated with the development of bone metastases in breast cancer patients. This bone metastasis signature is the first to be identified using a supervised classification approach in a large series of patients and will help forward research in this area towards clinical applications.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-016-3741-z) contains supplementary material, which is available to authorized users.
Keywords: Organotropism, Pathology, Site-specific relapse
Introduction
After the initial treatment of primary breast cancer, 20–30 % of patients develop distant metastases [17, 47]. The survival outcomes and sites at which distant metastases develop differ greatly among patients [24, 25, 29, 55]. Several studies have already reported gene expression profiles correlated with risk of distant metastasis, which are in the process of being validated with prospective studies [16, 45, 61]. Moreover, breast cancer’s propensity to spread to certain organs, so-called “non-random organ-specific metastasis”, has also been investigated [3, 10, 14, 30, 41]. There have been several important studies using animal models to unravel the mechanism of site-specific distant metastases in breast cancer [6, 7, 33, 36, 43, 44, 65, 66]. These studies focusing on organotropism of metastatic breast cancer have used human breast cancer cell lines which were injected in immune-compromised mice. By combining genomic profiling of organ-tropic metastatic variants selected in vivo from the animal models of metastatic disease with clinical genomic studies, Massague and his colleagues were able to identify gene expression signatures that were associated with metastasis to bone, lung and brain [7, 33, 43]. They have further explored the association between specific patterns of gene expression and metastatic pattern. The discovered candidate genes were then further investigated and their metastatic role was confirmed by means of overexpressing or inactivating their expression. Hereafter they have validated these gene expression signatures in several cohorts of primary breast tumours with known metastatic disease.
We have recently described the metastatic behaviour (organ-specific metastasis)-related immunophenotypic findings of the primary tumours in a retrospective study including 263 primary breast tumours with known metastatic disease [52]. We have shown that the time to distant metastasis was less than 5 year in 90 % of the hormone receptor negative breast cancer patients as compared to 66 % of hormone receptor-positive patients. The role of estrogen receptor (ER) positivity was found to be closely associated to the development of bone metastasis including bone-only and bone-first metastasis in the course of the disease, whereas ER negativity was found to be related to visceral (liver, lung or brain) metastasis. Along with the hormone status, tumour size and tumour grade, we found that patients who developed visceral metastasis had worse survival outcome, in terms of metastasis-specific survival and overall survival and additionally they frequently developed multiple metastasis during the course of the disease. We have concluded that tumour types were associated with survival and pattern of distant metastasis during the course of the disease. Gene expression profiling patterns predicting site-specific metastasis may aid in better understanding the mechanisms for the development of distant metastases.
In this study, we analysed the gene expression profile of 157 primary tumours that are all metastasized. In order to identify and validate tumour factors of metastatic breast cancer that are predictive of metastatic behaviour, gene expression profiling of primary tumours is correlated to metastasis pattern, and subsequently, gene expression signatures are investigated for prediction of the site-specific distant metastasis.
Materials and methods
The methodology for selection of patient and tumour samples, gene expression profiling experiments, microarray data analysis/bioinformatics and identification and validation of site-specific metastasis signature is described in details in a supplementary file (Supplementary file 1).
Results
For 157 primary invasive breast carcinomas from patients who all developed metastatic disease, mRNA expression signatures were assessed using microarray analysis. The patient characteristics and metastasis patterns are described in Table 1. Tumours were subdivided into 5 molecular subtypes using the PAM50 classifier [48]. Out of 157 cases, 67 (42.7 %) were identified as Luminal A, 46 (29.3 %) as Luminal B, 18 (11.5 %) as HER2-like and 25 (15.9 %) as basal type. One (0.6 %) of these tumours was identified as normal-like. For statistical purposes, the normal-like breast tumour was excluded from the multivariate analysis. Median follow-up time for patients who were alive was 11.5 years (range 6.2–17.3 years). 79.4 % of the patients with Luminal A, 72.5 % of Luminal B, 78.6 % HER2-like and 87.5 % of basal-type tumours received adjuvant therapy. None of the patients received trastuzumab as adjuvant therapy; a subgroup of patients (n = 10) received trastuzumab for treatment of metastatic disease.
Table 1.
Clinical and pathological characteristics of metastatic breast cancer patients
| N | % | |
|---|---|---|
| Age at diagnosis, years | ||
| <50 | 83 | 52.9 |
| >50 | 74 | 47.1 |
| Surgical procedure | ||
| None | 4 | 2.8 |
| Mastectomy | 73 | 51.8 |
| Breast conserving | 64 | 45.4 |
| Adjuvant therapy | ||
| None | 30 | 21.1 |
| Only CT | 50 | 35.2 |
| Only HT | 17 | 12.0 |
| CT + HT | 45 | 31.7 |
| Lymph node status | ||
| None | 43 | 29.3 |
| 1–3 positive | 48 | 32.7 |
| >3 positive | 56 | 38.1 |
| Histology | ||
| Ductal | 134 | 86.5 |
| Lobular | 14 | 9.0 |
| Other | 7 | 4.5 |
| Tumour grade | ||
| 1 | 13 | 8.6 |
| 2 | 84 | 55.3 |
| 3 | 55 | 36.2 |
| Time to distant metastasisa | ||
| Early | 117 | 77.0 |
| Late | 35 | 23.0 |
| Metastasis at first presentation | ||
| No | 141 | 92.8 |
| Yes | 11 | 7.2 |
| Multiple metastasis sites at first presentation | ||
| No | 97 | 64.2 |
| Yes | 54 | 35.8 |
| Multiple metastasis sites during follow-up | ||
| No | 37 | 24.5 |
| Yes | 114 | 75.5 |
CT chemotherapy, HT hormonal therapy
aCut-off point 5 years
Bone was the most frequent site of distant metastasis (71.5 %) followed by liver (51.7 %) and lung (34.4 %). 74.2 % of the patients developed visceral organ metastasis (lung, liver or brain).
