Abstract
Sleep has been shown to subtly disrupt the spatial organization of functional connectivity networks in the brain, but in a way that largely preserves the connectivity within sensory cortices. Here we evaluated the hypothesis that sleep does impact sensory cortices, but through alteration of activity dynamics. We therefore examined the impact of sleep on hemodynamics using a method for quantifying non-random, high frequency signatures of the blood-oxygen-level dependent (BOLD) signal (amplitude variance asymmetry; AVA). We found that sleep was associated with the elimination of these dynamics in a manner that is restricted to auditory, motor and visual cortices. This elimination was concurrent with increased variance of activity in these regions. Functional connectivity between regions showing AVA during wakefulness maintained a relatively consistent hierarchical structure during wakefulness and N1 and N2 sleep, despite a gradual reduction of connectivity strength as sleep progressed. Thus, sleep is related to elimination of high frequency non-random activity signatures in sensory cortices that are robust during wakefulness. The elimination of these AVA signatures conjointly with preservation of the structure of functional connectivity patterns may be linked to the need to suppress sensory inputs during sleep while still maintaining the capacity to react quickly to complex multimodal inputs.
Highlights
-
•
We examined time-domain dynamics of BOLD activity during sleep.
-
•
We used a method previously showing unique dynamics in sensory cortices during wakefulness.
-
•
We find elimination of wakefulness dynamics during N2 and N3 sleep, but not N1.
Introduction
Light sleep is associated with different modes of information processing and sensory responses relative to wakeful rest (Nashida et al., 2000, Atienza et al., 2001, Czisch et al., 2002). There are however notable similarities in the organization of brain activity during wakefulness and NREM sleep. Neuroimaging studies comparing wakefulness and sleep have documented similar magnitudes of evoked responses to sensory stimuli (e.g., evoked auditory responses; Issa and Wang 2011). In addition, intrinsic connectivity networks (ICN) such as the default mode network (DMN) are preserved in light sleep (Larson-Prior et al., 2009, Horovitz et al., 2008, Horovitz et al., 2009), with changes limited to a decoupling of the medial prefrontal regions and posterior regions of the DMN and other large functional networks during deeper (N3) slow wave sleep (Horovitz et al., 2009, Larson-Prior et al., 2011, Sämann et al., 2011). Increases in functional connectivity, e.g., between posterior DMN regions and parahippocampal, parietal, and posterior cingulate regions have also been documented (Larson-Prior et al. 2011) as well as highly similar patterns of resting state connectivity when defined from auditory, visual and motor regions (Larson-Prior et al. 2009). Network analyses documented preservation of functional modules, albeit with reduced inter-modular integration (Boly et al., 2012, Tagliazucchi et al., 2013a, Uehara et al., 2014). Electroencephalography (EEG) microstates are preserved across wakefulness and deep sleep (Brodbeck et al. 2012), and the association of ICNs with microstates (Britz et al. 2010) further supports the similarity of intrinsic brain activity during wakefulness and sleep. Furthermore, even the modest sleep-related changes in the topology of functional connectivity networks (see Tagliazucchi et al. 2013b for in depth discussion) largely exclude sensory cortex (e.g., Horovitz et al., 2008, Larson-Prior et al., 2009). Similarly, Uehara et al. (2014) documented local changes in nodal efficiency in transition from wakefulness to stage 1 sleep but these did not include any sensory region. To summarize, while sleep has been repeatedly shown to impact the magnitude of synchronization between brain regions, the actual topological structure of functional networks seen during wakeful rest is only moderately affected, particularly in sensory networks.
These prior findings raise the question we address in the current work, which pertains the activation regimes of sensory cortices during sleep. Specifically, we examined whether there exist activity patterns in sensory cortices that characterize the progression to deep sleep. In prior work (Davis et al. 2014) we identified rapid, non-random and non-oscillatory signatures in resting state fMRI data (amplitude variance asymmetry; AVA) that clustered around sensory systems in awake adults. AVA is a time-domain measure that is based on properties of the rapidly changing peak/pit structure of the BOLD time series. The magnitude and timing of these peaks and pits have been linked to functional processing (Skipper et al. 2009). In our prior work (Davis et al. 2014) we found that sensory regions showed a particular signature of the BOLD's signal local minima and maxima, which was reflected in the relative asymmetry of variances of the time series' peaks and pits. We also found that the magnitude of AVA correlated with IQ within a group of young children.
Formally, the AVA of a time series is given by [σ2(peaks)/σ2(pits)] of the time series. White noise has an AVA of 1, time series where the variance of the local maxima exceeds that of the local minima have an AVA greater than 1, and those showing the converse pattern have an AVA below 1. In this way AVA distinguishes between (i) ‘floor mode’ time series (AVA > 1) consistent with spontaneous activity of different magnitudes, and (ii) ‘ceiling mode’ time series (AVA < 1), consistent with diverse levels of spontaneous decreases in activity. AVA can therefore differentiate between time series with identical variance or entropy. Because in our prior work, adults' AVA patterns only showed floor-mode patterns and were limited to sensory cortices, we suggested they are related to the monitoring of the sensory environment during wakefulness.
