Abstract
Syntenin has crucial roles in cell adhesion, cell migration and synaptic transmission. Its closely linked postsynaptic density‐95, discs large 1, zonula occludens‐1 (PDZ) domains typically interact with C‐terminal ligands. We profile syntenin PDZ1‐2 through proteomic peptide phage display (ProP‐PD) using a library that displays C‐terminal regions of the human proteome. The protein recognizes a broad range of peptides, with a preference for hydrophobic motifs and has a tendency to recognize cryptic internal ligands. We validate the interaction with nectin‐1 through orthogonal assays. The study demonstrates the power of ProP‐PD as a complementary approach to uncover interactions of potential biological relevance.
Keywords: PDZ domain, peptide interaction, phage display, protein–protein interaction, short linear motif
Abbreviations
- AP‐MS
affinity purification coupled to mass spectrometry
- ELISA
enzyme‐linked immunosorbent assay
- PDZ
postsynaptic density‐95, discs large 1, zonula occludens‐1
- ProP‐PD
proteomic peptide phage display
- Y2H
yeast‐two‐hybrid
Syntenin is a small adaptor protein involved in a range of processes regulating cell–cell adhesion, transmembrane receptor trafficking and clustering, tumor cell metastasis and neuronal synaptic function 1. Among others, it plays important roles in the Arf6 dependent recycling of target proteins 2 and in loading of exosomes with cargo 3, 4. Syntenin is upregulated in different cancers and functions as a positive regulator of melanoma progression and metastasis 5, 6, 7. Its closely connected PDZ domains mediate protein–protein interactions and are flanked by intrinsically disordered N‐ and C‐terminal regions. The syntenin PDZ1‐2 tandem has a limited interdomain flexibility 8, 9 and has been suggested to function as a supramodule 10, 11 as the unfolding of the PDZ1‐2 tandem is cooperative 9 and both domains are commonly required for the interactions with target proteins. Syntenin's PDZ1 and PDZ2 domains are known to interact with C‐terminal peptides containing distinct sequence motifs, namely class 1 (x‐S/T‐x‐Φ‐COOH), class 2 (x‐Φ‐x‐Φ‐COOH) or the less common class 3 (x‐D/E‐x‐Φ‐COOH) 1, 12, with class 2 ligands typically being bound by PDZ2 and class 3 ligands by PDZ1. Syntenin PDZ1‐2 also interacts with internal binding motifs found in disordered regions of target proteins 13, 14 and with membrane phospholipids such as phosphatidylinositol‐4,5‐bisphosphate (PIP2) 2, 13, 15.
Most known syntenin ligands have been identified through yeast‐two‐hybrid (Y2H) experiments 1, 16, a method with a bias toward soluble proteins that can be translocated to the nucleus. More recently, high‐throughput affinity purification coupled to mass spectrometry (AP‐MS) has provided novel insights into syntenin's interactome 17. However, AP‐MS are ideal for the identification of strong protein–protein interactions but less suited for the identification of PDZ‐peptide interactions due to the moderate affinities of these interactions 18. We recently developed a dedicated approach for the identification of domain–motif interactions proteins on a proteome‐wide scale termed proteomic peptide phage display (ProP‐PD) 19, 20, 21. Essentially, we created a phage library that displays peptides that represent the C‐terminal regions of the human proteome. ProP‐PD can be used for the direct identification of ligands of potential biological relevance 20. This contrast with the commonly used combinatorial phage display, where highly diverse libraries are used to establish preferred binding motifs, which can then be used for genome‐wide scanning of potential ligands in the proteome 22. Such predictions often lead to tedious experimental validations due to the short length of consensus binding motifs and their subsequent over‐prediction in proteomic sequence data.
Here, we elucidate the binding preferences of syntenin through ProP‐PD and through combinatorial peptide phage display. We provide insights into its binding specificity and a list of targets of potential biological relevance. We confirm that peptides with class 2 motifs are preferred targets, but also find that syntenin appears to frequently recognize internal binding motifs. We validate an interaction between syntenin and the cell adhesion protein nectin‐1 through orthogonal methods including affinity measurement, Y2H and cell‐based assays.
Experimental procedures
Protein expression and purification
Syntenin PDZ1‐2 (amino acids 107–275 in the pETM‐11 expression vector (EMBL)), PDZ1 (107–199) and PDZ2 (198–275) was expressed in Escherichia coli (E. coli) ER2566 cells and purified through nickel affinity chromatography as described elsewhere 23. The purity of the protein was confirmed by SDS/PAGE.
ProP‐PD display selections
Phage display selections were carried out as described 19. The selections were successful as judged by pooled phage enzyme‐linked immunosorbent assays (ELISA) and resultant phage pools from rounds 4 and 5 were analyzed by next‐generation sequencing (NGS) on the Illumina platform. The data were processed as described elsewhere 19, 24.
Combinatorial phage display and clonal analysis
To identify carboxyl‐terminal and internal peptide ligand specificities of the syntenin PDZ1‐2 supramodule, we used highly diverse combinatorial libraries consisting of either random internal dodecamer peptides or random carboxyl terminus heptamer peptides fused to the M13 gene‐8 major coat protein 25. The heptamer and dodecamer phage libraries were incubated with syntenin PDZ1‐2 tandem domain immobilized on a 96‐well Maxisorp plate (Thermo Scientific, Waltham, MA, USA), and the unbound phages were washed off with PT buffer (phosphate buffered saline, 0.05% Tween 20). Bound phage were eluted and amplified by infection of E. coli XL1‐Blue cells in medium supplemented with M13 KO7 helper phage to facilitate phage production. The amplified pools were cycled through five rounds of selection. After five rounds of selection, 48 individual clones from each binding selection were subjected to ELISAs with immobilized syntenin PDZ1‐2 domain and control GST to detect phage‐displayed peptides that exhibited positive binding signals. Phage that bound to the syntenin PDZ domain, but not to a control GST were subjected to DNA sequence analysis. All PDZ1‐2 binding clones were individually tested by phage ELISAs for interactions with immobilized PDZ1‐2 tandem, PDZ1 and PDZ2.