Survival analysis revealed that luminal-type tumours had better outcomes in terms of metastasis-specific and overall survival compared to basal-type tumours and HER2-like tumours (p < 0.000). Median time to develop metastasis was 37, 27, 19 and 15 months for Luminal A, Luminal B, HER2-like tumours and basal-type tumours, respectively. 88.3 % of basal-type and HER2-like tumours developed metastases within 5 years versus 72.7 % of luminal A and 76.7 % of Luminal B tumours.
Among luminal subtype 80.5 % of the tumours developed bone metastasis as opposed to, respectively, 41.7 and 55.6 % of basal-type and HER2-like tumours (p 0.001). This group of tumours also composed the 81.8 % of the tumours which metastasized to bone as initial site of metastasis (p 0.001). The rates of development of visceral metastasis were 70.4 % in luminal-type tumours, 87.5 % in basal-type tumours and 77.8 % in HER2-like tumours. Of basal-type tumours, 66.7 % developed visceral metastasis as first metastasis site and 29.2 % of these tumours had only visceral site metastasis during the course of disease (p 0.061 and p 0.034).
The tumour samples from all patients were assigned to the poor prognostic group according to the 70-gene signature [61]. Based on recently published epithelial mesenchymal transition (EMT) gene classifiers [26], 100 of the tumours allocated as EMT-activated and the rest, n = 51, as EMT-non-activated.
Validation of a previously identified gene signature for bone-specific metastasis
First, we have studied the predictive value of the previously published bone metastasis signature of Kang et al. [33]. This signature was assessed as positive in 110 of the tumours in the current study set. All (100 %) Luminal A tumours and 90.7 % of the Luminal B tumours were found be positive for the signature, whereas 33 % of the HER2-like tumours were positive. None of the basal-type tumours were found to be positive for this site-specific metastasis signature. Within this site-specific signature positive subgroup of tumours, 80 % had clinically identified bone metastasis (n = 88, p 4.26e−04). Kang et al’ s 102-gene expression signature for bone metastasis was able to identify 81.5 % of the tumours with bone metastasis, 84.1 % of the tumours which had bone as initial site of metastasis and 100 % (n = 18) of the tumours which had bone-only metastasis in the training set (p values < 0.001, <0.001 and 0.002, respectively. Sensitivity: 81.5 % and specificity: 48.8 %). When tested in ER-positive (n = 108) and ER-negative (n = 43) groups separately, 61.1 % (n = 66) of the ER-positive tumours and 60.4 % (n = 26) of the ER-negative tumours were tested to be positive with this 102-gene expression signature. Out of positively tested ER-positive tumours (n = 66), 83.3 % had clinically evident bone metastasis (p 0.456). Of the 26 bone signature positive tested ER-negative tumours, 50 % had bone metastatic disease (p 1.000).
Supervised classification of bone (specific) metastasis-related genes
To identify site-specific metastasis genes, differentially expressed genes between tumours with bone metastasis and the ones without bone metastasis were explored. A t test was conducted with a p value of <0.01. After application of filtering criteria, differentially expressed genes were identified between two subgroups of tumours with and without bone metastasis. The group of differentially expressed genes were subsequently validated in the training dataset as well as in the independent dataset with the help of K-means and t testing.
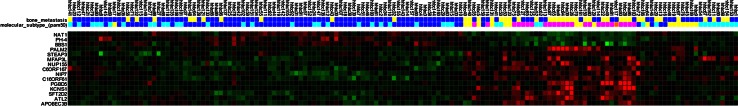
We identified 15 differentially expressed genes between tumours with bone metastasis and the ones without bone metastasis (Table 2). The heat map with gene expression pattern of these 15 genes is displayed in Fig. 1. None of the genes in this set overlapped with the bone signature of Kang et al. Three genes, namely NAT1, PH-4 and BBS1, were up-regulated and the other genes were found to be down-regulated. Mapping into the Gene Ontology and Kyoto Encyclopaedia of Genes and Genomes databases showed an overrepresentation of membrane-bound molecules with molecular function of protein binding (APOPEC3B, ATL2, BBS1, MMS22L, KCNS1, MFAP3L, NIP7, NUP155, PALM2, PH-4 and STEAP3).
Table 2.