On the basis of prior sleep and AVA work, we hypothesized that progression to deep sleep would be associated with a reduction in the robust AVA signatures found in sensory systems during wakeful rest. In contrast, on the basis of prior work we also expected that functional connectivity among sensory regions would largely maintain its topological structure during NREM sleep, potentially accompanied by an increase in BOLD variance (Horovitz et al., 2008, Tagliazucchi et al., 2013c). Finding such a pattern would indicate (1) that sleep is specifically linked to reduction in AVA even though it is linked to increased overall variance, and (2) that functional connectivity patterns and AVA are dissociable and therefore driven by different factors.
Methods
EEG–fMRI acquisition and artifact correction
We analyzed a data set collected during a synchronous EEG–fMRI acquisition protocol, which was approved by the ethical board of Goethe University (Kommission des Fachbereichs Medizin der J. W. Goethe-Universität Frankfurt am Main as of January 10, 2008). This data has been analyzed in several prior studies that examined identification of K-complex correlates in N2 sleep (Jahnke et al. 2012), use of functional connectivity for sleep staging (Tagliazucchi et al. 2012a), and the impact of sleep on serial autocorrelation of BOLD time series (Tagliazucchi et al. 2013c). In the current study, the EEG data were only used for purposes of determining sleep stages. EEG was sampled at an initial sampling rate of 5 kHz, low pass filtered at 1 kHz, and down-sampled to 250 Hz for artifact cleaning and sleep staging and further analysis (see below). Data were recorded using a BrainCapMR (Easycap) EEG cap (Herrsching, Germany) with 30 recording channels. The MR-compatible amplifiers were BrainAmp MR +, BrainAmp ExG; Brain Products (Gilching, Germany). Data was recorded using Brain Products' “Recorder” and analyzed using Brain Products' “Analyzer”. Analysis steps included: MRI and pulse artifact correction performed based on the average artifact subtraction (AAS) method (Allen et al., 1998) as implemented in Vision Analyzer2 (Brain Products, Germany) followed by objective (CBC parameters, Vision Analyzer) ICA-based rejection of residual artifact-laden components after AAS (Laufs et al., 2008). We obtained good quality EEG, which allowed for sleep staging by an expert, according to the criteria of the American Academy of Sleep Medicine (AASM 2007). Sleep staging was based on scoring 30 s blocks of the EEG data. Based on this scoring we ignored sections associated with transitions between sleep stages, maintaining those without transitions. BOLD time series sections matching these were spliced from the recorded time series.
Functional MRI scans were acquired on a 3 T system (Siemens Trio, Erlangen, Germany) using single-shot T2*-weighted EPI (32 slices, repetition time/echo time = 2080 ms/30 ms, matrix = 64 × 64, voxel size = 3 × 3 × 2 mm3, distance factor = 50%). To correct for physiological noise, physiological responses (cardiac, respiratory) were recorded through sensors from the MR scanner (sampling rate = 50 Hz) and MR-compatible devices (BrainAmp MR +, BrainAmp ExG; Brain Products). Sixty-three healthy non-sleep-deprived participants (thirty-six females, mean ± SD age of 23.4 ± 3.3 years) were scanned in the evening (starting from 8:00 PM). Data from those 55 participants who reached at least sleep stage N1 was used in our analysis.
BOLD time series selection
Each of the 55 participants provided epochs of functional imaging data during wakefulness (W) and at least the N1 sleep stage. From these epochs, we set the minimal time series length at 135.2 s (65 volumes), amounting to 293 total epochs. In practice, most of the epochs were 5–10 times as long (see Supplementary Table 1 for epoch descriptives). We conducted a quality control procedure in which we examined the functional time series for loss of signal in any volume, and derived temporal SNR histograms for each epoch to identify whether any epoch was associated with a shifted distribution indicating low quality data. Authors B.D. and U.H. conducted this procedure jointly.
fMRI preprocessing
We used AFNI for preprocessing and physiological noise correction (PN-correction). De-spiking of the time series was carried out as part of the PN-correction workflow. The physiological (cardiac, respiratory) data were down-sampled to the acquisition rate of single volume slices (15.4 Hz). We used AFNI's retroTS.m procedure to create slice-based regressors from these data. The utility produces (for each physiological regressor) a time series that is phase shifted to match the timing of each slice's acquisition. From the cardiac and respiration recordings we derived 13 such slice-based regressors (4 for the cardiac series and its harmonics, 4 for the respiratory series and its harmonics, and 5 for respiration variation over time and its harmonics, Birn et al. 2006). We removed the variance explained by these 13 regressors from the BOLD time series using the RETROICOR procedure (Glover et al. 2000) as implemented in AFNI.