Bioinformatics
Published syntenin interactions were retrieved from Biogrid 26. The Database for Annotation, Visualization and Integrated Discovery (DAVID) 27 was used to identify enrichments of GO terms among the host proteins of the identified ligands using the default human proteome as a background and the GO FAT annotations with medium stringency setting. The significances of the enrichments were evaluated by the Benjamini‐Hochberg false discovery rate corrected P‐values.
Surface plasmon resonance
Freshly prepared proteins were dialyzed into the running buffer (10 mm HEPES, 150 mm NaCl, 0.005% Tween20, pH 7.4) and perfused over the surface at a flow of 30 μL·min−1 in a range of concentrations. The surface was regenerated by 30 s pulses of 1 m NaCl, 50 mm NaOH. N‐terminal bitoinylated nectin‐1 C‐terminal peptide (AENMVSQNDGSFISKKEWYV‐COOH) (GeneCust Europe, Luxembourg) was immobilized to 50–60 RUs on a SA chip (GE Healthcare, Uppsala, Sweden). The reference channel was immobilized with a nonbinding peptide, the biotinylated phosphorylated cytoplasmic domain of syndecan2 (Biot‐RMRKKDEGSYDLGERKPSSAAYQKAPTKEFpYA‐COOH). Data were double reference subtracted and apparent K d values were estimated by plotting the equilibrium response against the protein concentration and fitting the data to a 1 : 1 Langmuir binding isotherm using graphpad prism (San Diego, CA, USA).
Y2H
Competent MaV203 yeast cells were successively transformed with pDBa‐PDZ‐nectin‐1 and 100 μg of human fetal brain cDNA library in pEXP‐AD502 vector, and ~ 5 × 105 transformants were plated onto minimal selective medium lacking tryptophane, leucine and histidine, were supplemented with 20 mm 3‐amino‐1,2,4‐triazole, and were incubated at 30 °C for 5 days. Positive clones were then tested for two other two‐hybrid reporters, uracil and β‐galactosidase, as described 28. Positive colonies were PCR‐amplified, and phenotypes were retested by reintroducing PCR products and pEXP‐AD502 vector linearized into fresh yeast cells containing the bait. Colonies giving rise to more than one PCR product were eliminated. pEXP‐AD502 inserts of positive clones after the second round of two‐hybrid screening were sequenced and identified. The two clones with ORF sequences in‐frame with the GAL4AD coding sequence were considered for further investigation.
Co‐immunoprecipitation
Cos7 cells were co‐transfected with C‐terminally Myc‐tagged full‐lenght nectin‐1, ΔPDZ‐nectin‐1, full‐length syntenin or truncated variant of syntenin. Transfected cells were lysed in MES buffer (50 mm 2‐N‐morpholino ethanesulfonic acid pH 6.8, 70 mm NaCl, 1 mm EGTA, 1 mm MgCl2, 1% NP‐40) complemented with protease inhibitor cocktail (Roche, Mannheim, Germany). 500 μg of lysate were immunoprecipitated with anti‐nectin‐1 antibody or anti‐syntenin antibody 29. After gel migration and transfer, membranes were blocked in PBS containing 0.5% BSA and hybridized with anti‐myc, anti‐syntenin or anti‐nectin‐1 antibodies as indicated in Figs 2 and 3. For co‐immunoprecipitation experiments with endogenous proteins, MCF‐7 cells were seeded in heparin‐coated plate for 2 days and then lysed in MES buffer (50 mm 2‐N‐morpholino ethanesulfonic acid pH 6.8, 70 mm NaCl, 1 mm EGTA, 1 mm MgCl2, 1% NP‐40) complemented with protease inhibitor cocktail (Roche). 500 μg of lysate were immunoprecipitated with anti‐nectin‐1 antibodies (R1.302) 29. After gel migration and membranes transfer, membranes were blocked in PBS containing 0.5% BSA and hybridized with anti‐syntenin antibody (SySy) and anti‐nectin‐1 (R154) (generous gift of Eisenberg RJ) 30. Blots were revealed with peroxidase coupled secondary reagent (Jackson Immunoresearch, West Grove, PA, USA) followed by ECL.
Figure 2.