The list of differentially expressed genes in bone metastatic disease
| Accession number | HUGO | Description | R value | p value | Level of expression* | |
|---|---|---|---|---|---|---|
| 1 | NM_004900 | APOBEC3B | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B (APOBEC3B), mRNA | −0.407 | 3.55e−03 | < |
| 2 | NM_153485 | NUP155 | Nucleoporin 155 kDa (NUP155), transcript variant 1, mRNA | −0.385 | 8.43e−03 | < |
| 3 | NM_021647 | MFAP3L | Microfibrillar-associated protein 3-like (MFAP3L), transcript variant 1, mRNA | −0.382 | 6.77e−03 | < |
| 4 | NM_016101 | NIP7 | Nuclear import 7 homolog (S. cerevisiae) (NIP7), mRNA | −0.375 | 8.67e−03 | < |
| 5 | NM_198468 | C6orf167 | Chromosome 6 open reading frame 167 (C6orf167), mRNA | −0.371 | 7.22e−03 | < |
| 6 | NM_002251 | KCNS1 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (KCNS1), mRNA | −0.368 | 7.41e−03 | < |
| 7 | NM_001258311 | PGBD5 | PiggyBac transposable element derived 5 (PGBD5), mRNA | −0.364 | 7.88e−03 | < |
| 8 | NM_182915 | STEAP3 | STEAP family member 3 (STEAP3), transcript variant 1, mRNA | −0.364 | 8.77e−03 | < |
| 9 | NM_020188 | C16orf61 | Chromosome 16 open reading frame 61 (C16orf61), mRNA | −0.357 | 9.84e−03 | < |
| 10 | NM_053016 | PALM2 | Paralemmin 2 (PALM2), transcript variant 2, mRNA | −0.356 | 9.02e−03 | < |
| 11 | NM_022374 | ATL2 | Atlastin GTPase 2 (ATL2), mRNA | −0.354 | 9.68e−03 | < |
| 12 | NM_199344 | SFT2D2 | SFT2 domain containing 2 (SFT2D2), mRNA | −0.353 | 9.66e−03 | < |
| 13 | NM_001160170 | NAT1 | N-acetyltransferase 1 (arylamine N-acetyltransferase) (NAT1), mRNA | 0.352 | 9.24e−03 | > |
| 14 | NM_177938 | PH-4 | Hypoxia-inducible factor prolyl 4-hydroxylase (PH-4), transcript variant 2, mRNA | 0.357 | 9.20e−03 | > |
| 15 | NM_024649 | BBS1 | Bardet-Biedl syndrome 1 (BBS1), mRNA | 0.372 | 8.29e−03 | > |
* > up-regulated, < down-regulated
Fig. 1.
The gene expression pattern of 15 genes of bone metastasis gene signature. Heat map shows the gene expression profiling pattern of 15-genes among 151 patients. Primary tumours with clinically evident bone metastasis are illustrated in blue and the ones without bone metastasis are in yellow. For each primary tumour, the expression level of the specific gene is exhibited as red, if up-regulated and green, if down-regulated
In order to validate this gene expression signature, conjointly with our training set, an independent large combined microarray dataset of four studies was analysed. This combined dataset was previously published by Harrell et al. [27]. With the help of K-means clustering method, we have grouped our training dataset and independent dataset into two groups based on their expression levels for our newly developed bone metastasis gene expression signature and subsequently these two groups were compared using a t test.
The 15-gene bone metastasis gene signature was found to be present in 103 tumours in the training dataset. With the help of this signature, 82.4 % of the tumours with known metastatic disease, 85.2 % of the tumours which had bone metastasis as first metastasis site and 100 % of the ones with bone metastasis only were identified (p 9.99e−09, sensitivity: 82.4 % and specificity: 67.4 %). When analysed in the independent dataset, the 15-gene expression signature was found to be present in 160 tumours (total n = 376) and 81.2 % of these positive tested tumours had also clinically evident bone metastatic disease (p 4.28e−10, sensitivity 54.6 % and specificity: 78.2 %). The independent database of Harrell et al. was also utilized to test the bone-specific metastasis of Kang et al. The 102-gene expression signature was assessed as present in 201 tumours (total = 376) and 72.6 % of these tumours reported to have bone metastasis (p 6.92e−05, sensitivity: 61.3 % and specificity: 60.1 %).
In addition, the independent dataset was analysed separately in ER-positive and ER-negative tumours. Among ER-positive tumours (n = 245), the 15-gene expression signature was found to be present in 136 tumours and 83.1 % of these tumours had known bone metastasis; 38.5 % of the negatively tested tumours had no bone metastasis (p 2.38e−04, sensitivity: 79.3 and specificity: 57.1 %). Out of 139 ER-positive tumours which were tested to be positive for the 102-gene expression signature, 75.5 % had bone metastatic disease and 29.2 % of the negatively tested tumours had no bone metastasis (p 0.466, sensitivity: 63.2 % and specificity: 47.6 %). Within the ER-negative subgroup (n = 128), 74 tumours were tested positive for the 15-gene expression signature and 56.8 % these tumours had bone metastasis; 70.4 % of negatively tested tumours had no evidence of bone metastasis (p 3.83e−03, sensitivity: 72.4 % and specificity: 56.8 %). Out of 56 ER-negative tumours which were tested positive for 102-gene expression signature, 55.4 % had clinically bone metastasis; 62.5 % of negatively tested tumours had no bone metastasis (p 0.05, sensitivity: 53.5 % and specificity 64.3 %). Table 3 summarizes the validation of gene signatures in training and independent datasets.