Following PN-correction, we discarded the first 5 volumes of each epoch from the analysis to allow for T1 stabilization effects, and then performed slice timing correction (3dTshift), motion correction (3dvolreg), and spatial smoothing (3dmerge, 6 mm FWHM Gaussian Kernel) on all the images. We then removed several sources of variance from the time series data via linear regression. These included (i) 6 motion parameters estimated during the head motion correction, and (ii) linear, second-order and third-order polynomial trends. Because we partialed out physiological-related variance from direct measurements we did not use proxy measures typically used for this purpose such as data from CSF or white matter voxels. While these are sometimes used as proxies for physiological effects, they are only moderately correlated (see Chang & Glover, 2009). Furthermore, given the impact of global-mean correction on functional connectivity estimates (see Murphy et al., 2009, Schölvinck et al., 2010), we did not implement this procedure. The residuals of the regression procedure were used in all subsequent analyses. Following preprocessing, the time series of all voxels were analyzed using the “R” software (R Core Team 2012).
Defining BOLD amplitude variance asymmetry
After fMRI preprocessing, we created amplitude variance asymmetry (AVA) maps for each epoch using the R software (R Core Team 2012), following the procedures detailed in Davis et al. (2014). Briefly, we used the Pastecs library (Ibanez and Etienne 2006) to identify the location of turning points in the BOLD time series. Turning points are deflections in the slope of the time series that occur when the slope changes from positive to negative (a local maxima, i.e., “peak”), or from negative to positive (a local minima, i.e., “pit”). We saved the amplitude values for the identified peaks and pits and calculated separately the variance of the peak amplitudes and the variance of the pit amplitudes. We refer to the ratio of these variance terms, σ2(peaks)/σ2(pits), as the variance ratio (VR). For the inferences we draw, a VR of 3/2 is as significant a departure from 1 (the null hypothesis) as is a VR of 2/3. Therefore, significance tests are only performed after a natural log transform of the VR since this transformation assigns the same absolute value to a ratio and its inverse; e.g., | log(3/2)| = | log(2/3)| changing only the sign. The logarithmic transform of VR,log[σ2(peaks)/σ2(pits)], returns 0 when VR equals 1, returns positive numbers when VR > 1, and returns negative numbers when VR < 1. We defined AVA as the logarithmic transform of the VR.
In order to reduce the impact of high frequency fluctuations, before calculating the turning points and their variance, the time series X(t) were temporally smoothed using a moderate three-point temporal averaging kernel:
This temporal smoothing was aimed at reducing the impact of very small deviations (see Davis et al. 2014 for discussion) and has been used previously in similar analyses of point processes in time series (Morgan et al. 2008). We note that peaks and pits occur on a temporal scale that is faster than those thought to mediate functional connectivity, with our previous work identifying peaks every 8 s on average, in functional scans with a repetition time (TR) of around 2 s. We refer to time series where AVA significantly exceeds 0 as ‘floor mode’ patterns, because the distribution of local maxima is more varied, and to time series where AVA is significantly below 0 as ‘ceiling mode’ patterns.
fMRI group-level analyses
MNI registration
Following AVA analysis, we obtained a transformation between each EPI epoch to its corresponding anatomical image using FSL's epi_reg script. The most important steps in this procedure are FAST's (FMRIB's Automated Segmentation Tool; Zhang et al. 2001) histogram based segmentation of the T1 structural scans to derive white matter maps, and the use of the boundaries of these white matter maps to perform Boundary-Based co-Registration of the EPIs to their corresponding T1 structural images (BBR; Greve and Fischl 2009). We then performed nonlinear normalization (FNIRT), of each subject's T1 images into 2 × 2 × 2 mm MNI space. Importantly, we concatenated the two transformations (EPI to T1; Linear, guided by white matter boundaries) and T1 to MNI (nonlinear warp) to derive a single transformation from EPI space to MNI space. We used this transformation to align the AVA maps from original space to the MNI template in a single step.
Constructing single-participant AVA maps
Because participants had more than one AVA map per sleep stage (max = 6, see Supplementary Table 1), after MNI registration, these were averaged (within sleep stage) to construct a single AVA map per sleep stage per participant. We implemented weighted averaging in order to account for different epoch lengths as follows: we created weighted averages of the AVA values by (1) multiplying each AVA image by the total number of volumes in the epoch used to derive it, (2) summing all the AVA volumes maps in a specific participant's sleep stage and (3) dividing the result by the total number of volumes in all the epochs in that participant's sleep stage. This procedure weights the contributions to an individual's sleep stage AVA map in proportion to the length of the time series, relatively down weighting shorter series and up weighting longer series (i.e., the weights reflect percentage of time within condition, and sum to 1). Note that epoch length may have some effect on AVA estimation (though no relation to AVA magnitude) as longer epochs will offer a more precise sampling of the peak/pit variance and their ratio. That said, most of the epochs were very long (see Supplementary Table 1) and we further weighted the contribution of each epoch to AVA estimation by its length as described above.