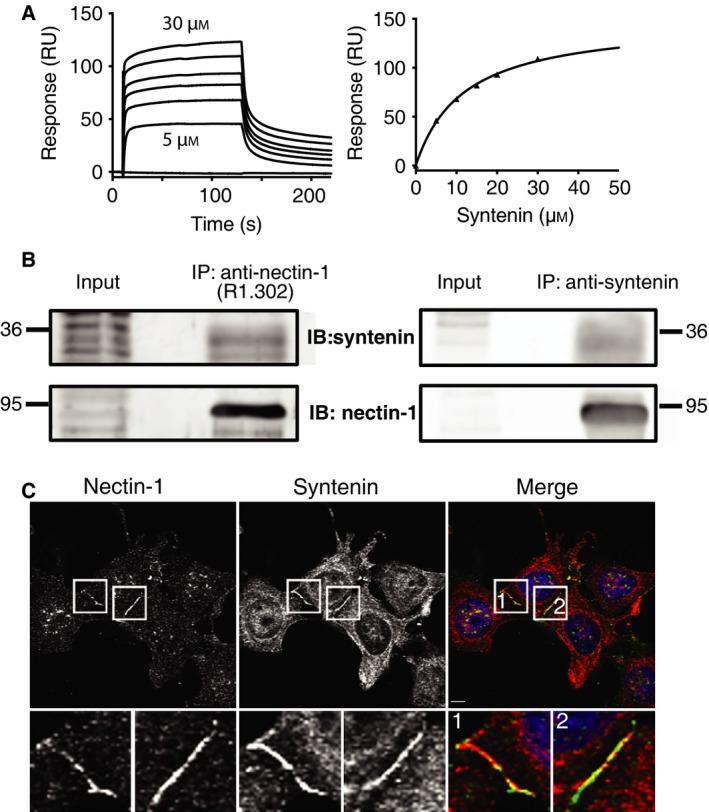
Analysis of the nectin‐1‐syntenin interaction through SPR, Y2H and colocalization experiments. (A) Double reference subtracted sensograms of full‐length syntenin injected over the biotinylated C‐terminus of nectin‐1 (left, representative example). The apparent K d value of the interaction was determined to be 14 ± 3 μm (n = 3) by fitting a 1 : 1 binding isotherm to the plot of the RUmax against protein concentration (right). (B) Co‐immunoprecipitation of syntenin and nectin‐1 in MCF‐7 cells. Nectin‐1 was immunoprecipitated using a specific antibody (R1.302) and detected using another antibody (R1.54) (left). Coprecipitated syntenin was revealed using a rabbit anti‐syntenin antibody. Syntenin was immunoprecipitated using an anti‐syntenin antibody and detected using the same antibody (right). Coprecipitated Nectin‐1 was revealed using a rabbit anti‐nectin‐1 antibody (R1.54). (C) Colocalization of endogenous nectin‐1 and syntenin. MCF‐7 cells were grown for 5 h on coverslips precoated with fibronectin and processed for immunofluorescence using anti‐nectin‐1 antibody (green in merge) and anti‐syntenin antibody (red in merge). Inserts are enlarged at the bottom. Scale bar: 4 μm.
Figure 3.
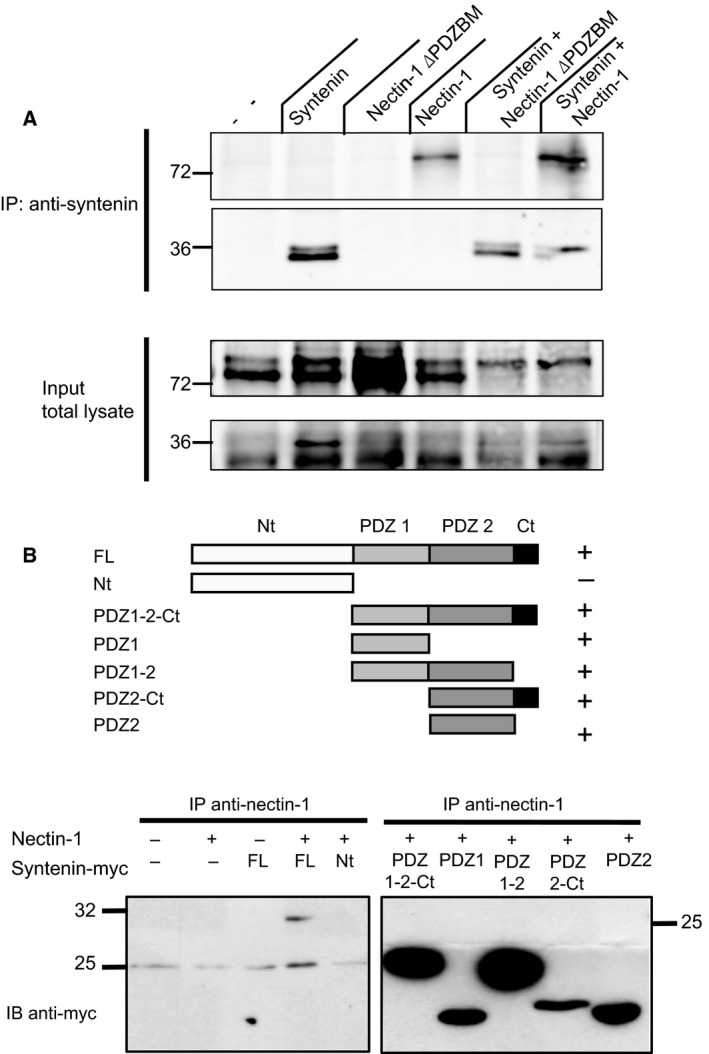
Characterization of the interaction between syntenin and nectin‐1. (A) Co‐IPs of overexpressed wild‐type and truncated (deleted PDZ binding motif, ΔPDZBM) nectin‐1 and myc‐tagged syntenin in Cos cells. Syntenin was immunoprecipitated using anti‐syntenin1 antibody, and detected with the same antibody, and bound nectin was detected with an anti‐nectin antibody. Truncation of the PDZBM of nectin‐1 confers a loss of interaction. (B) Top: Schematic representation of C‐terminally Myc‐tagged full‐length syntenin and truncated constructs thereof that were used for co‐IPs of overexpressed nectin‐1. ‘Nt’ and ‘Ct’ indicate ‘N‐terminal region and C‐terminal region’. ‘+’ indicates that the co‐IPs of nectin‐1 and syntenin was positive with the indicated construct. Bottom: Co‐IP of nectin‐1 in and truncated C‐terminally Myc‐tagged syntenin constructs.