Table 3.
Performance of the gene expression signatures
| Gene expression signatures | Bone metastasis | |||||
|---|---|---|---|---|---|---|
| Signature | Training dataset | Independent dataset | ||||
| Yes | No | p | Yes | No | p | |
| 102-gene expression signature a | ||||||
| All | ||||||
| Present | 88 | 22 | 4.26e−04 | 146 | 55 | 6.92e−05 |
| Absent | 20 | 21 | 92 | 83 | ||
| ER-positive | ||||||
| Present | 55 | 11 | 0.456 | 105 | 34 | 0.466 |
| Absent | 32 | 10 | 75 | 31 | ||
| ER-negative | ||||||
| Present | 13 | 13 | 1.000 | 27 | 45 | 0.051 |
| Absent | 8 | 9 | 31 | 25 | ||
| 15-gene expression signature b | ||||||
| All | ||||||
| Present | 89 | 14 | 9.99e−09 | 130 | 30 | 4.28e−10 |
| Absent | 19 | 29 | 108 | 108 | ||
| ER-positive | ||||||
| Present | 69 | 9 | 1.99e−03 | 113 | 23 | 2.38e−04 |
| Absent | 18 | 12 | 67 | 42 | ||
| ER-negative | ||||||
| Present | 14 | 6 | 0.015 | 42 | 32 | 3.83e−03 |
| Absent | 7 | 16 | 16 | 38 | ||
ER estrogen receptor
aThe 102-gene signature by Kang et al
bThe 15-gene expression signature developed in this study
In addition, in a subsequent study a subset of 50-genes (out of initially identified 102 genes) was selected by Massague’s group [44]; this subset of 50 genes was also analysed in our training and in the independent datasets for its predictive value for bone-specific metastasis The 50-gene signature was able to identify the patients with bone metastasis in the training set (p 1.14e−03) and the independent dataset (p 0.014). When tested in the ER-positive and the ER-negative tumours separately, this 50-gene signature was not predictive for bone metastatic disease.
When tested among all patients with metastatic and not-metastatic disease in the independent dataset (n = 855), the 15-gene signature was able to identify the patients with bone metastasis (p 5.48e04, sensitivity: 54.6 % and specificity: 58.7 %). This gene expression signature remained statistically significant for identification of bone metastasis when separately analysed in ER-positive and ER-negative tumours (p 3.45e−04, sensitivity: 63.9 % and specificity: 52.2 %; p 3.82e-03, sensitivity: 75.9 % and specificity: 45.5 %, respectively).
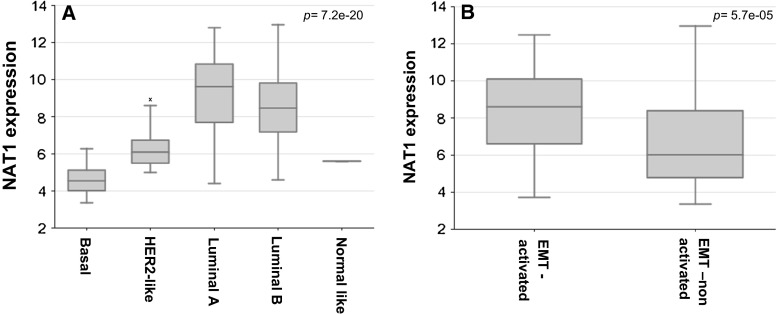
The up-regulated genes and their correlation with molecular subtypes and known prognostic gene signatures were further explored. NAT1 was identified to be expressed at the highest levels in Luminal A followed by Luminal B, HER2-like group and being least expressed in the basal-type group. NAT1 expression was also correlated with the EMT-activated group, being overexpressed in this group of tumours compared to the EMT-non-activated group (p 5.7e−05) (Fig. 2). Similarly the other up-regulated genes, BBS1 and PH-4, were also found to be significantly correlated with the EMT-activated group of tumours (p: 5.8e−04 and p 0.01, respectively).
Fig. 2.
The expression levels (log2) of NAT1 among molecular subtypes (a) and in EMT-activated and EMT-non-activated group (b). The box plots show that NAT1 expression was higher in Luminal-type tumours compared to the other molecular subtypes (p 7.2e−20). NAT1 expression was also found to be higher in the EMT (epithelial to mesenchymal transition)-activated group (p 5.7e−05)
The 15-gene bone metastasis signature was positive in 96.9 % of the Luminal A tumours, in 76.7 % of luminal B tumours and in 38.9 % of HER2-like tumours. Similar to Kang’s bone metastasis signature, none of the basal-like tumours were found to be positive for this signature.
Univariate analyses showed that our bone metastasis signature was significantly correlated to the development of bone metastasis especially in the group of patients who developed only bone metastasis in the course of their disease (p < 0.001). As expected, ER status and molecular subtypes were the parameters that were closely related to bone metastasis status (p < 0.001). Subsequently, multivariate analyses were applied in order to further explore the link between our gene signature and these parameters. Table 4 displays the multivariate analyses results for ER status, molecular subtypes and two separate gene datasets (training and independent) for bone-specific metastasis. As shown, the 15-gene signature was the only parameter that was significantly correlated to bone metastasis status in the training dataset (p < 0.001, 95 % CI 3.86–48.02). In the independent dataset, together with the molecular subtype, the 15-gene signature was significantly correlated to bone metastasis status (p 0.001, 95 % CI 1.54–5.00).