Group-level statistical analysis
For wakefulness and each sleep stage (W, N1, N2, N3), in order to determine the brain regions in which the AVA reliably differed from chance (i.e., 0) on the group level, we conducted group-level whole-brain voxel wise t-test analyses of the AVA maps using AFNI's 3dttest ++ utility. We note that only 14 of the 55 participants contributed data to all four conditions (W, N1, N2, N3). The within-participant analysis based on this sub-group alone is likely to be underpowered, particularly given the strong potential for low correlations across the repeated measures, if indeed AVA indicates different processes across stages. For this reason we conducted two types of group-level analyses. The first focused on contrasts against the chance AVA value (i.e., 0), which we define as a baseline value. Here we used data from all those participants who provided data for a given stage (for W n = 50, for N1 n = 44, for N2 n = 36, and for N3 n = 17). This constituted the most sensitive test against baseline, and was based on one-sample t-tests evaluating difference from the null hypothesis, AVA = 0. The second analysis focused on differences between the experimental conditions (i.e., sleep stages and wakefulness). To maximize power, here we used paired t-tests, with each test relying on the maximal number of participants with data for the two conditions contrasted.
In all analyses we controlled for Family-Wise Error (FWE) using cluster-based thresholding, following the simulation procedures described by Forman et al. (1995) as implemented in AFNI's Clustsim utility. For contrasts against baseline, the following single-voxel thresholds were used (W: p < .001, N1, N2, N3: p < .005 we used different thresholds owing to different degrees of freedom in W vs. sleep stages), and these were corrected for FWE using cluster-based thresholding (p < .05). For contrasts between conditions, we used two cluster-forming thresholds: a very liberal (p < .05) single-voxel threshold that we corrected for FWE using cluster-based thresholding (p < .05), as well as a less-liberal single-voxel threshold of p < .005 (FWE corrected using cluster-based thresholding). The former was used to evaluate whether clusters found at the less-liberal threshold were isolated peaks or parts of larger clusters.
Comparison of AVA changes during sleep with changes in standard deviation during sleep
To evaluate whether decreases in AVA were accompanied by a specific pattern of changes in the standard deviation of the BOLD time series we first quantified the temporal standard deviation of the BOLD time series (SD) in each sleep stage. In cases of multiple epochs per sleep stage, these were averaged using the weighted mean procedure as described above for the AVA maps, after registration to MNI space. We calculated SD from the same time series for which AVA values were calculated.
We evaluated patterns of SD changes in areas showing significant changes in AVA. Within the regions showing higher AVA in W than N2 we identified 10 local difference maxima. Around each maxima we defined a sphere (33 voxels each 1 mm3), and also verified the spheres were at least 30 mm distant from each other. Within each sphere we analyzed whether SD values varied between the W and the N2 or N3 sleep stages.
Using the SD and AVA statistical parameter maps defined from the W vs. N2 contrast (n = 33), we calculated several metrics to understand the spatial relation between these two measures. We used the DICE overlap (Dice, 1945) to quantify the spatial similarity of SD and AVA statistical maps, and this was done across a complete parameter sweep of significant T values by which the SD map was thresholded. (The DICE overlap was computed as the number of voxels in the intersection of a thresholded AVA and a thresholded SD map, divided by total number of voxels in these maps, multiplied by 2.) Our starting minimum T value for the parameter sweep was 2.04 (uncorrected p < 0.1) and the maximum was 13.24 (increment = 0.01). We also derived another measure, which captured the proportion of voxels showing significant SD changes that also showed significant AVA changes (for a given T value threshold in the SD map).
AVA in thalamic subregions
To examine sleep-related impact on AVA patters in the thalamus we obtained a high-resolution thalamic atlas in MNI space (Krauth Atlas, Krauth et al. 2010), and used it to define 8 thalamic subregions (anterior, posterior, lateral, and medial thalamus bilaterally; 8 regions in all) per participant. This was based on the grouping of subregions into 8 sets, as presented in Krauth et al. (2010). Using the 14 participants for whom we had data in each condition, we examined AVA patterns in the 4 experimental conditions within these 8 regions using repeated measure ANOVAs.