Immunofluorescence staining and confocal microscopy
Cultured MCF‐7 cells on fibronectin‐coated coverslips were fixed with MetOH for 3 min, washed in PBS and then incubated with the indicated in house made antibodies 31, 32 in PBS containing 0.3% BSA and 0.05% saponin. Coverslips were mounted on DABCO/Mowiol and observed with a Zeiss Meta confocal microscope (LSM 510 META; Zeiss, Marly‐le‐Roi, France) with a 63× objectives.
Results
Syntenin PDZ1‐2 was used as a bait in selections against a ProP‐PD library that displays C‐terminal peptides of the human proteome 19. The retained phage pools were analyzed by NGS and the retrieved data was filtered for peptides that corresponded to C‐terminal peptides of proteins with reviewed Uniprot entries. A cut‐off value of at four sequencing counts was assigned (Fig 1A. Table S1) leaving a set of 302 peptides, of which the top 5% are listed in Table 1. The selection was dominated by two class 2 peptides: the GGDDFWF peptide of the potassium channel KCNV1 and the SKKEWYV peptide of the cell adhesion protein PVRL1, also called nectin‐1 (Table 1). These two peptides together constituted 84% of the sequencing counts listed in Table S1.
Figure 1.
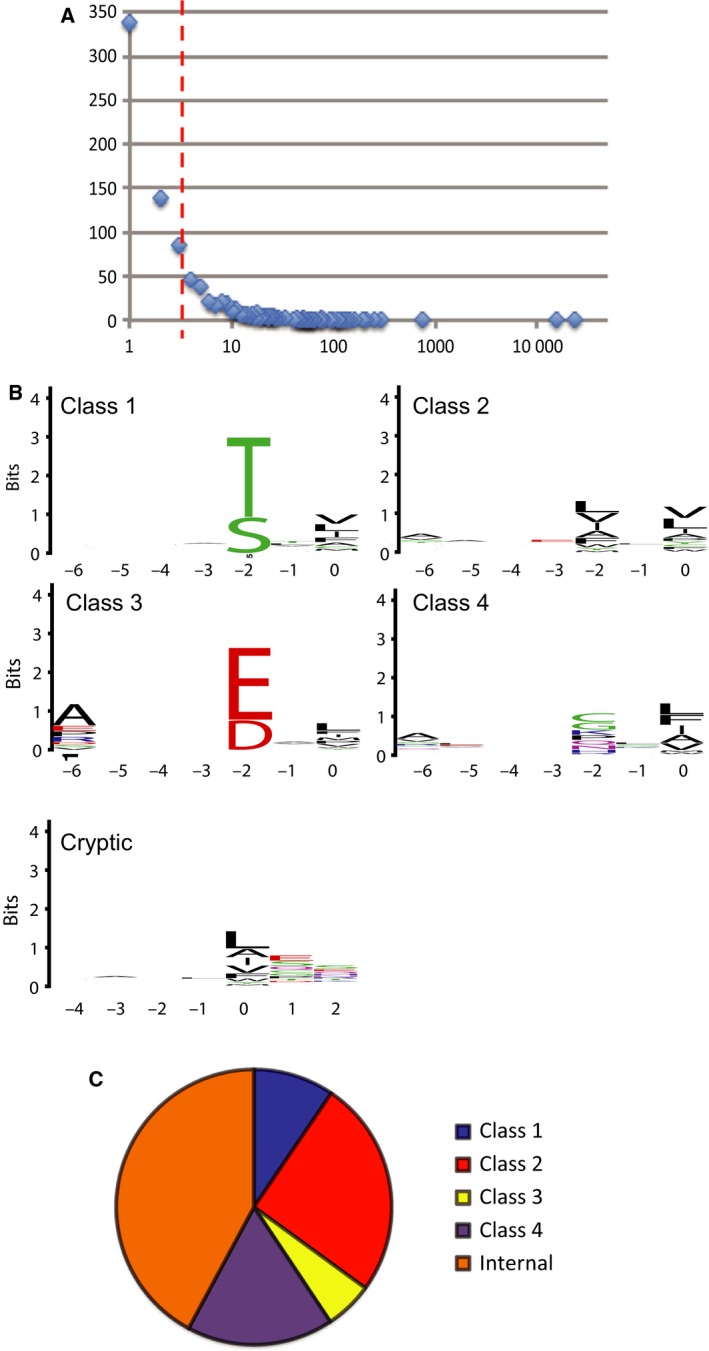
Analysis of the ProP‐PD selection data. (A) Histogram of the next‐generation sequencing data of the phage pools after the fourth and fifth round of selections, with the number of sequencing counts on the x‐axis and the number of peptides receiving a certain number of counts on the y‐axis. (B) PWMs representing the peptide binding specificity of syntenin PDZ1‐2. The numbers on the x‐axis indicate the position in the peptide following the typical PDZ ligand nomenclature, with p0 typically being the C‐terminal residue. The relative sized of the letters indicate their occurrences among the aligned sequences. The high of the stacked letters indicate the information content at the position given in bits. (C) Pie chart of the binding motifs assigned among syntenin ligands identified through ProP‐PD.
Table 1.
