Abstract
Background
Wolf–Hirschhorn syndrome (WHS) is a contiguous gene deletion syndrome involving variable size deletions of the 4p16.3 region. Seizures are frequently, but not always, associated with WHS. We hypothesised that the size and location of the deleted region may correlate with seizure presentation.
Methods
Using chromosomal microarray analysis, we finely mapped the breakpoints of copy number variants (CNVs) in 48 individuals with WHS. Seizure phenotype data were collected through parent-reported answers to a comprehensive questionnaire and supplemented with available medical records.
Results
We observed a significant correlation between the presence of an interstitial 4p deletion and lack of a seizure phenotype (Fisher's exact test p=3.59e-6). In our cohort, there were five individuals with interstitial deletions with a distal breakpoint at least 751 kbp proximal to the 4p terminus. Four of these individuals have never had an observable seizure, and the fifth individual had a single febrile seizure at the age of 1.5 years. All other individuals in our cohort whose deletions encompass the terminal 751 kbp region report having seizures typical of WHS. Additional examples from the literature corroborate these observations and further refine the candidate seizure susceptibility region to a region 197 kbp in size, starting 368 kbp from the terminus of chromosome 4.
Conclusions
We identify a small terminal region of chromosome 4p that represents a seizure susceptibility region. Deletion of this region in the context of WHS is sufficient for seizure occurrence.
Keywords: Epilepsy and seizures, Clinical genetics, Microarray, Neurology
Introduction
Wolf–Hirschhorn syndrome (WHS; OMIM #194190) is a genetic disorder occurring in 1:20 000 to 1:50 000 births.1 Females are approximately twice as likely as males to be affected.2 The syndrome was first described by Hirschhorn and Cooper in a preliminary report in 1961 and later formalised with back-to-back publications by Wolf et al and Hirschhorn et al in Humangenetik in 1965.3 WHS is characterised by a specific pattern of craniofacial features including a wide nasal bridge that extends to the forehead, widely spaced eyes, distinct mouth, short philtrum, micrognathia, prenatal and postnatal growth delay, intellectual disability (ID) and seizures.2–12 Following identification of these features, WHS has historically been diagnosed by karyotype and/or FISH. Submicroscopic deletions associated with this disorder have more recently been identified by chromosomal microarray analysis (CMA).
In addition to the core features of WHS listed above, additional highly variable clinical features of WHS include, but are not limited to, feeding difficulties, congenital heart defects, hearing loss, skeletal anomalies, kidney and urinary tract malformations, and ophthalmological and dental abnormalities.2 Terminal deletion resulting in partial monosomy of chromosome 4p is the most common cause of WHS. Interstitial deletions, unbalanced translocations, ring chromosomes and other complex genetic rearrangements can also give rise to WHS.2 4 5 As a result, deletions associated with WHS are highly variable in size and genetic content, potentially causing or contributing to the variability in presentation of this disorder.
Two adjacent regions, located approximately 1.8–2.0 Mbp from the 4p terminus, are each proposed to be the minimal region of deletion necessary to observe the core WHS features. These regions were identified based on determination of the smallest region of overlap (SRO) of individuals with WHS. The first critical region described was a 165 kbp interval encompassing part of the WHSC1 gene and all of the WHSC2 (NELFA) gene.6 These genes play a role in the regulation of key bone differentiation genes7 and regulation of DNA replication and cell-cycle progression.8 The identification of two patients with the WHS phenotype who have more distal deletions led to the proposal that the critical region (designated WHSCR2) lies in an adjacent, 300–600 kbp interval that includes the 5′ end of WHSC1 and the entirety of LETM1, a candidate seizure gene9–11 (figures 1 and 2).
Figure 1.
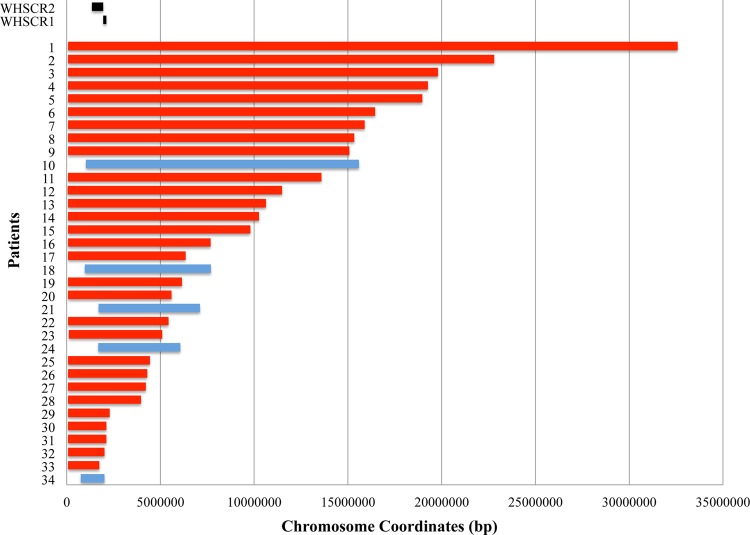
Size and relative locations of 4p deletions of 34 patients with no other clinically reportable CNV findings (henceforth designated as ‘individuals with only 4p deletions’). The deletions of individuals with seizures are shown in red. Deletions of individuals without seizures are shown in blue. The Wolf–Hirschhorn syndrome (WHS) critical regions 1 and 2 (WHSCR1 and WHSCR2) are shown in black; all patients with the exception of patient 33 have deletion encompassing both critical regions. Patient 33's deletion partially overlaps with WHSCR2 only and excludes LETM1. Patient 34's deletion starts 751 kbp from the 4p terminus and is the patient deletion that lies closest to the 4p terminus. All chromosome coordinates for this patient group are given in online supplementary table S1.
Figure 2.