Functional connectivity between regions associated with AVA, during wakefulness and sleep
Quantifying connectivity of brain regions showing AVA during wakefulness
From the AVA maps in the wakefulness condition we defined 11 regions of interest (ROIs) to be used as seed regions for functional connectivity analyses. These regions were defined as follows. On the group level, from the wakefulness analysis we automatically identified local maxima of AVA values (in 2 × 2 × 2 mm MNI space) within significant clusters. We then constructed a 3 mm radius spherical ROI around each maximum with the constraint that no two peaks were closer than 30 mm to each other. From these peaks, we manually chose the peaks that most closely matched regions reported in our prior work (Davis et al. 2014), in order to evaluate whether the functional connectivity structure replicated our prior findings and whether its features would change with sleep (we note that the peaks chosen this way tended to be the strongest within the clusters). These included bilaterally the superior temporal gyrus (STG), postcentral gyrus, lingual gyrus, middle occipital gyrus, as well as (unilaterally) the left inferior occipital gyrus, right calcarine gyrus and right supplementary motor area (SMA; see Supplementary Table 2 for locations). Because the purpose of this analysis was to see if we could replicate the wakefulness connectivity structure we documented in our prior work (Davis et al. 2014), we selected local maxima of AVA patterns in the current data rather than peak AVA locations from our prior work.
To determine if the functional connectivities of these regions (in W, N1, N2 and N3) were indicative of a single source of fluctuation, we constructed single-participant and group-level dendrograms for each condition, following the procedure we outlined in Davis et al. (2014). Group-level dendrograms were created from the data of the 14 participants that contributed data to all conditions. For each participant, the time series selected for each condition was matched to the length of shortest of the 4 conditions.
The following steps were implemented for each condition separately: 1) for each participant we constructed a pairwise correlation matrix of the connectivity of the 11 regions; 2) this matrix was subjected to hierarchical clustering that generated a dendrogram reflecting hierarchical connectivity per participant (using the hclust function in the R clue package with Euclidian distance for defining dissimilarity and complete-linkage method for hierarchical clustering); 3) the set of the 14 participants' dendrograms was then subjected to a ‘consensus cluster analysis’ that returns a centroid solution that optimally represents the set of individual level solutions. Furthermore, following Davis et al. (2014; see details wherein) we determined whether the mean dissimilarity of participants' dendrograms within condition differed from what would be expected by chance, using 10,000 permutations, where for each permutation, the labels of the ROIs were shuffled for each participant and the mean dissimilarity established for that permutation.
Given that we found that the connectivity structure of the set of regions defined by the W AVA map tended to maintain across several sleep stages, we wanted to know whether stronger changes in connectivity would be found for a different set of regions — that defined by the W vs. N2 AVA contrast. For this reason we also evaluated the functional connectivity between brain regions identified by that contrast. The procedures for defining these networks and group-level dendrograms were identical to those described above.
Analysis of pairwise regional correlation values
We also analyzed the magnitude of the pairwise correlation values for the connectivity of the 11 regions. In this analysis, we conducted pairwise contrasts between conditions, and used the number of participants that contributed data to both conditions. This was done, for each region pair, by Fisher-Z transforming each participant's correlation value and then conducting a paired t-test between conditions on the group level. For comparisons between W and N1 we used data from the 41 participants that contributed data to both, and for comparisons between the W and N2 conditions we used data from the 33 participants who contributed data to both. For comparisons between the N2 and N3 conditions we used data from the 14 participants that contributed data to both conditions and we matched the length of the time series within each participant across these conditions.
These analyses were meant to identify whether, in the functional connectivity patterns we studied, there is reduced connectivity with deeper sleep. Note that for any of the contrasts, the time series in the two conditions were matched for length within each participant by selecting the shortest time series of the two, and trimming the longer to match (this minimum value could vary by participant). Hence, this parameter was not confounded with sleep stage. All tests were paired-sample t-tests. The aim of these series of tests was to identify data patterns (i.e., whether sleep is generally associated with reduced connectivity among these regions) rather than document which specific reductions were significant. For this reason we report the number of significant statistical tests, but do not control for Family-Wise Error.
Evaluating motion differences between conditions
In order to determine if head movement motion parameters differed between conditions, for each participant's sleep epoch, we derived the average root-mean-square (RMS) deviation between each volume of the epoch and the reference volume used in motion correction. Then on the group level, we conducted paired t-tests between conditions to examine differences in this motion parameter. Motion was estimated for those time series used for dendrogram construction and functional connectivity analysis.
Evaluating peak-to-peak timing differences between conditions
The number of peaks and pits in a time series is orthogonal to the ratio of the variance of the peaks over the pits of the time series. It is thus possible that changes in peak-to-peak timings (i.e., the relative ‘pace’ of this time-domain measure) would be independent of changes in AVA. We therefore evaluated the relative frequency of the peaks and the pits in each condition (we thank an anonymous reviewer for this suggestion). To do so we simply computed for each voxel the number of pits in a given time series and then transformed this number into units of peaks per second to allow merging of data across series. These data (peak-to-peak interval) were processed on the single participant and group level identically to the AVA data.