The top 5% of peptides identified as syntenin ligands through ProP‐PD. Motifs were assigned based on the canonical classes 1, 2 and 3, and the general class 4. Peptides assigned to have cryptic internal binding motifs are indicated by ‘cryptic’. Putative cyptic motifs are underlined when applicable
| Peptide | Motif | Total count | Gene | Uniprot | Comment/Reference |
|---|---|---|---|---|---|
| GGDDFWF | 2 | 30 205 | KCNV1 | Q6PIU1 | |
| SKKEWYV | 2 | 15 632 | PVRL1 | Q15223 | |
| AALNGPV | 4 | 765 | PPP6R2 | O75170 | 33 |
| SRREWYV | 2 | 300 | PVRL3 | Q9NQS3 | |
| IRDFLRW | 2 | 252 | GAL3ST1 | Q99999 | |
| LYAADKH | cryptic | 213 | OTUB2 | Q96DC9 | |
| EEEPMNL | 2 | 165 | HDAC7 | Q8WUI4 | |
| QDAKLIS | cryptic | 146 | AFF4 | Q9UHB7 | |
| KPQTMER | cryptic | 132 | ABCB6 | Q9NP58 | |
| SQFPPGF | 4 | 125 | TBC1D3H | P0C7X1 | |
| ASGDLCV | 2 | 114 | IFNL3 | Q8IZI9 | Secreted |
| GSGTDDE | cryptic | 113 | SMARCA2 | P51531 | 16 |
| GHRNGLE | cryptic | 110 | SP8 | Q8IXZ3 | |
| SVGKDVF | 3 | 101 | EGLN1 | Q9GZT9 | |
| ETGKLCS | cryptic | 92 | BRINP2 | Q9C0B6 | Secreted |
We investigated the presence of known binders by comparing the data set with the ligands reported in the BioGrid database, which identified three known interactions among the ligands above the cut‐off value, (Table S1). Two of these are among the top 5% of ligands, namely the transcriptional coactivator SMARCA2, which has recently been identified as a syntenin ligand through high‐throughput Y2H 16 and a regulatory subunit of protein phosphatase 6, PPP6R2, which has been identified trough high‐throughput AP‐MS 33. In the large set of ligands below the cut‐off, we identified an additional set of 6 established syntenin ligands (IL5RA 34; GRIA4A 35; SULTB1, KRTAP5‐9, PNMA2 and INO80E 16.
We analyzed the distribution of PDZ binding motifs among the peptides. As illustrated by the PWMs (Fig 1B,C), the motifs are degenerate, which is in line with previous studies 36. A total of 10% of the ligands have a class 1 motif, 24% have a class 2 motif and 6% have a C‐terminal class 3 motif. There appears to be a slight preference for class 3 ligands with alanine in p‐6. In addition, 17% of the peptides have degenerate PDZ binding motifs, with hydrophobic residues at the ultimate p0 and variable resides at p‐2. We use ‘class 4’ to indicate this broad category of peptides. Strikingly, 43% of the peptides lack a canonical C‐terminal PDZ binding motif. These peptides frequently contain cryptic internal PDZ binding motifs upstream of the extreme C‐terminus, followed by an acidic residue or a proline. For example, the LYAADKH peptide of OTUB2 contains a class 2 motif at the p‐3 to p‐6 position. Such cryptic internal PDZ binding motifs have been reported also for other PDZ domains, such as PICK1 37 but have not been described for syntenin PDZ‐1 until now. Examination of the 194 syntenin ligands listed in BioGrid (September 2015) revealed that only 36% of these ligands have C‐terminal class 1–4 PDZ binding motifs. This can be in part explained by other parts of syntenin engaging in the interactions, as for example the syntenin‐Alix interaction that engage LYPX(n)L motifs in syntenin's unstructured N‐terminal region 3, but also point toward the importance of noncanonical PDZ interactions.
Combinatorial phage display
To gain further insights into the binding preferences of syntenin PDZ1‐2 we performed a phage display experiment using two highly diverse combinatorial peptide phage libraries, in the first case displaying highly diverse heptamer C‐terminal peptides, and in the second case displaying dodeca‐peptides flanked by flexible Gly/Ser linker regions. Sequencing of confirmed binding clones derived from C‐terminal library revealed one unique class 2 sequence (SHWTIDI, 11 clones), supporting a dominating preference for C‐terminal ligands of class 2. Although this may be an optimal syntenin binder, there is no peptide in the proteome that matches this sequence. Clonal ELISA of phage displaying the C‐terminal SHWTIDI peptide against the baits PDZ1, PDZ2 or PDZ1‐2 suggest that both PDZ domains are necessary for binding to this peptide (Table 2).
Table 2.
Ligands for syntnin‐1 PDZ1‐2 obtained through combinatorial peptide phage display. Bold letters in the sequence indicate the hydrophobic cluster. The numbers indicate the number of clones of a specific sequence. ‘+’ or ‘−’ indicate if the specific sequence was positive or negative in clonal phage ELISAs against PDZ1, PDZ2 or PDZ1‐2
| Peptide | Number | PDZ1 | PDZ2 | PDZ1‐2 |
|---|---|---|---|---|
| C‐terminal | ||||
| SHWTIDI | 11 | − | − | + |
| Internal | ||||
| GHFNGVRWIMGE | 10 | − | + | + |
| FGHWWETHLAEY | 5 | − | + | + |
| YYWFWTGENVYS | 2 | + | + | + |
| YYWAATGENVYS | 0 | − | − | − |
We identify three unique ligands from the selection against the internal peptide library. The first two peptides appear to only interact with PDZ2, but the third interacts with both domains, as judged by clonal phage ELISA (Table 2). Both PDZ1 and PDZ2 thus recognize internal peptide ligands. The sequence identities between the three internal peptides are low, but they share a hydrophobic patch of two or three residues followed by small, polar or charged residues that may be crucial for exiting the C‐terminal binding groove. Such binding specificity is consistent with the fact that syntenin PDZ2 can accomplish peptide binding by utilizing three consecutive binding pockets for hydrophobic residues 36. Substitution of the ‘WF’ sequence to ‘AA’ in the YYWFWTGENVYS peptide conferred a loss of binding in phage ELISAs, confirming that the hydrophobic cluster is required for binding.