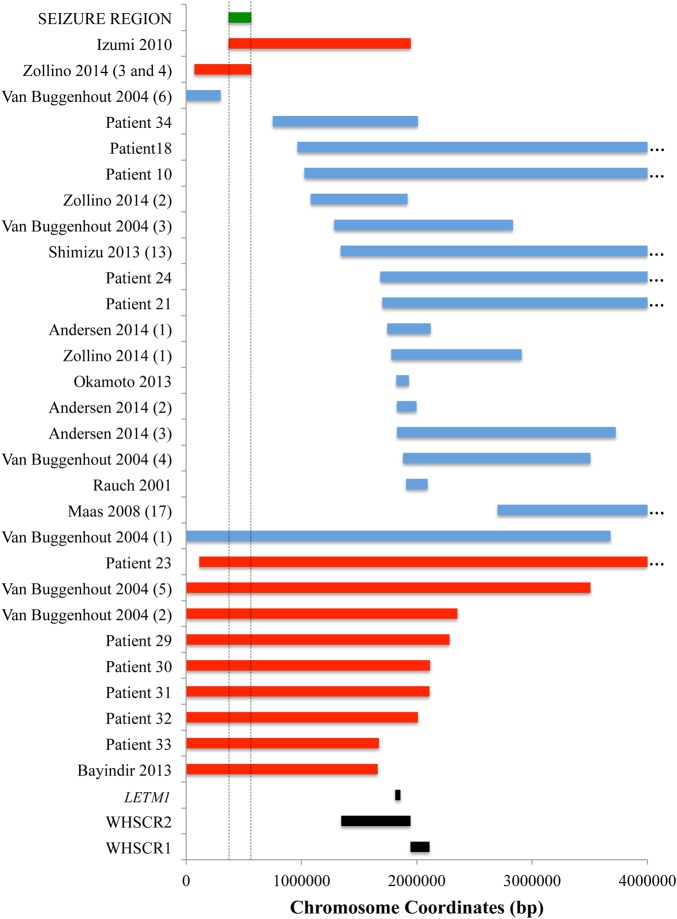
Mapping a candidate seizure propensity region on chromosome 4. Bars show deletion sizes and locations of small 4p terminal or interstitial deletions in the 4p region that help define a 197 kbp seizure susceptibility region. The smallest region of overlap between three patients with seizures is shown as a green bar, ‘SEIZURE REGION’. This region is supported by patients from our cohort (patient numbers labelled on Y-axis) as well as from the literature who have deletions excluding the seizure region and lack seizures (blue indicates no seizures) and patients who have deletions including the seizure region who have seizures (red indicates a seizure phenotype). Patient data from the literature are indicated along the Y-axis by citation followed by the number of the patient as assigned in the citation in parentheses. Correspondingly, ‘Zollino 2014 (3 and 4)’ labels the size and location of the deletion shared by siblings, patients 3 and 4, in Zollino et al 27. Landmarks such as the Wolf–Hirschhorn syndrome (WHS) critical regions 1 and 2 (WHSCR1 and WHSCR2) are shown (black), as well as the location of the LETM1 gene (black). Coordinates are given in base pairs (bps) along the X-axis. Ellipses (…) indicate that the deletion extends further than shown. Chromosome coordinates for all deletions and regions shown in this figure are given in online supplementary table S4.
Epilepsy represents a major clinical challenge during early years, with significant impact on quality of life. Seizures occur in over 90% of individuals with WHS with onset typically within the first 3 years of life and are often induced by low-degree fever.12 The most frequently occurring seizure types are generalised tonic–clonic seizures, tonic spasms, complex partial seizures and clonic seizures. Unilateral/generalised clonic or tonic–clonic status epilepticus occurs in 50% of individuals with WHS.12
A significant challenge to understanding the genetics of WHS is the identification of a gene or genes that, when in hemizygous state, give rise to the core features and variable co-morbidities of WHS. Because WHS is a contiguous gene deletion syndrome, loss of one copy of a single gene or the synergistic effects of loss of two or more genes could give rise to the features of WHS. One such gene, LETM1, falls within WHSCR2 and has been proposed as a candidate seizure gene,9 13–15 due to the suggested pathogenic link between mitochondrial dysfunction and epilepsy.16 The protein encoded by LETM1 localises to mitochondria and functions in Ca2+ homeostasis, oxidative stress prevention and ATP generation.17–19 Consistent with the hypothesis that LETM1 is a seizure susceptibility gene, heterozygous Letm1± mice, as well as rats with a lentiviral-mediated Letm1 knockdown, demonstrate increased seizure susceptibility in response to kainic acid or pilocarpine seizure induction.15 19
Despite this evidence, LETM1 is not likely to be the sole seizure susceptibility gene in the 4p region. In recent years, increased awareness of the diagnostic features of WHS within the medical community, coupled with the advent of high-resolution cytogenetic methods, has led to the identification and characterisation of submicroscopic 4p deletions. Some of these deletions suggest that LETM1 deletion is neither necessary nor sufficient for the expression of a seizure phenotype in individuals with WHS4 5 20–26 and have led to the proposal of alternative candidate seizure genes.27
Here, we present the identification of a seizure-susceptibility region by the use of high-density microarray analysis combined with parent-reported seizure phenotypes. A relatively large, 48-individual cohort was recruited through partnership with the 4p- Support Group.28 Evaluation of deletion coordinates and seizure phenotypes in this cohort identified a likely seizure susceptibility region within the 751 kbp terminal region of chromosome 4p. Combining these data with cases described in the literature, we narrowed this seizure susceptibility region to a region 197 kbp in size that includes two genes and one pseudogene. We also describe the types of seizures associated with WHS observed in our cohort and the response to antiepileptic medications reported by our cohort. Our study demonstrates the potential value of using high-resolution CMA for the diagnosis and medical management of seizures associated with WHS.
Methods
This retrospective study correlated clinical genetic testing results from high-resolution CMA with clinical traits related to WHS. Clinical feature data were collected using a comprehensive parent-completed questionnaire (provided in online supplementary materials), coupled with a review of available medical records. This study was approved by the University of Utah Institutional Review Board. Informed consent and/or parental authorisation, as appropriate, was obtained for each patient.
Patient cohort
Forty-eight individuals with a diagnosis of WHS, along with their parents, consented to this study during one of two national meetings of the 4p- Support Group held in July 2012 in Indianapolis, Indiana, and July 2014 in Harrisburg, Pennsylvania.28 In total, 28 females and 20 males with WHS, with an average age of 11.2 years, were recruited into this study (table 1).