Results
Quality control for BOLD time series
On the basis of quality control procedures (see Methods) we rejected 15 epochs (6 W, 4 N1, 4 N2, 1 N3). The remaining data consisted of 278 epochs in W, 79 epochs in N1, 70 epochs in N2, and 22 epochs in N3. We also examined the head motion signatures (RMS of each functional volume) derived from the registration of the BOLD time series in the W, N1, N2 and N3 stages, for the 14 participants that contributed data to all conditions. The N1 condition was associated with less motion (N1 < W, t(13) = 2.01, p = .03; N1 < N2, t(13) = 2.29, p = .02; N1 < N3, t(13) = 2.17, p = .03), and no other pairwise comparison between conditions approached significance. The absolute RMS magnitudes for the different conditions were W M = 24.5 ± 8.15, N1 M = 20.07 ± 4.87, N2 M = 24.59 ± 5.98, N3 M = 27.38 ± 10.87). Given the relatively small differences in RMS magnitude, we still included N1 in all analyses.
AVA changes in NREM sleep
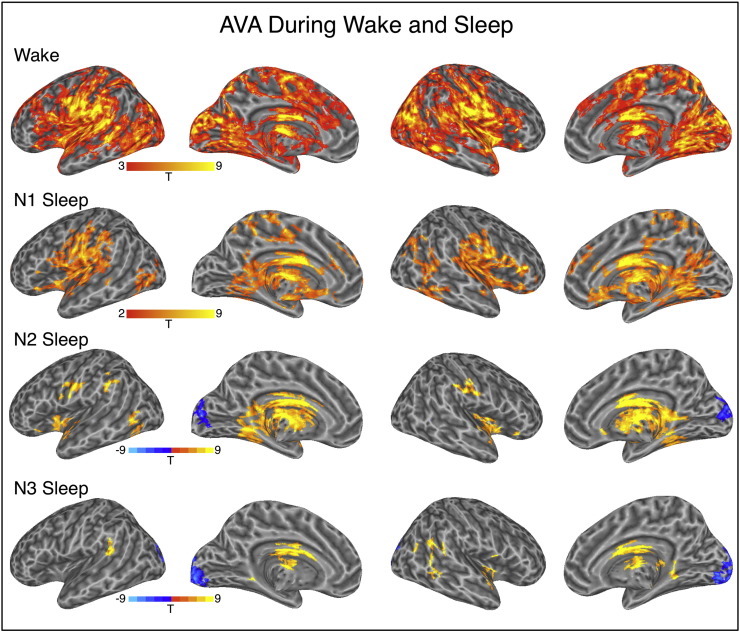
During wakeful rest, AVA patterns were significant bilaterally in visual clusters with a dorsal stream orientation that extended to the parietal lobe, straddling the central sulcus, large clusters in the lateral temporal lobe, and prefrontal cortex. This distribution with a dominant floor mode (variance peaks > variance pits, Fig. 1, n = 50 in t-tests against 0) corresponds to regions where we had previously found AVA to depart from chance during wakeful rest (Davis et al. 2014).
Fig. 1.
AVA maps for the four experimental conditions. Warm/cold colors indicate regions where AVA was significantly above/below zero. All analyses corrected for multiple comparisons using cluster extent thresholding (Family-Wise Error, p < .05). Uncorrected single-voxel thresholds used; wake: p < .001, all other conditions, p < .005.
During the initial N1 phase of NREM sleep, the spatial extent of statistically significant AVA clusters was more limited, but the distribution was similar (Fig. 1) and a direct contrast between W and N1, based on 41 participants with data in both conditions, produced a null finding. (Here and in all other between-condition contrasts: p < .005 single-voxel threshold, corrected for FWE using cluster extent thresholding at p < .05. A similar analysis probing for more diffuse clusters employed a very liberal single-voxel threshold of p < .05, similarly controlled for FWE at p < .05, but revealed no differences of W vs. N1; see Methods.)
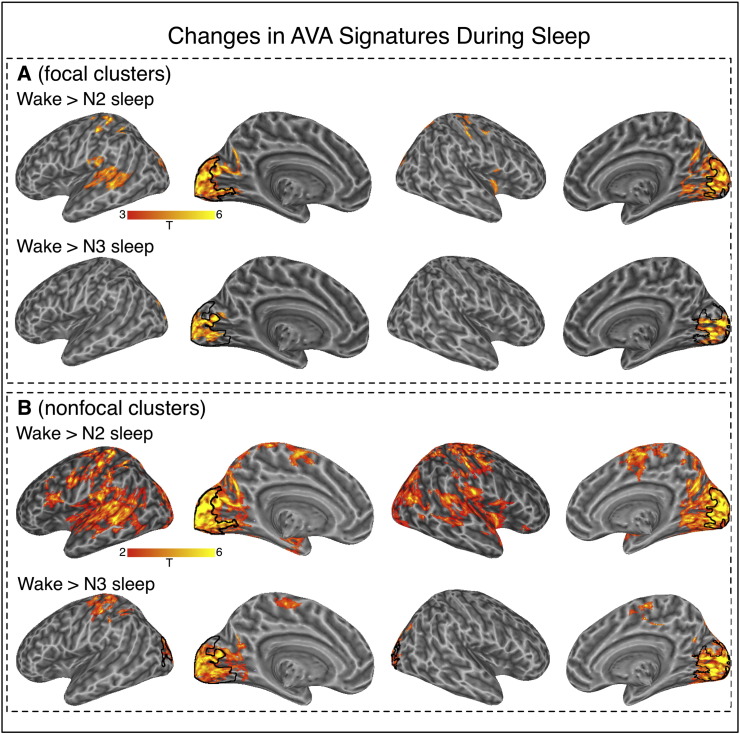
AVA patterns during N2 sleep strongly departed from wakefulness. The extent of the positive, floor-mode AVA profile was reduced, with a few surviving clusters mostly in the bilateral fusiform gyri, motor cortex and insula (see Fig. 1; n = 36 in t-tests against 0). Notably, we also found an occipital cluster with a strong ceiling mode profile (variance peaks < variance pits, blue clusters in Fig. 1). A direct contrast between W and N2 based on 33 participants with data in both conditions confirmed these observations statistically (see Fig. 2a).