Bioinformatics of ProP‐PD ligands
We explored the potential biological relevance of identified ProP‐PD ligands by a GO term 38 enrichment analysis (Table S2). The highest enrichment scores are related to molecular functions such as ‘hormone binding’ and ‘peptide receptor activity, G‐protein coupled’, to biological processes such as ‘sphingolipid biosynthetic process’ and ‘cyclic‐nucleotide‐mediated signaling’, and to cellular compartments such as ‘nuclear pore’ and ‘anchored to membrane’. Although their statistical significance remains low due to the small amount of binders falling in these categories, they appear relevant based on the established biological functions of syntenin. The apparent connection to the nuclear pore complex may suggest a novel function of the protein.
Validation of nectin‐1‐syntenin interactions
To examine the relevance of ProP‐PD ligands in the context of full‐length proteins we focused on the cell adhesion protein PVRL1, also called nectin‐1. Nectin‐1 is one of the top ranked candidates based on sequencing counts and mediate cell–cell adhesion in several different tissues such as epithelia, endothelia and neural tissue. Nectins are also involved in the regulation of cell movement, proliferation, survival, differentiation and apico‐basal polarity 39. Nectin‐1 interacts with other PDZ proteins, such as MPP3 40, afadin 41, PAR3 42 and PICK1 43. Nectin‐1 is further known to homodimerize and to heterodimerize with nectin‐3, which is also identified as a syntenin ligand in this study.
To validate the interaction we performed surface plasmon resonance (SPR) experiment where the interaction between the biotinylated cytoplasmic domain of nectin‐1 and full‐length syntenin was assessed. It was established that the interaction occurs with a K d of 14 μm (Fig. 2A). We then investigated if syntenin is a preferred binding partner of nectin‐1 through Y2H experiments using the C‐terminus of nectin‐1 as bait and a human fetal brain library as prey. Among nine positive clones sequenced, five corresponded to syntenin, two to the adaptor molecule GRIP2 and one to PICK1, an already described nectin partner 43. The identification of a known partner validates the Y2H screening and the predominance of syntenin among the nectin‐1 binders suggests that syntenin is a preferred nectin‐1 binder in this assay.
To investigate if the interaction between nectin‐1 and syntenin is effective in cells, we performed co‐IPs of the proteins in MCF‐7 cells. The immunoprecipitation of nectin‐1, using a nectin‐1 specific antibody, followed by immunoblotting with an anti‐syntenin antibody confirmed an interaction between the endogenous proteins (Fig. 2B). We further established that endogenous syntenin colocalizes with nectin‐1 at the sites of cell–cell contacts in freshly plated MCF‐7 cells (Fig. 2C).
To confirm that the interaction relies on the C‐terminus of nectin‐1 we performed co‐IPs of over‐expressed nectin‐1 (wild‐type or truncated form) and syntenin (Fig. 3). Truncation of the nectin PDZ binding motif abrogated the interaction, confirming the importance of the C‐terminus for the interaction. To characterize which domain of syntenin is implicated in the interaction, we co‐expressed distinct truncated forms of syntenin with nectin‐1 in Cos cells (Fig. 3B). Co‐immunoprecipitation (co‐IP) with nectin‐1 demonstrated that all the constructs containing at least one of the PDZ domains of syntenin are able to interact with nectin‐1. Thus, we identified nectin‐1 as an interaction partner of syntenin through ProP‐PD and confirmed the interaction through a combination of in vitro binding experiments and cell‐based assays.
Discussion
We explored the peptide binding of syntenin PDZ1‐2 through phage display. We established that syntenin is a multi‐tasking adaptor protein that can bind C‐terminal as well as internal binding sites, which suggests that a yet unknown set of ligands may reside within the intrinsically disordered regions of the human proteome. In this study, we identified more than 300 potential ligands in the C‐terminal regions. GO term enrichment analysis of these ligands revealed no strong enrichments, which might be expected from a selection against a bait protein with promiscuous binding specificity. Importantly, ProP‐PD identifies interactions that might occur in a biological context, but identified interactions may also be irrelevant, as there are no constrains to certain cellular compartments or cell types. Thus, care should be taken when selecting candidates for validation. Indeed, we found that about 10% of the retrieved ligands matched extracellular and secreted proteins (Table S1) and are unlikely physiological targets. However, it cannot be excluded that they still interact with syntenin during some stage of their export, as syntenin is known to load exosomes with cargo 3 and is linked to the secretion of angiopotin‐2 through exosomes 44. In a biological context, specificity is obtained through the use of multiple additional cues such as phospholipids, through ligand dimierization and by regulation through phosphorylation 8, 13, 15, and is further determined by the availability of the ligands. The archetypical syntenin ligand syndecan is, for example, present in one million copies per cell 45.
Through our analysis, we found that nectin‐1 is one of the ligands with the highest sequencing counts among the ligands from the ProP‐PD selections. It might be a relevant target based on its adhesive role at the adhere junctions in epithelial cells and at the synapses. We therefore validated the syntenin‐nectin interaction through orthogonal methods. We found that syntenin is the favored ligand of the C‐terminus of nectin‐1 in a Y2H assay and show that the two proteins colocalize in freshly plated MCF‐7 cells. We further confirm that the interaction is dependent on the PDZ domains of syntenin and the extreme C‐terminus of nectin‐1. However, the biological relevance and role of this interaction remain to be elucidated.
At this stage, we have only started to appreciate the power of ProP‐PD for interaction analysis through the use of dedicated C‐terminal peptide phage libraries. The development of more extensive ProP‐PD libraries that cover larger proportions of the disordered regions of target proteomes will allow identification of potential ligands of a large number of peptide‐binding proteins in the human proteome.