Table 1.
Clinical and molecular cytogenetic findings of the study cohort
| Total participants | 48 | Female:male | 28:20 |
|---|---|---|---|
| Average age | 11.2 years | Range | 0.9–38 years |
| Initial diagnosis by karyotype/FISH | 88% (30/34) | Initial diagnosis by CMA | 12% (4/34) |
| Size range of 4p deletion | 1.7–33.9 Mbp | Number of genes deleted | 28–207 |
| Individuals with a second CNV | 29% (14/48) | Average size of second CNV | 3.2 Mbp (range 51.3 kbp to 8.3 Mbp) |
| Individuals with only a 4p deletion by deletion type | Interstitial: 5 | Terminal: 29 |
CMA, chromosomal microarray analysis.
Clinical and molecular cytogenetic studies
All cytogenetic analyses were performed through regular clinical services in clinical laboratory improvement amendments (CLIA)-certified laboratories. All genomic coordinates for CNVs are reported herein using human reference sequence hg19/GRCh37. All patients (exceptions noted below) were physician referred for clinical microarray testing to Lineagen (Salt Lake City, Utah, USA). Testing for these patients was done using Lineagen's custom 2.8M probe SNP-based microarray. The Affymetrix Chromosome Analysis Suite (ChAS) software was used for CNV detection (Affymetrix, Santa Clara, California, USA). Exceptions to the above were as follows: a 2.7M probe Cytogenetics Array (Affymetrix) was performed by Lineagen on patients 35 and 40. Patients 12, 17 and 45 obtained prior clinical CMA from other CLIA laboratories, and these patients provided a copy of their laboratory reports for analysis.
Phenotype analysis
Phenotype data were collected through parent-reported answers to a comprehensive questionnaire developed by Battaglia et al29 (see online supplementary materials). This questionnaire captures the health, medical profile, developmental history, and treatment responses of individuals with WHS. For the present study, we focused our attention on the presence or absence of seizures, age of seizure onset, types of seizures, antiepileptic drugs (AEDs) used and responses to these AEDs, as well as responses to the ketogenic diet. For cases with incomplete, contradictory or unclear parental responses, medical records of patients were consulted. When available medical records were also incomplete, ‘no answer’ is indicated in the relevant text and tables.
Statistical methods
Two-tailed Fisher's exact test was used for comparing the group of individuals with interstitial 4p deletions to the group with terminal deletions and their seizure phenotypes. Significance was defined as p<0.01.
Results
Table 1 shows the age and gender characteristics of this study cohort. Prior to this study, the initial diagnosis of WHS was made by individuals’ physicians using clinical assessment and a combination of G-banded karyotyping and FISH, or CMA (table 1). Fourteen individuals did not indicate which method(s) were used in their initial diagnosis.
Physician-ordered CMA was performed on the 44 individuals comprising the cohort who had not already had chromosomal microarray testing done as part of their diagnostic work up. The array used was a custom 2 784 985-probe chromosomal microarray to achieve high-resolution mapping of the 4p deletion breakpoints, as well as to define the breakpoints of any other clinically reportable CNVs that could be detected (see online supplementary table S1).
Twenty-nine per cent of our cohort had a second deletion or duplication involving either chromosome 4 or another chromosome. This percentage is in keeping with previous studies of chromosomal rearrangements associated with WHS4 (table 1).
Some of the second CNVs in our cohort are pathogenic, while others are of unknown clinical significance. The pathogenic CNVs are associated with developmental delay, ID, autism spectrum disorder, dysmorphic features and seizures. The breakpoints of all patients’ 4p deletions, as well as the breakpoints of the second CNV if present, and the association of this second CNV to any clinical features are shown in online supplementary tables S1 and S2.
Consistent with previous studies,30–33 we found that 90% (43/48) of our cohort had seizures, which were of early onset (see online supplementary tables S1 and S2), were often brought on by fever (25/41 individuals reported having febrile seizures) and tended to wane in frequency during the preteen years. All seizure types surveyed (tonic–clonic, tonic, clonic, myoclonic, absence, atonic, complex partial, simple partial, atypical and status epilepticus) were detected in this cohort. The seizure types most commonly reported in our WHS cohort are shown in table 2.
Table 2.
Most frequently reported seizure types
| Type | Individuals with only 4p deletion | Individuals with 4p deletion and an additional CNV |
|---|---|---|
| Tonic–clonic | 19/24 (79%) | 9/13 (69%) |
| Absence | 12/24 (50%) | 8/13 (62%) |
| Status epilepticus | 10/24 (42%) | 7/13 (54%) |
| Complex partial | 8/24 (33%) | 3/13 (23%) |
| Myoclonic | 5/24 (21%) | 5/13 (38%) |
Note that data from the following are not included in the table: five individuals who have only 4p deletions do not have seizures. An additional five individuals in the cohort with only 4p deletions do have seizures but did not specify the type of seizures they had, and so could not be included in this table. One individual with multiple CNVs had seizures but also did not specify kind.
Mapping a seizure susceptibility candidate region
To identify a region conferring a genetic susceptibility to seizures, we evaluated the 34 patients in our cohort with only 4p deletions. Figure 1 shows the deletions of this group aligned by size and location. All individuals in this group have deletions that encompass both critical regions WHSCR1 and WHSCR2 except for patient 33, whose deletion only overlaps WHSCR2 but not WHSCR1.
We asked whether 4p deletion size and genetic content correlate with seizure severity by first examining the records of the five individuals with the smallest terminal deletions in our cohort, patients 29–33 (figures 1 and 2). Their deletions range in size from 1.7 to 2.2 Mbp. Typically, individuals with small 4p terminal deletions less than 3.5–6 Mbp in size exhibit the mildest phenotypes, including seizure phenotypes.1 34–36 Notably, four of these five individuals (patients 29, 31, 32 and 33) reported having severe seizure phenotypes, indistinguishable in terms of seizure types, frequency or response to AEDs (see online supplementary table S1) from the rest of the cohort with larger deletion sizes. Patient 33 is noteworthy because her deletion does not remove LETM1, the purported candidate seizure gene, yet her seizures are consistent with WHS. We thus observe that in our cohort, small terminal 4p deletions including one that does not include LETM1 can result in severe seizure phenotypes.