Table 4.
Multivariate analyses results of predictive factors among the gene datasets
| B | Wald x 2 | p | Odds ratio | 95 % CI | |
|---|---|---|---|---|---|
| Training dataset | |||||
| ER status | −0.48 | 0.53 | .468 | 0.620 | 0.17–2.25 |
| Molecular subtype | 0.53 | 0.07 | .793 | 1.05 | 0.71–1.57 |
| 15-gene signaturea | 2.61 | 16.49 | <.000 | 13.61 | 3.86–48.03 |
| Independent dataset | |||||
| ER status | 0.25 | 0.06 | .939 | 1.02 | 0.54–1.96 |
| Molecular subtype | 0.30 | 10.70 | .001 | 1.36 | 1.13–1.64 |
| 15-gene signaturea | 2.62 | 11.54 | .001 | 2.78 | 1.54–5.00 |
ER estrogen receptor, CI confidence interval
aNovel gene expression signature
Discussion
The metastatic potential of the primary tumour revolves around multistep biological processes within host tissue and microenvironment of the distant organ site [20]. In addition to the early origin of genetic instability [4, 19, 20] and hence the metastatic potential of the tumour cells, several intrinsic and extrinsic factors are recognized as potential promoters of metastatic relapse [11, 46, 53]. Upon sustaining the elementary steps of dissemination, the circulating tumour cells can colonize a new organ, forming a detectable metastasis [10, 20].
Experimental models of metastasis yielded distinct sets of genes that mediated site-specific metastasis in breast cancer [7, 33, 36, 43]. Kang et al. identified a bone metastasis signature composed of 102 genes mostly encoding cell surface and secretory proteins, with functions including bone marrow homing and extravasation, pericellular proteolysis and invasion, angiogenesis, osteoclastogenesis, growth factor regulation and extracellular matrix alteration [33]. The authors concluded that this gene set was superimposed on a poor-prognosis gene signature to provide additional functions in order to achieve an overt bone-specific metastasis.
Despite these interesting findings from mouse model system and validation of the results from the mouse models in human breast cancer, no clinical application or follow-up research has emerged since these first findings. Here we present results of the largest study to date on the association between gene expression profiling of primary breast cancer and the development of bone metastases, and the first study in which supervised classification has been used to identify a bone metastasis associated gene expression signature. This gene expression signature was composed of 15 genes, with 3 (NAT1, PH-4 and BBS1) of them being up-regulated in the primary tumour samples. The overexpressed genes in this bone-specific metastasis signature were associated with metabolic (NAT1) and oxidation–reduction (PH-4) processes, and protein transport (BBS1), in agreement with previous works hypothesizing their potential role in altering the host tissue environment in order to achieve a bone metastasis [11, 28, 49, 53].
N-acetyltransferase 1 (NAT1) was first reported to be associated with enhanced growth and survival of breast epithelial cells by Adam et al. [1], and later reported to be a potential biomarker for breast cancer [15, 18, 37, 59, 60]. In several studies, inhibiting NAT1 resulted in cell morphology change, a loss of surface filopodia and subsequent reduction of invasive potential both in vitro and in vivo [60]. Likewise, knockdown of this gene led to inhibition of invasion and metastasis, by means of modification/rearrangement of filopodia (intracellular) actin [58, 59]. In agreement with other gene expression profiling studies in human cancer samples, here we showed that NAT1 clusters close to the estrogen receptor with higher expression levels in luminal-type tumours [1, 5, 56]. Tiang et al. also showed that the loss of NAT1 resulted in alteration of cell-to-cell contact and up-regulation of E-cadherin. Based on aforementioned cell-line studies, a possible association between this gene and EMT/MET has been speculated [58]. Interestingly, in our dataset overexpression of this gene was significantly correlated to the so-called EMT-activated group (p = 5.7e−05). To our knowledge, this is the first study pointing to the association between NAT1 and EMT in human female breast cancer samples. Along with the considerations of the potentiality of this gene as a drug target [57, 58], we believe that further studies in human breast cancer samples are indicated to explore this link.
The extracellular matrix (ECM) plays important role in diverse pathological and physiological processes including cancer invasion and metastasis [22, 32]. Collagens compose the major component of ECM. Increased expression of collagens, thereupon increase in deposition and stiffening in ECM, is associated with tumour progression [38, 50]. Collagen prolyl 4-hydroxylase (PH-4), a member of post-transcription modification enzyme family, is required in collagen biosynthesis and angiogenesis. Hypoxia-induced collagen prolyl 4-hydroxylase expression is reported to be associated with increased progression and mortality in breast cancer [12, 21, 50]. Indeed, animal studies showed that knockdown of PH-4 resulted in inhibition of tumour growth and lung metastasis [23, 62]. With gene expression profiling of breast cancer samples, we have found that PH-4 was positively correlated with site-specific metastasis to bone. This finding confirms the observations by others [22, 32, 38, 50] and advocates for the importance of extracellular matrix alterations in disease progression.