Fig. 2.
Regions where AVA differed for the wake condition as compared to the N2 and N3 conditions. Panel A: More focal clusters identified by cluster-based thresholding with single-voxel type-I error of p < .005. Panel B: Non-focal clusters identified by cluster-based thresholding with single-voxel type-I error of p < .05. In both panels, the black outline in occipital regions marks areas with AVA < 0 in the N2 and N3 conditions. No significant differences were found for the contrast between wake and N1.
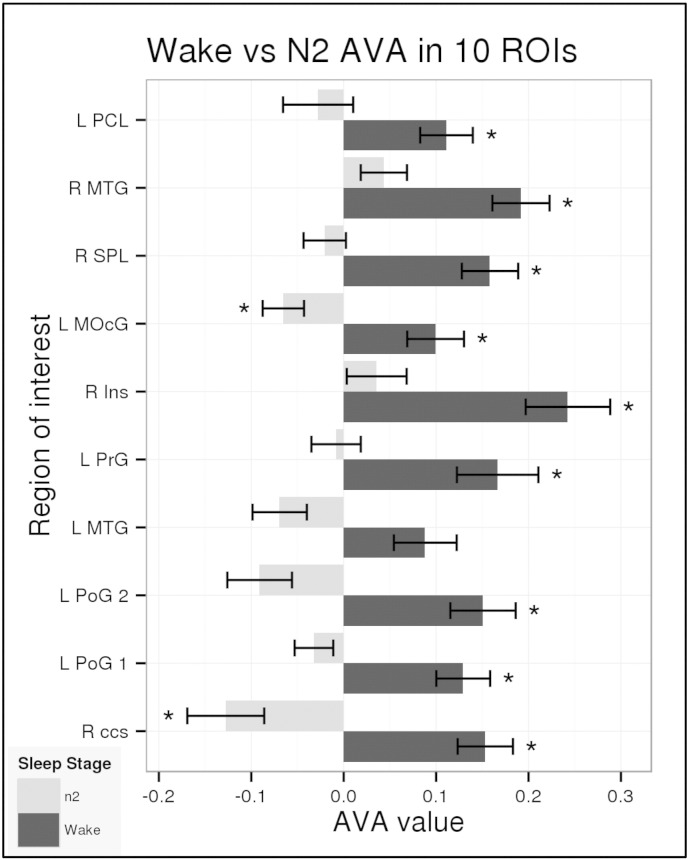
To better characterize the change in AVA patterns from W to N2 we identified 10 local maxima in this contrast map, positioned spheres around those points and treated those as functional regions of interest (fROIs; see Table 1). Note that the fact that these regions were identified in a contrast does not speak to the magnitude of their absolute AVA values in W and N2, because both values may be positive, negative, or a mixture of the two. The change in AVA from W to N2 in these fROIs is presented in Fig. 3.
Table 1.
Locations of local maxima (MNI coordinates) within clusters surviving the W vs. N2 contrast.
| T value | x | y | z | Anatomical name |
|---|---|---|---|---|
| 7.25 | 10 | − 84 | 6 | R_calcerine (R ccs) |
| 6.15 | − 56 | − 16 | 50 | L_postcentral (L PoG1) |
| 5.89 | − 34 | − 38 | 66 | L_postcentral (L PoG2) |
| 5.68 | 40 | − 28 | 64 | L_midtemporal (L MTG) |
| 5.32 | − 58 | 10 | 34 | L_precentral (L Prg) |
| 5.23 | 40 | 6 | − 12 | R_insula (R Ins) |
| 5.23 | − 26 | − 86 | 4 | L_mid_occipital (L MOcG) |
| 5.15 | 14 | − 68 | 66 | R_superior_parietal (R SPL) |
| 4.96 | 46 | − 68 | 18 | R_mid_temporal (R MTG) |
| 4.87 | − 4 | − 34 | 74 | L_paracentral_lobule (L PCL) |
Fig. 3.