Author contributions
SGU performed co‐IPs and Y2H; PG performed phage display; RG & FL performed SPR and colocalization experiments; RA performed bioinformatics analysis; all authors analyzed results. SGU, ML, PZ, SSS and YI conceived the experiments. SGU, RG, RA & YI wrote the manuscript.
Supporting information
Table S1. Summary of syntenin ligands identified through ProP‐PD.
Table S2. GO term enrichment analysis of identified ligands.
Acknowledgements
This study was supported by grants from the Swedish Research Council (YI: C0509201), Ake Wiberg foundation (YI: 3773397), the Fund for Scientific Research ‐ Flanders (PZ: FWO, G.0479.12), the Concerted Actions Program of KU Leuven (PZ: GOA/12/016), the Canadian Institutes for Health Research (SSS: MOP‐93684) and by INSERM. RG was the recipient of a postdoctoral fellowship of the ARC French Foundation for Cancer Research.
[The copyright line for this article was changed on 1st February, 2016 after original online publication.]
References
- 1. Beekman JM and Coffer PJ (2008) The ins and outs of syntenin, a multifunctional intracellular adaptor protein. J Cell Sci 121, 1349–1355. [DOI] [PubMed] [Google Scholar]
- 2. Zimmermann P, Zhang Z, Degeest G, Mortier E, Leenaerts I, Coomans C, Schulz J, N'Kuli F, Courtoy PJ and David G (2005) Syndecan recycling [corrected] is controlled by syntenin‐PIP2 interaction and Arf6. Dev Cell 9, 377–388. [DOI] [PubMed] [Google Scholar]
- 3. Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E et al (2012) Syndecan‐syntenin‐ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14, 677–685. [DOI] [PubMed] [Google Scholar]
- 4. Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N, Slavik J, Machala M and Zimmermann P (2014) Syntenin‐ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun 5, 3477. [DOI] [PubMed] [Google Scholar]
- 5. Helmke BM, Polychronidis M, Benner A, Thome M, Arribas J and Deichmann M (2004) Melanoma metastasis is associated with enhanced expression of the syntenin gene. Oncol Rep 12, 221–228. [PubMed] [Google Scholar]
- 6. Lin JJ, Jiang H and Fisher PB (1998) Melanoma differentiation associated gene‐9, mda‐9, is a human gamma interferon responsive gene. Gene 207, 105–110. [DOI] [PubMed] [Google Scholar]
- 7. Kegelman TP, Das SK, Emdad L, Hu B, Menezes ME, Bhoopathi P, Wang XY, Pellecchia M, Sarkar D and Fisher PB (2015) Targeting tumor invasion: the roles of MDA‐9/Syntenin. Expert Opin Ther Targets 19, 97–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grembecka J, Cierpicki T, Devedjiev Y, Derewenda U, Kang BS, Bushweller JH and Derewenda ZS (2006) The binding of the PDZ tandem of syntenin to target proteins. Biochemistry 45, 3674–3683. [DOI] [PubMed] [Google Scholar]
- 9. Kang BS, Cooper DR, Jelen F, Devedjiev Y, Derewenda U, Dauter Z, Otlewski J and Derewenda ZS (2003) PDZ tandem of human syntenin: crystal structure and functional properties. Structure 11, 459–468. [DOI] [PubMed] [Google Scholar]
- 10. Feng W and Zhang M (2009) Organization and dynamics of PDZ‐domain‐related supramodules in the postsynaptic density. Nat Rev Neurosci 10, 87–99. [DOI] [PubMed] [Google Scholar]
- 11. Grootjans JJ, Reekmans G, Ceulemans H and David G (2000) Syntenin‐syndecan binding requires syndecan‐synteny and the co‐operation of both PDZ domains of syntenin. J Biol Chem 275, 19933–19941. [DOI] [PubMed] [Google Scholar]
- 12. Ivarsson Y (2012) Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett 586, 2638–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wawrzyniak AM, Vermeiren E, Zimmermann P and Ivarsson Y (2012) Extensions of PSD‐95/discs large/ZO‐1 (PDZ) domains influence lipid binding and membrane targeting of syntenin‐1. FEBS Lett 586, 1445–1451. [DOI] [PubMed] [Google Scholar]
- 14. Das SK, Bhutia SK, Sokhi UK, Azab B, Su ZZ, Boukerche H, Anwar T, Moen EL, Chatterjee D, Pellecchia M et al (2012) Raf kinase inhibitor RKIP inhibits MDA‐9/syntenin‐mediated metastasis in melanoma. Cancer Res 72, 6217–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G and Gettemans J (2002) PIP(2)‐PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell 9, 1215–1225. [DOI] [PubMed] [Google Scholar]
- 16. Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R et al (2014) A proteome‐scale map of the human interactome network. Cell 159, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K et al (2015) The bioplex network: a systematic exploration of the human interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perkins JR, Diboun I, Dessailly BH, Lees JG and Orengo C (2010) Transient protein‐protein interactions: structural, functional, and network properties. Structure 18, 1233–1243. [DOI] [PubMed] [Google Scholar]
- 19. Ivarsson Y, Arnold R, McLaughlin M, Nim S, Joshi R, Ray D, Liu B, Teyra J, Pawson T, Moffat J et al (2014) Large‐scale interaction profiling of PDZ domains through proteomic peptide‐phage display using human and viral phage peptidomes. Proc Natl Acad Sci USA 111, 2542–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sundell GN and Ivarsson Y (2014) Interaction analysis through proteomic phage display. Biomed Res Int 2014, 176172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blikstad C and Ivarsson Y (2015) High‐throughput methods for identification of protein‐protein interactions involving short linear motifs. Cell Commun Signal 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teyra J, Sidhu SS and Kim PM (2012) Elucidation of the binding preferences of peptide recognition modules: SH3 and PDZ domains. FEBS Lett 586, 2631–2637. [DOI] [PubMed] [Google Scholar]