In contrast, we identified four individuals, patients 18, 21, 24 and 34, who did not have seizures as well as one additional individual, patient 10, who is considered as not having seizures, as explained below. All of these individuals have interstitial deletions that leave, minimally, the terminal 751 kbp of chromosome 4p intact (blue bars, figure 1). Patient 10 had the largest interstitial 4p deletion, 14.6 Mb in size, who had one febrile seizure at age 1.5 years associated with a kidney infection. Having an isolated febrile seizure is an unusual presentation for WHS-associated epilepsy; in accordance with his medical records and parent answers on our survey, we scored him as not having WHS-related seizures. Taken together, these data show that deletion of the terminal 751 kbp of chromosome 4p, not monosomy of LETM1, correlates with an epileptic phenotype (p=3.59e-6) using a two-tailed Fisher's exact test (see online supplementary table S3).
We turned to the literature to determine if other rare interstitial deletions or small terminal deletions would support or refute the hypothesis that the deletion of the terminal region of 4p correlates with a seizure phenotype. Nine additional cases of non-related individuals with WHS and without seizures have been previously described in the literature.1 22 27 34 37 38 Their reported deletion sizes and locations are shown in figure 2, along with the deletions of patients from our cohort who lack seizures (blue bars). Also included in figure 2 are three small interstitial deletions described by Andersen et al,20 all of which encompass at least portions of the WHSC1 and LETM1 genes. The three individuals with these deletions show features of WHS but do not meet the minimal diagnostic criteria for the syndrome and do not have seizures.20 Strikingly, 16 out of 17 individuals without seizures have interstitial deletions, most of which result in monosomy of LETM1 while leaving the terminal 751 kbp intact. The exception to this observed correlation was an 11-year-old girl without seizures who had a ∼3.7 Mbp terminal deletion that also removes LETM1 (Van Buggenhout 2004, patient 1) (figure 2). The corresponding chromosome coordinates for all these patients are given in online supplementary table S4.
One individual described by Van Buggenhout et al22 was a clinically normal patient with a history of multiple miscarriages and no seizures. This patient was found to have a 0.3 Mbp terminal deletion (Van Buggenhout 2004 patient 6, figure 2) using a BAC array. While the lower resolution of BAC arrays must be taken into account, the deletion in this individual nevertheless suggests that a deletion encompassing approximately 0.3 Mbp of the 4p terminus does not contribute to the seizure phenotype or any other characteristic traits of WHS.
Next, we searched the literature for examples of individuals with seizures who had the smallest described terminal and interstitial deletions of chromosome 4p. The deletions of 12 such individuals, including five from our cohort, are shown (figure 2, red bars). Eight individuals in this group have terminal deletions and four have interstitial deletions, all of which affect at least the distal-most 500 kbp of chromosome 4p. Most notably, Zollino et al27 have recently described two siblings, with a paternally inherited 564 kbp terminal deletion (figure 2, Zollino 2014, patients 3 and 4). Both siblings, as well as their father, have a history of seizures.
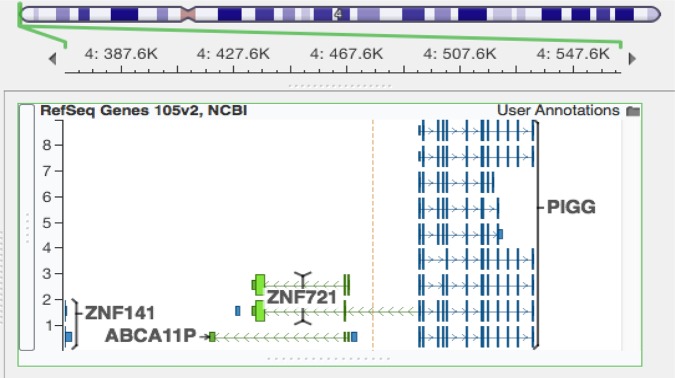
A 1.58 Mbp interstitial deletion of a 33-month-old girl overlaps with the deletions of patients 3 and 4 from Zollino et al.27 This patient, described by Izumi et al,25 presented with a typical WHS seizure phenotype. The SRO shared by the deletions of these three patients can therefore be used to define a seizure susceptibility region 197 kbp in length, starting with the distal coordinate defined by the Izumi patient and the proximal coordinate defined by the two Zollino siblings (figures 2 and 3). There are two genes and one pseudogene in this region: ZNF721, encoding a zinc-finger containing protein of unknown function, PIGG, a member of the phosphatidylinositol glycan anchor biosynthetic pathway, and ABCA11P, a pseudogene with sequence similarity to ATP-binding cassette, subfamily A genes (figure 3).
Figure 3.
Two genes and a pseudogene lie within the 197 kbp seizure candidate region, PIGG, ZNF721, and pseudogene ABCA11P. The location of this region on Chromosome 4 is shown with the green bracket. hg19/GRCh37 coordinates for this region: chr4:367691–564593. (Screenshot is from Golden Helix GenomeBrowse visualisation tool V.2.1.0 by GoldenHelix, Inc.46)
As our cohort and cases described in the literature have shown, individuals with interstitial 4p deletions that leave this candidate region intact (with the exception of patient 1 from Van Buggenhout et al22) do not have seizures. Conversely, deletion of this region gives rise to seizures. These observations suggest that deletion of this region is both necessary and sufficient for the seizure phenotype in individuals with WHS.
Treatment responses
Study participants reported 19 different AEDs, as well as the ketogenic diet and homeopathic approaches, to control seizures, with varying degrees of success (table 3, see online supplementary tables S1 and S2). The responses of the four most commonly used seizure medications in this cohort are shown in table 3, with levetiracetam and valproic acid showing the most positive responses within this group. These observations are consistent with previous studies reporting that valproic acid, used alone or in combination with ethosuximide, is the effective treatment for atypical absences common to individuals with WHS.32 39
Table 3.