Twelve out of 15 genes were found to be down-regulated in the primary tumours of breast cancer patients who developed bone metastasis. One of these genes, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like-3B (APOBEC3B), is reported to be up-regulated in a large proportion of breast tumours and high levels of APOBEC3B were found to be associated with worse disease-free and overall survival [8, 9, 51, 54]. Recently, several independent genome-wide association studies have shown a deletion resulting in complete elimination of the APOBEC3B gene-encoding region [34, 40, 63]. This deletion has been indicated to be associated with decreased expression of APOBEC3B in breast cancer cells [34]. In this study, we have also shown that APOBEC3B was significantly down-regulated in the group of tumours with bone metastatic disease (p 3.55e−03). We believe that further copy number variations studies are required to explore such an association between APOBEC3B deletion and site-specific metastasis. Six-transmembrane epithelial antigen of prostate 3 (STEAP3), which is thought be involved in apoptosis and cell-cycle progression [2, 39, 64], is also found to be down-regulated in the bone metastatic group of primary breast tumours in our study. STEAP3 expression is shown to be diminished in hepatocellular carcinoma nodules compared to cirrhotic peritumoral tissue and healthy liver [13]. Another family member of these proteins, STEAP1, has already shown to be overexpressed in breast cancer cells [31, 35, 42]. However, we could not retrieve any similar data pointing STEAP3 expression levels in breast cancer tissues.
In order to determine the validity of the experimentally derived 102 gene bone metastasis signature, Kang et al. have utilized a cohort of 63 primary breast carcinomas to test this signature. The authors have selected a subset of 50 genes to carry on their validation studies and they have shown that this gene set was not able to identify the group of tumours with bone metastasis. When the authors restricted their analyses to 25 breast tumours with known metastatic disease, they were able to distinguish the tumours preferentially metastasized to bone rather than other distant organs [44]. In this current study along with new identified 15-gene expression signature, we have shown that the 102-gene expression signature and the subset of 50 genes as reported by Kang et al. were informative in identifying likelihood of developing bone metastasis in the training and the independent datasets. However, when datasets subdivided into two groups according to their ER status, the 102-gene expression signature as well as the 50-gene signature were not effective in predicting bone metastasis, whereas herein identified 15-gene expression signature remained associated with the likelihood of bone metastasis development in ER-positive and ER-negative tumour groups.
Notably, the bone-specific metastasis signature presented in this study did not include any of the genes from already published Kang’s bone signature [33]. The absence of overlap between these gene sets could be justified with the fact that in the former study tumour cells from the metastasis site were utilized to generate gene signatures in contrast to primary tumours in the current study. Considering that tumour progression and development of metastasis requires compiled steps of modification, we may assume that these two different gene signature sets play a complementary role in separate levels of this multi-complex process.
Notwithstanding several well-received studies focusing on the biology of metastatic breast cancer, little progress has been made over the past years to identify a robust gene expression signature for site-specific metastasis. Moreover, the experimentally derived gene expression signatures when tested in human breast carcinomas were not as strongly associated with site-specific metastasis as in the experimental conditions. A reproducible gene expression signature associated with the development of bone metastases in breast cancer will have clinical utility in two ways: first, the knowledge of the specific gene expressed at higher or lower levels in the metastatic disease will lead to the investigation of targeted therapy options directed to the altered mechanism related to this gene, and second, reliable identification of the patients at high risk of developing bone metastases may lead to therapeutic interventions specifically aimed at preventing the development of bone metastases, for example treatment with bisphosphonates.
In summary, we present the largest study to date revealing the association between the gene expression profiling patterns and bone-specific metastasis in breast carcinomas. The identification of novel 15-gene expression signature will forward this area of research, including subsequent exploration of the underlying mechanisms of metastatic behaviour and ultimately help improve outcome for breast cancer patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by the Center for Translational Molecular Medicine (BreastCARE).
Abbreviations
- ER
Estrogen receptor
- PR
Progesterone receptor
- HER2
Human epidermal growth factor receptor 2
- MSS
Metastasis-specific survival
- OS
Overall survival
- EMT
Epithelial to mesenchymal transition
- ECM
Extracellular matrix
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
This study material was strictly handled after coding of the data according to national ethical guidelines of ‘Code for Proper Secondary Use of Human Tissue’ developed by Federation of Medical Societies (FMWV) in the Netherlands. Therefore, the need for obtaining informed consent was waived by the Medical Ethical Committee of the Academic Medical Center.
Contributor Information
C. Dilara Savci-Heijink, Phone: +31-20-566 4021, Email: c.d.savciheijink@amc.uva.nl.
Hans Halfwerk, Email: j.b.halfwerk@amc.uva.nl.
Jan Koster, Email: jankoster@amc.uva.nl.