- 23. Ivarsson Y, Wawrzyniak AM, Wuytens G, Kosloff M, Vermeiren E, Raport M and Zimmermann P (2011) Cooperative phosphoinositide and peptide binding by PSD‐95/discs large/ZO‐1 (PDZ) domain of polychaetoid, Drosophila zonulin. J Biol Chem 286, 44669–44678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McLaughlin ME and Sidhu SS (2013) Engineering and analysis of Peptide‐recognition domain specificities by phage display and deep sequencing. Methods Enzymol 523, 327–349. [DOI] [PubMed] [Google Scholar]
- 25. Tonikian R, Zhang Y, Boone C and Sidhu SS (2007) Identifying specificity profiles for peptide recognition modules from phage‐displayed peptide libraries. Nat Protoc 2, 1368–1386. [DOI] [PubMed] [Google Scholar]
- 26. Chatr‐Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L et al (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res 43, D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang da W, Sherman BT and Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 28. Walhout AJ and Vidal M (2001) High‐throughput yeast two‐hybrid assays for large‐scale protein interaction mapping. Methods 24, 297–306. [DOI] [PubMed] [Google Scholar]
- 29. Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P and Campadelli‐Fiume G (1998) The V domain of herpesvirus Ig‐like receptor (HIgR) contains a major functional region in herpes simplex virus‐1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA 95, 15700–15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krummenacher C, Nicola AV, Whitbeck JC, Lou H, Hou W, Lambris JD, Geraghty RJ, Spear PG, Cohen GH and Eisenberg RJ (1998) Herpes simplex virus glycoprotein D can bind to poliovirus receptor‐related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol 72, 7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zimmermann P, Tomatis D, Rosas M, Grootjans J, Leenaerts I, Degeest G, Reekmans G, Coomans C and David G (2001) Characterization of syntenin, a syndecan‐binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol Biol Cell 12, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cocchi F, Menotti L, Mirandola P, Lopez M and Campadelli‐Fiume G (1998) The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol 72, 9992–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K et al (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Geijsen N, Uings IJ, Pals C, Armstrong J, McKinnon M, Raaijmakers JA, Lammers JW, Koenderman L and Coffer PJ (2001) Cytokine‐specific transcriptional regulation through an IL‐5Ralpha interacting protein. Science 293, 1136–1138. [DOI] [PubMed] [Google Scholar]
- 35. Hirbec H, Perestenko O, Nishimune A, Meyer G, Nakanishi S, Henley JM and Dev KK (2002) The PDZ proteins PICK1, GRIP, and syntenin bind multiple glutamate receptor subtypes. Analysis of PDZ binding motifs. J Biol Chem 277, 15221–15224. [DOI] [PubMed] [Google Scholar]
- 36. Kang BS, Cooper DR, Devedjiev Y, Derewenda U and Derewenda ZS (2003) Molecular roots of degenerate specificity in syntenin's PDZ2 domain: reassessment of the PDZ recognition paradigm. Structure 11, 845–853. [DOI] [PubMed] [Google Scholar]
- 37. Erlendsson S, Rathje M, Heidarsson PO, Poulsen FM, Madsen KL, Teilum K and Gether U (2014) Protein interacting with C‐kinase 1 (PICK1) binding promiscuity relies on unconventional PSD‐95/discs‐large/ZO‐1 homology (PDZ) binding modes for nonclass II PDZ ligands. J Biol Chem 289, 25327–25340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rikitake Y, Mandai K and Takai Y (2012) The role of nectins in different types of cell‐cell adhesion. J Cell Sci 125, 3713–3722. [DOI] [PubMed] [Google Scholar]
- 40. Dudak A, Kim J, Cheong B, Federoff HJ and Lim ST (2011) Membrane palmitoylated proteins regulate trafficking and processing of nectins. Eur J Cell Biol 90, 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A et al (1999) Nectin/PRR: an immunoglobulin‐like cell adhesion molecule recruited to cadherin‐based adherens junctions through interaction with Afadin, a PDZ domain‐containing protein. J Cell Biol 145, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takekuni K, Ikeda W, Fujito T, Morimoto K, Takeuchi M, Monden M and Takai Y (2003) Direct binding of cell polarity protein PAR‐3 to cell‐cell adhesion molecule nectin at neuroepithelial cells of developing mouse. J Biol Chem 278, 5497–5500. [DOI] [PubMed] [Google Scholar]
- 43. Reymond N, Garrido‐Urbani S, Borg JP, Dubreuil P and Lopez M (2005) PICK‐1: a scaffold protein that interacts with Nectins and JAMs at cell junctions. FEBS Lett 579, 2243–2249. [DOI] [PubMed] [Google Scholar]
- 44. Ju R, Zhuang ZW, Zhang J, Lanahan AA, Kyriakides T, Sessa WC and Simons M (2014) Angiopoietin‐2 secretion by endothelial cell exosomes: regulation by the phosphatidylinositol 3‐kinase (PI3K)/Akt/endothelial nitric oxide synthase (eNOS) and syndecan‐4/syntenin pathways. J Biol Chem 289, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yanagishita M and Hascall VC (1992) Cell surface heparan sulfate proteoglycans. J Biol Chem 267, 9451–9454. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of syntenin ligands identified through ProP‐PD.
Table S2. GO term enrichment analysis of identified ligands.