Responses to the four most commonly reported seizure medications
| Phenobarbital (n=13) | Levetiracetam (n=13) | Topiramate (n=11) | Valproic acid (n=11) | |
|---|---|---|---|---|
| Negative reports | 5 | 1 | 4 | 2 |
| Positive reports | 0 | 4 | 1 | 2 |
In table 3, we summarise the reported responses. AEDs were scored as positive if the patient's parents reported without prompting that the drug gave a significant and observable increase in control over seizures. AED responses were scored as negative if the patients’ parents reported a negative reaction (allergic reaction or other) without prompting that caused them to stop using that drug, or if the drug conferred no control over seizures.
Discussion
Genotype–phenotype correlation studies of patients with WHS have met with limited success primarily because (1) the prevalence of the disorder is low and therefore assembling a study cohort large enough to achieve statistical power to find significant correlations is difficult; (2) the phenotypic presentation of WHS is highly variable and likely influenced by a number of both genetic and environmental factors and (3) accurate breakpoint mapping has only become possible within the last decade, and the majority of individuals with a diagnosis of WHS available for such studies have not had CMA as part of their diagnostic workup. In this study, we have attempted to address these challenges by (1) partnering with the world's largest support group for WHS, the 4p- Support Group, in order to assemble a relatively large cohort; (2) collecting phenotypic information from parents using a highly detailed questionnaire and (3) employing high-resolution clinical CMA to map deletion breakpoints as well as identify any additional CNVs that could contribute to phenotype.
Because seizures affect approximately 90% of all individuals with WHS and can greatly influence the quality of life for these individuals, we focused our analysis on seizures. By fine mapping the 4p deletion breakpoints of our cohort, we describe a 751 kbp terminal 4p candidate seizure region. The deletion of this region correlates strongly with the presence of seizures, and its preservation, as in cases of the interstitial WHS deletions we described, correlates with the absence of seizures. Rare interstitial and submicroscopic terminal deletions described in the literature not only support the idea that deletion of this region is necessary for seizure phenotype but also support the idea that its deletion is sufficient for predisposition to seizures. In particular, three individuals described in the literature, two of whom are siblings, allowed us to further refine the boundaries of the candidate seizure susceptibility region to a locus 197 kbp in size, starting 368 kbp from the terminal end of chromosome 4.
This 197 kbp region encompasses two genes and one pseudogene. ZNF721 encodes a zinc-finger-containing protein of unknown function, PIGG encodes a member of the phosphatidylinositol glycan anchor biosynthetic pathway and ABCA11P is a pseudogene with sequence similarity to ATP-binding cassette, subfamily A. While not much is known about the biological function of ZNF721, several intriguing lines of evidence indicate PIGG as an excellent candidate seizure susceptibility gene.
PIGG encodes one of 26 members of a biosynthetic pathway involved in assembling and attaching the phosphatidylinositol glycan (GPI) anchor to a group of over 150 proteins.40 The GPI anchor serves to attach these proteins to the outer leaflet of the plasma membrane where they carry out various signalling and extracellular functions. Deficiencies in GPI anchor synthesis have been linked to disorders of congenital glycosylation, all of which are autosomal recessive and are associated with infantile encephalopathy, ID, and/or seizures.40–42 Further work is necessary to characterise PIGG's role as a candidate seizure susceptibility gene. We note that if its deletion alone is sufficient to cause seizures, it would be the first description of haploinsufficiency for a GPI anchor biosynthetic gene. This may be consistent with the proposed importance of stoichiometry in the PIGG protein's role in the biosynthetic pathway, in which it functions as a catalytic component and competes with phosphatidylinositol glycan anchor biosynthesis protein, class O (PIGO) for binding to phosphatidylinositol glycan anchor biosynthesis protein, class F (PIGF) in order to add an ethanolamine-phosphate side chain to a mannose moiety.40 Alternatively, deletion of one copy of PIGG always occurs in the context of the deletion of other 4p terminal genes in cases of WHS; it may be that the deletion of a combination of genes in the WHS region acts synergistically to predispose individuals to seizures.
There are significant similarities shared between the two conditions, WHS and Dravet syndrome. Dravet syndrome is characterised by early-onset seizures including febrile, afebrile, generalised/unilateral clonic, myoclonic, focal, and atypical absence seizures. These seizures can be prolonged and often are intractable to pharmacotherapies, leading to cognitive, motor and behavioural impairment.43 Individuals with WHS display a distinctive electroclinical pattern resembling the severe myoclonic epilepsy of infancy or Dravet syndrome.30 In addition, some patients with a milder presentation of WHS-related dysmorphologies are sometimes first suspected of having Dravet syndrome, as attested by published studies in which SCN1A sequencing was conducted and found to be negative in at least two cases21 27 until the true cause, a deletion of the 4p terminus, was identified. Furthermore, carbamazepine and lamotrigine have been shown to exacerbate seizures in both individuals with WHS as well as individuals with Dravet syndrome.2 44
In zebrafish, there is an ortholog of SCN1A that corresponds to human SCN1B that has also been linked to Dravet syndrome, designated scn1bb. The Rohon–Beard neurons of zebrafish require functional Scn1bb protein, as well as the phosphatidylinositol biosynthetic pathway, for touch sensitivity. Nakano et al45 showed that zebrafish mutants that lack functional members of the phosphatidylinositol biosynthetic pathway, or morpholino knockdown of members of this pathway, result in the failure of the sodium channel Scn1bb to localise correctly to the plasma membrane. This observation could provide an intriguing mechanistic link between seizures in WHS and Dravet syndrome.43
Our study includes some limitations. To assess seizure phenotypes, we relied almost solely on parental answers to our questionnaire, with limited contribution from medical records. A follow-up study in which EEG recordings are analysed and correlated with genetic findings would be a valuable extension of the observations we present here. Our data on AED responses suggest that certain AEDs may be more effective than others at achieving early seizure control and warrants further study. Our interpretation of the seizure susceptibility region is based on five individuals whose lack of a seizure phenotype can change with time. Despite this fact, now that the average age of these five individuals (6.8 years) is well beyond the typical average age of seizure onset, we remain optimistic that seizure presentation in this group has already distinguished itself from typical WHS seizure presentation. Patient 1 described in Van Buggenhout et al22 who lacks both the candidate seizure susceptibility terminal region described here as well as LETM1 and yet who does not have seizures, highlights the complexity of this region and emphasises that the knowledge of genetic contributions to seizures is incomplete. It is highly likely that there are multiple seizure susceptibility genes in the 4p region, and that final seizure presentation is a result of the lack or presence of the unique genetic and environmental combinations that can result.