Marc J. van de Vijver, Phone: +31-20-566 4100, Email: m.j.vandevijver@amc.uva.nl
References
- 1.Adam PJ, Berry J, Loader JA, Tyson KL, Craggs G, Smith P, De Belin J, Steers G, Pezzella F, Sachsenmeir KF, Stamps AC, Herath A, Sim E, O’Hare MJ, Harris AL, Terrett JA. Arylamine N-acetyltransferase-1 is highly expressed in breast cancers and conveys enhanced growth and resistance to etoposide in vitro. Mol Cancer Res. 2003;1:826–835. [PubMed] [Google Scholar]
- 2.Aerbajinai W, Giattina M, Lee YT, Raffeld M, Miller JL. The proapoptotic factor Nix is coexpressed with Bcl-xL during terminal erythroid differentiation. Blood. 2003;102:712–717. doi: 10.1182/blood-2002-11-3324. [DOI] [PubMed] [Google Scholar]
- 3.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 4.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 5.Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6:R252–R263. doi: 10.1186/bcr784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos PD, Nguyen DX, Massague J. Modeling metastasis in the mouse. Curr Opin Pharmacol. 2010;10:571–577. doi: 10.1016/j.coph.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 11.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Borresen-Dale AL, Giaccia A, Longaker MT, Hastie T, Yang GP, van de Vijver MJ, Brown PO. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, Friedl A, Keely PJ. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulouarn C, Derambure C, Lefebvre G, Daveau R, Hiron M, Scotte M, Francois A, Daveau M, Salier JP. Global gene repression in hepatocellular carcinoma and fetal liver, and suppression of dudulin-2 mRNA as a possible marker for the cirrhosis-to-tumor transition. J Hepatol. 2005;42:860–869. doi: 10.1016/j.jhep.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 14.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM, Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolled-Filhart M, Ryden L, Cregger M, Jirstrom K, Harigopal M, Camp RL, Rimm DL. Classification of breast cancer using genetic algorithms and tissue microarrays. Clin Cancer Res. 2006;12:6459–6468. doi: 10.1158/1078-0432.CCR-06-1383. [DOI] [PubMed] [Google Scholar]
- 16.Drukker CA, van Tinteren H, Schmidt MK, Rutgers EJ, Bernards R, van de Vijver MJ, Van’t Veer LJ. Long-term impact of the 70-gene signature on breast cancer outcome. Breast Cancer Res Treat. 2014;143:587–592. doi: 10.1007/s10549-013-2831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Early Breast Cancer Trialists’ Collaborative G Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 18.Endo Y, Yamashita H, Takahashi S, Sato S, Yoshimoto N, Asano T, Hato Y, Dong Y, Fujii Y, Toyama T. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer. 2014;14:990. doi: 10.1186/1471-2407-14-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 20.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 21.Fox SB, Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Bersiga A, Allevi G, Milani M, Aguggini S, Mele T, Dogliotti L, Bottini A, Harris AL. The prolyl hydroxylase enzymes are positively associated with hypoxia-inducible factor-1alpha and vascular endothelial growth factor in human breast cancer and alter in response to primary systemic treatment with epirubicin and tamoxifen. Breast Cancer Res. 2011;13:R16. doi: 10.1186/bcr2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Giordano SH, Buzdar AU, Smith TL, Kau SW, Yang Y, Hortobagyi GN. Is breast cancer survival improving? Cancer. 2004;100:44–52. doi: 10.1002/cncr.11859. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg PA, Hortobagyi GN, Smith TL, Ziegler LD, Frye DK, Buzdar AU. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J Clin Oncol. 1996;14:2197–2205. doi: 10.1200/JCO.1996.14.8.2197. [DOI] [PubMed] [Google Scholar]
- 26.Groger CJ, Grubinger M, Waldhor T, Vierlinger K, Mikulits W. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One. 2012;7:e51136. doi: 10.1371/journal.pone.0051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, Anders C, Ewend M, Perou CM. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132:523–535. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 29.Hayes DF, Van Zyl JA, Hacking A, Goedhals L, Bezwoda WR, Mailliard JA, Jones SE, Vogel CL, Berris RF, Shemano I, et al. Randomized comparison of tamoxifen and two separate doses of toremifene in postmenopausal patients with metastatic breast cancer. J Clin Oncol. 1995;13:2556–2566. doi: 10.1200/JCO.1995.13.10.2556. [DOI] [PubMed] [Google Scholar]
- 30.Hu G, Kang Y, Wang XF. From breast to the brain: unraveling the puzzle of metastasis organotropism. J Mol Cell Biol. 2009;1:3–5. doi: 10.1093/jmcb/mjp005. [DOI] [PubMed] [Google Scholar]
- 31.Hubert RS, Vivanco I, Chen E, Rastegar S, Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, Jakobovits A, Saffran DC, Afar DE. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/S1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 34.Komatsu A, Nagasaki K, Fujimori M, Amano J, Miki Y. Identification of novel deletion polymorphisms in breast cancer. Int J Oncol. 2008;33:261–270. [PubMed] [Google Scholar]
- 35.Korkmaz CG, Korkmaz KS, Kurys P, Elbi C, Wang L, Klokk TI, Hammarstrom C, Troen G, Svindland A, Hager GL, Saatcioglu F. Molecular cloning and characterization of STAMP2, an androgen-regulated six transmembrane protein that is overexpressed in prostate cancer. Oncogene. 2005;24:4934–4945. doi: 10.1038/sj.onc.1208677. [DOI] [PubMed] [Google Scholar]