We find that the use of whole genome CMA for the genetic characterisation of individuals with WHS is valuable, since it provides a significantly higher resolution of breakpoint coordinates than does karyotyping. Additional CNVs frequently occur in this population,4 yet on average are smaller than would be detectable even by high-resolution karyotyping (see online supplementary table S2), and can therefore be easily missed. In addition, the presence or absence of a terminal 197 kbp deletion is most effectively detected using CMA. Further investigation of the relationship of genetics to the clinical manifestations of WHS using high-resolution mapping techniques as well as whole-genome sequencing will lead to a deeper understanding of the molecular underpinnings of this complex disorder as well as an improvement of medical treatments for these individuals.
On a final note, the identification of a relatively small candidate seizure region now affords the opportunity to create loss-of-function knockouts of candidate genes in model organisms to confirm that haploinsufficiency of such genes is sufficient to increase seizure susceptibility and also to perform functional studies that will further elucidate the mechanism of these genes’ functions in health and disease. Using such an approach, precision medicine for complex genetic disorders such as contiguous gene disorders becomes possible.
Supplementary Material
Acknowledgments
This work is dedicated to the bright memories of Trisha York and Isabella Kirch, as well as all the members of the 4p- Support Group, who generously participated in this research. We thank Dr E Robert Wassman for critical reading of the manuscript.
Footnotes
Twitter: Follow Karen Ho at @picturewing
Contributors: All authors on this paper meet the four criteria for authorship as set forth by The International Committee of Medical Journal Editors Recommendations for the Conduct, Reporting, Editing and Publication of Scholarly Work in Medical Journals. In specific, JCC and RJV conceived the idea for a phenotype–genotype study of WHS individuals; KSH obtained IRB approval for the study, led the research effort, performed the data interpretation, drafted the manuscript and serves as the guarantor of the paper; STS interpreted microarray data; AL, MRS, RJV, MMM, AC, AP and JCC assisted with data collection and medical records ascertainment; CHH, JCC and AB interpreted the data, drawing upon their respective and extensive expertise in human genetics, medical genetics and epilepsy; CGL conducted the statistical analysis and AB also developed the survey instrument. All authors participated in all aspects of manuscript preparation.
Funding: Lineagen, Inc.
Competing interests: KSH, CHH, MRS, RJV and MMM are employees of Lineagen, Inc. and have received stock options. STS was an employee of ARUP Laboratories during the course of this work; she is now an employee of 23andMe. STS, CGL and AP received consultant fees from Lineagen for clinical work. STS received consultant fees from Affymetrix for work unrelated to this work. All other authors have nothing to declare.
Ethics approval: University of Utah IRB.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Maas NMC, Van Buggenhout G, Hannes F, Thienpont B, Sanlaville D, Kok K, Midro A, Andrieux J, Anderlid B-M, Schoumans J, Hordijk R, Devriendt K, Fryns J-P, Vermeesch JR. Genotype-phenotype correlation in 21 patients with Wolf-Hirschhorn syndrome using high resolution array comparative genome hybridisation (CGH). J Med Genet 2008;45:71–80. 10.1136/jmg.2007.052910 [DOI] [PubMed] [Google Scholar]
- 2.Battaglia A, Carey JC, South ST, et al. Wolf-Hirschhorn syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al., eds. GeneReviews. Seattle, WA: University of Washington, Seattle, 1993. –2015:1–18. http://www.ncbi.nlm.nih.gov/books/NBK1183/ (accessed 29 May 2015). [Google Scholar]
- 3.Hirschhorn K. A short history of the initial discovery of the Wolf-Hirschhorn syndrome. Am J Med Genet C Semin Med Genet 2008;148C:244–5. 10.1002/ajmg.c.30186 [DOI] [PubMed] [Google Scholar]
- 4.South ST, Whitby H, Battaglia A, Carey JC, Brothman AR. Comprehensive analysis of Wolf-Hirschhorn syndrome using array CGH indicates a high prevalence of translocations. Eur J Hum Genet 2008;16:45–52. 10.1038/sj.ejhg.5201915 [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, Hermetz KE, Jackson JM, Mulle JG, Dodd A, Tsuchiya KD, Ballif BC, Shaffer LG, Cody JD, Ledbetter DH, Martin CL, Rudd MK. Diverse mutational mechanisms cause pathogenic subtelomeric rearrangements. Hum Mol Genet 2011;20:3769–78. 10.1093/hmg/ddr293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright TJ, Ricke DO, Denison K, Abmayr S, Cotter PD, Hirschhorn K, Keinanen M, Mcdonald-Mcginn D, Somer M, Spinner N, Yang-Feng T, Zackai E, Altherr MR. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet 1997;6:317–24. 10.1093/hmg/6.2.317 [DOI] [PubMed] [Google Scholar]
- 7.Lee YF, Nimura K, Lo WN, Saga K, Kaneda Y. Histone H3 lysine 36 methyltransferase Whsc1 promotes the association of Runx2 and p300 in the activation of bone-related genes. PLoS One 2014;9:e106661 10.1371/journal.pone.0106661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerzendorfer C, Hannes F, Colnaghi R, Abramowicz I, Carpenter G, Vermeesch JR, O'driscoll M. Characterizing the functional consequences of haploinsufficiency of NELF-A (WHSC2) and SLBP identifies novel cellular phenotypes in Wolf-Hirschhorn syndrome. Hum Mol Genet 2012;21:2181–93. 10.1093/hmg/dds033 [DOI] [PubMed] [Google Scholar]
- 9.Endele S, Fuhry M, Pak SJ, Zabel BU, Winterpacht A. LETM1, a novel gene encoding a putative EF-hand Ca(2+)-binding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics 1999;60:218–25. 10.1006/geno.1999.5881 [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez L, Zollino M, Climent S, Mansilla E, López-Grondona F, Martínez-Fernández ML, Murdolo M, Martínez-Frías ML. The new Wolf-Hirschhorn syndrome critical region (WHSCR-2): a description of a second case. Am J Med Genet A 2005;136:175–8. 10.1002/ajmg.a.30775 [DOI] [PubMed] [Google Scholar]
- 11.Zollino M, Lecce R, Fischetto R, Murdolo M, Faravelli F, Selicorni A, Buttè C, Memo L, Capovilla G, Neri G. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet 2003;72:590–7. 10.1086/367925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battaglia A, Carey JC, South ST. Wolf-Hirschhorn syndrome: a review and update. Am J Med Genet C Semin Med Genet 2015;169:216–23. 10.1002/ajmg.c.31449 [DOI] [PubMed] [Google Scholar]
- 13.Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science 2009;326:144–7. 10.1126/science.1175145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlickum S, Moghekar A, Simpson JC, Steglich C, O'brien RJ, Winterpacht A, Endele SU. LETM1, a gene deleted in Wolf-Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics 2004;83:254–61. 10.1016/j.ygeno.2003.08.013 [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Chen G, Lu Y, Liu J, Fang M, Luo J, Cao Q, Wang X. Association of mitochondrial letm1 with epileptic seizures. Cereb Cortex 2014;24:2533–40. 10.1093/cercor/bht118 [DOI] [PubMed] [Google Scholar]
- 16.Zsurka G, Kunz WS. Mitochondrial dysfunction and seizures: the neuronal energy crisis. Lancet Neurol 2015;14:956–66. 10.1016/S1474-4422(15)00148-9 [DOI] [PubMed] [Google Scholar]
- 17.Doonan PJ, Chandramoorthy HC, Hoffman NE, Zhang X, Cárdenas C, Shanmughapriya S, Rajan S, Vallem S, Chen X, Foskett JK, Cheung JY, Houser SR, Madesh M. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. FASEB J 2014;28:4936–49. 10.1096/fj.14-256453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart L, Rauch A, Carr AM, Vermeesch JR, O'driscoll M. LETM1 haploinsufficiency causes mitochondrial defects in cells from humans with Wolf-Hirschhorn syndrome: implications for dissecting the underlying pathomechanisms in this condition. Dis Model Mech 2014;7:535–45. 10.1242/dmm.014464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D, Zhao L, Clish CB, Clapham DE. Letm1, the mitochondrial Ca2+/H+ antiporter, is essential for normal glucose metabolism and alters brain function in Wolf-Hirschhorn syndrome. Proc Natl Acad Sci U S A 2013;110:E2249–54. 10.1073/pnas.1308558110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen EF, Carey JC, Earl DL, Corzo D, Suttie M, Hammond P, South ST. Deletions involving genes WHSC1 and LETM1 may be necessary, but are not sufficient to cause Wolf-Hirschhorn Syndrome. Eur J Hum Genet 2014;22:464–70. 10.1038/ejhg.2013.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayindir B, Piazza E, Della Mina E, Limongelli I, Brustia F, Ciccone R, Veggiotti P, Zuffardi O, Dehghani MR. Dravet phenotype in a subject with a der(4)t(4;8)(p16.3;p23.3) without the involvement of the LETM1 gene. Eur J Med Genet 2013;56:551–5. 10.1016/j.ejmg.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 22.Van Buggenhout G, Melotte C, Dutta B. Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet 2004;41:691–8. 10.1136/jmg.2003.016865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engbers H, Van Der Smagt JJ, Van ‘T Slot R, Vermeesch JR, Hochstenbach R, Poot M. Wolf-Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur J Hum Genet 2009;17:129–32. 10.1038/ejhg.2008.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faravelli F, Murdolo M, Marangi G, Bricarelli FD, Di Rocco M, Zollino M. Mother to son amplification of a small subtelomeric deletion: a new mechanism of familial recurrence in microdeletion syndromes. Am J Med Genet A 2007;143A:1169–73. 10.1002/ajmg.a.31723 [DOI] [PubMed] [Google Scholar]
- 25.Izumi K, Okuno H, Maeyama K, Sato S, Yamamoto T, Torii C, Kosaki R, Takahashi T, Kosaki K. Interstitial microdeletion of 4p16.3: contribution of WHSC1 haploinsufficiency to the pathogenesis of developmental delay in Wolf-Hirschhorn syndrome. Am J Med Genet A 2010;152A:1028–32. 10.1002/ajmg.a.33121 [DOI] [PubMed] [Google Scholar]
- 26.Misceo D, Barøy T, Helle JR, Braaten O, Fannemel M, Frengen E. 1.5Mb deletion of chromosome 4p16.3 associated with postnatal growth delay, psychomotor impairment, epilepsy, impulsive behavior and asynchronous skeletal development. Gene 2012;507:85–91. 10.1016/j.gene.2012.07.021 [DOI] [PubMed] [Google Scholar]
- 27.Zollino M, Orteschi D, Ruiter M, Pfundt R, Steindl K, Cafiero C, Ricciardi S, Contaldo I, Chieffo D, Ranalli D, Acquafondata C, Murdolo M, Marangi G, Asaro A, Battaglia D. Unusual 4p16.3 deletions suggest an additional chromosome region for the Wolf-Hirschhorn syndrome-associated seizures disorder. Epilepsia 2014;55:849–57. 10.1111/epi.12617 [DOI] [PubMed] [Google Scholar]