- 36.Landemaine T, Jackson A, Bellahcene A, Rucci N, Sin S, Abad BM, Sierra A, Boudinet A, Guinebretiere JM, Ricevuto E, Nogues C, Briffod M, Bieche I, Cherel P, Garcia T, Castronovo V, Teti A, Lidereau R, Driouch K. A six-gene signature predicting breast cancer lung metastasis. Cancer Res. 2008;68:6092–6099. doi: 10.1158/0008-5472.CAN-08-0436. [DOI] [PubMed] [Google Scholar]
- 37.Laurieri N, Crawford MH, Kawamura A, Westwood IM, Robinson J, Fletcher AM, Davies SG, Sim E, Russell AJ. Small molecule colorimetric probes for specific detection of human arylamine N-acetyltransferase 1, a potential breast cancer biomarker. J Am Chem Soc. 2010;132:3238–3239. doi: 10.1021/ja909165u. [DOI] [PubMed] [Google Scholar]
- 38.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/MCB.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long J, Delahanty RJ, Li G, Gao YT, Lu W, Cai Q, Xiang YB, Li C, Ji BT, Zheng Y, Ali S, Shu XO, Zheng W. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105:573–579. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorusso G, Ruegg C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin Cancer Biol. 2012;22:226–233. doi: 10.1016/j.semcancer.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Maia CJ, Socorro S, Schmitt F, Santos CR. STEAP1 is over-expressed in breast cancer and down-regulated by 17beta-estradiol in MCF-7 cells and in the rat mammary gland. Endocrine. 2008;34:108–116. doi: 10.1007/s12020-008-9113-7. [DOI] [PubMed] [Google Scholar]
- 43.Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, Kreike B, Zhang Y, Wang Y, Ishwaran H, Foekens JA, van de Vijver M, Massague J. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, Ponomarev V, Gerald WL, Blasberg R, Massague J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Investig. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mook S, Knauer M, Bueno-de-Mesquita JM, Retel VP, Wesseling J, Linn SC, Van’t Veer LJ, Rutgers EJ. Metastatic potential of T1 breast cancer can be predicted by the 70-gene MammaPrint signature. Ann Surg Oncol. 2010;17:1406–1413. doi: 10.1245/s10434-009-0902-x. [DOI] [PubMed] [Google Scholar]
- 46.Nuyten DS, Hastie T, Chi JT, Chang HY, van de Vijver MJ. Combining biological gene expression signatures in predicting outcome in breast cancer: an alternative to supervised classification. Eur J Cancer. 2008;44:2319–2329. doi: 10.1016/j.ejca.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Shaughnessy J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10(Suppl 3):20–29. doi: 10.1634/theoncologist.10-90003-20. [DOI] [PubMed] [Google Scholar]
- 48.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/S1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 50.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savci-Heijink CD, Halfwerk H, Hooijer GK, Horlings HM, Wesseling J, van de Vijver MJ. Retrospective analysis of metastatic behaviour of breast cancer subtypes. Breast Cancer Res Treat. 2015;150:547–557. doi: 10.1007/s10549-015-3352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 54.Sieuwerts AM, Willis S, Burns MB, Look MP, Meijer-Van Gelder ME, Schlicker A, Heideman MR, Jacobs H, Wessels L, Leyland-Jones B, Gray KP, Foekens JA, Harris RS, Martens JW. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer. 2014;5:405–413. doi: 10.1007/s12672-014-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sledge GW, Jr, Hu P, Falkson G, Tormey D, Abeloff M. Comparison of chemotherapy with chemohormonal therapy as first-line therapy for metastatic, hormone-sensitive breast cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol. 2000;18:262–266. doi: 10.1200/JCO.2000.18.2.262. [DOI] [PubMed] [Google Scholar]
- 56.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugamori KS, Wong S, Gaedigk A, Yu V, Abramovici H, Rozmahel R, Grant DM. Generation and functional characterization of arylamine N-acetyltransferase Nat1/Nat2 double-knockout mice. Mol Pharmacol. 2003;64:170–179. doi: 10.1124/mol.64.1.170. [DOI] [PubMed] [Google Scholar]
- 58.Tiang JM, Butcher NJ, Cullinane C, Humbert PO, Minchin RF. RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One. 2011;6:e17031. doi: 10.1371/journal.pone.0017031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiang JM, Butcher NJ, Minchin RF. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer Med. 2015;4:565–574. doi: 10.1002/cam4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiang JM, Butcher NJ, Minchin RF. Small molecule inhibition of arylamine N-acetyltransferase Type I inhibits proliferation and invasiveness of MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun. 2010;393:95–100. doi: 10.1016/j.bbrc.2010.01.087. [DOI] [PubMed] [Google Scholar]
- 61.van de Vijver MJ, He YD, van’t Veer LI, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van de Vijver, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 62.Wottawa M, Leisering P, Ahlen M, Schnelle M, Vogel S, Malz C, Bordoli MR, Camenisch G, Hesse A, Napp J, Alves F, Kristiansen G, Farhat K, Katschinski DM. Knockdown of prolyl-4-hydroxylase domain 2 inhibits tumor growth of human breast cancer MDA-MB-231 cells by affecting TGF-beta1 processing. Int J Cancer. 2013;132:2787–2798. doi: 10.1002/ijc.27982. [DOI] [PubMed] [Google Scholar]
- 63.Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, Shu XO, Kelley MC, Zheng W, Long J. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013;34:2240–2243. doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW., 2nd Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 65.Zhang XH, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, Smid M, Foekens JA, Massague J. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.