- 28.Vanzo RJ, Lortz A, Calhoun ARUL, Carey JC. Academia, advocacy, and industry: a collaborative method for clinical research advancement. Am J Med Genet A 2014;164A:1619–21. 10.1002/ajmg.a.36509 [DOI] [PubMed] [Google Scholar]
- 29.Battaglia A, Filippi T, Carey JC. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: experience with 87 patients and recommendations for routine health supervision. Am J Med Genet C Semin Med Genet 2008;148C:246–51. 10.1002/ajmg.c.30187 [DOI] [PubMed] [Google Scholar]
- 30.Battaglia A, Carey JC. Seizure and EEG patterns in Wolf-Hirschhorn (4p-) syndrome. Brain Dev 2005;27:362–4. 10.1016/j.braindev.2004.02.017 [DOI] [PubMed] [Google Scholar]
- 31.Battaglia A, Carey JC, Cederholm P, Viskochil DH, Brothman AR, Galasso C. Natural history of Wolf-Hirschhorn syndrome: experience with 15 cases. Pediatrics 1999;103:830–6. 10.1542/peds.103.4.830 [DOI] [PubMed] [Google Scholar]
- 32.Battaglia A, Filippi T, South ST, Carey JC. Spectrum of epilepsy and electroencephalogram patterns in Wolf-Hirschhorn syndrome: experience with 87 patients. Dev Med Child Neurol 2009;51:373–80. 10.1111/j.1469-8749.2008.03233.x [DOI] [PubMed] [Google Scholar]
- 33.Battaglia D, Zampino G, Zollino M, Mariotti P, Acquafondata C, Lettori D, Pane M, Vasta I, Neri G, Dravet C, Guzzetta F. Electroclinical patterns and evolution of epilepsy in the 4p- syndrome. Epilepsia 2003;44:1183–90. 10.1046/j.1528-1157.2003.63502.x [DOI] [PubMed] [Google Scholar]
- 34.Shimizu K, Wakui K, Kosho T, Okamoto N, Mizuno S, Itomi K, Hattori S, Nishio K, Samura O, Kobayashi Y, Kako Y, Arai T, Oh-Ishi T, Kawame H, Narumi Y, Ohashi H, Fukushima Y. Microarray and FISH-based genotype-phenotype analysis of 22 Japanese patients with Wolf-Hirschhorn syndrome. Am J Med Genet A 2014;164A:597–609. 10.1002/ajmg.a.36308 [DOI] [PubMed] [Google Scholar]
- 35.Zollino M, Di Stefano C, Zampino G, Mastroiacovo P, Wright TJ, Sorge G, Selicorni A, Tenconi R, Zappalà A, Battaglia A, Di Rocco M, Palka G, Pallotta R, Altherr MR, Neri G. Genotype-phenotype correlations and clinical diagnostic criteria in Wolf-Hirschhorn syndrome. Am J Med Genet 2000;94:254–61. [DOI] [PubMed] [Google Scholar]
- 36.Zollino M, Murdolo M, Marangi G. On the nosology and pathogenesis of Wolf-Hirschhorn syndrome: genotype-phenotype correlation analysis of 80 patients and literature review. Am J Med Genet C Semin Med Genet 2008;148C:257–69. 10.1002/ajmg.c.30190 [DOI] [PubMed] [Google Scholar]
- 37.Okamoto N, Ohmachi K, Shimada S, Shimojima K, Yamamoto T. 109kb deletion of chromosome 4p16.3 in a patient with mild phenotype of Wolf-Hirschhorn syndrome. Am J Med Genet A 2013;161A:1465–9. 10.1002/ajmg.a.35910 [DOI] [PubMed] [Google Scholar]
- 38.Rauch A, Schellmoser S, Kraus C, Dörr HG, Trautmann U, Altherr MR, Pfeiffer RA, Reis A. First known microdeletion within the Wolf-Hirschhorn syndrome critical region refines genotype-phenotype correlation. Am J Med Genet 2001;99:338–42. 10.1002/ajmg.1203 [DOI] [PubMed] [Google Scholar]
- 39.Battaglia A, Carey JC. Wolf-Hirschhorn syndrome and the 4p-related syndromes. Am J Med Genet C Semin Med Genet 2008;148C:241–3. 10.1002/ajmg.c.30189 [DOI] [PubMed] [Google Scholar]
- 40.Kinoshita T. Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc Jpn Acad Ser B Phys Biol Sci 2014;90:130–43. 10.2183/pjab.90.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiyonobu T, Inoue N, Morimoto M, Kinoshita T, Murakami Y. Glycosylphosphatidylinositol (GPI) anchor deficiency caused by mutations in PIGW is associated with West syndrome and hyperphosphatasia with mental retardation syndrome. J Med Genet 2014;51:203–7. 10.1136/jmedgenet-2013-102156 [DOI] [PubMed] [Google Scholar]
- 42.Ilkovski B, Pagnamenta AT, O'grady GL, Kinoshita T, Howard MF, Lek M, Thomas B, Turner A, Christodoulou J, Sillence D, Knight SJ, Popitsch N, Keays DA, Anzilotti C, Goriely A, Waddell LB, Brilot F, North KN, Kanzawa N, Macarthur DG, Taylor JC, Kini U, Murakami Y, Clarke NF. Mutations in PIGY: expanding the phenotype of inherited glycosylphosphatidylinositol deficiencies. Hum Mol Genet 2015;24:6146–59. 10.1093/hmg/ddv331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chopra R, Isom LL. Untangling the dravet syndrome seizure network: the changing face of a rare genetic epilepsy. Epilepsy Curr Am Epilepsy Soc 2014;14:86–9. 10.5698/1535-7597-14.2.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunklaus A, Ellis R, Reavey E, Forbes GH, Zuberi SM. Prognostic, clinical and demographic features in SCN1A mutation-positive Dravet syndrome. Brain J Neurol 2012;135:2329–36. 10.1093/brain/aws151 [DOI] [PubMed] [Google Scholar]
- 45.Nakano Y, Fujita M, Ogino K, Saint-Amant L, Kinoshita T, Oda Y, Hirata H. Biogenesis of GPI-anchored proteins is essential for surface expression of sodium channels in zebrafish Rohon-Beard neurons to respond to mechanosensory stimulation. Development 2010;137:1689–98. 10.1242/dev.047464 [DOI] [PubMed] [Google Scholar]
- 46.Golden Helix GenomeBrowse® visualization tool (Version 2.0)[Software] Bozeman, MT: Golden Helix, Inc. http://www.goldenhelix.com [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.