AVA values for W and N2 conditions in ROIs showing differences between the two. Mean AVA values for the 33 participants with data in the W and N2 sleep stage in the spherical fROIs centered at the local maxima for the W > N2 contrast. Asterisks indicate significant tests against zero (p < 0.01) and are independent of the contrast procedure used to define each region. Abbreviations match region names in Table 1.
As Fig. 3 shows, during wakefulness, these fROIs showed significant floor-mode AVA, with the left middle temporal gyrus (L MTG) being the only exception. During N2, AVA was around zero (chance level) in most of the regions, and was significantly below zero in two regions: R.ccs and L MOcG. This shows that the floor-mode AVA profile observed during wakefulness is reduced with progression to sleep and that the visual cortex switches to a strong ceiling mode profile.
During N3, AVA retained a strong negative ceiling mode profile in the visual cortex, similar to that seen in N2 (variance peaks < variance pits, Fig. 1, n = 17 in t-tests against zero). Posterior lateral temporal clusters showed floor-mode AVA as observed for N2, N1 and W. Of these clusters, the visual cluster survived the direct W > N3 contrast, as did a cluster near the hand area of the central sulcus, which was identified when searching for clusters using a more liberal cluster-forming threshold (see Fig. 2 and Table 2, based on 14 participants contributing data to both conditions).
Table 2.
Locations of local maxima (MNI coordinates) within clusters surviving the W vs. N3 contrast.
| T value | x | y | z | Anatomical name |
|---|---|---|---|---|
| 7.28 | − 8 | − 90 | 2 | L_calcerine |
| 8.08 | − 54 | − 22 | 50 | L_postcentral |
Finally, we examined how AVA patterns in wakefulness and rest were reflected in thalamic subregions. Using a high-resolution thalamic atlas (see Methods) we divided each participant's left and right thalamus to anterior, posterior, lateral and medial sections, and examined the effect of sleep stage on AVA signatures in these 8 regions. Surprisingly, we found no impact of sleep on the AVA signatures in the thalamus. An omnibus 8 (Subregion) × 4 (Condition) analysis on mean regional AVA values revealed only a main effect of Subregion, with no effects of Condition or an interaction. When examining each subregion separately via 4-level repeated-measures ANOVA, none showed an effect of stage. In short, AVA signatures in the thalamus appeared relatively unaffected by sleep stage (see Supplementary Fig. 2).
Peak-to-peak intervals in wake and sleep
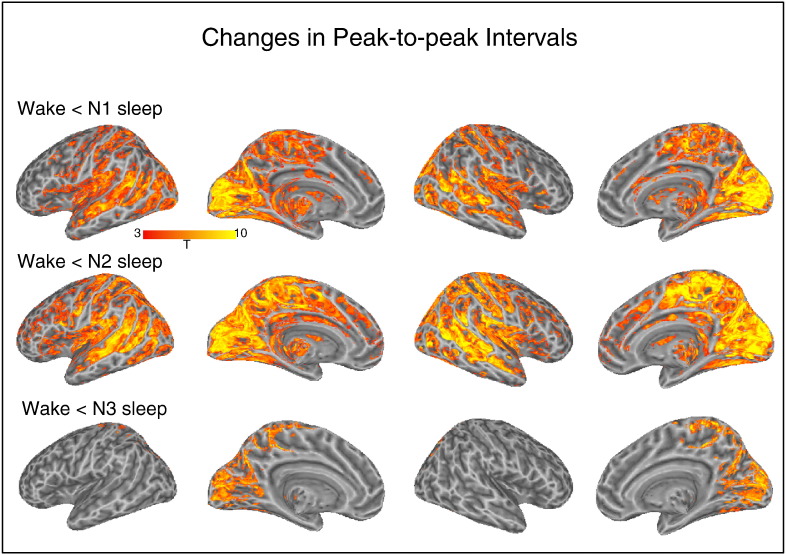
We observed significant increases in peak-to-peak interval during each of the sleep stages as compared to wakefulness (see Fig. 4). For the W vs. N1 contrast, increased intervals were found in sensory regions very similar to where AVA was positive in the W condition (but recall that we did not document AVA changes between W and N1). A similar set of regions was identified in the W vs. N2 contrast. For the W vs. N3 contrast, increased intervals were restricted to the visual and parietal cortex. Note that these comparisons relied on the maximal number of subjects in each condition and the more modest patterns in N3 could reflect the lower number of participant that contributed data to that condition. In absolute numbers, the average magnitude of the significant effect indicated around 0.5 s increase in peak-to-peak timing from Wake to sleep. The maximum of any voxels was a 1.5 s increase found in primary visual cortex (~ 11 s in wake vs. ~ 12.5 s during N1 sleep).
Fig. 4.
Regions where the peak-to-peak interval of the BOLD signal differed for the Wake condition as compared to the N1, N2 and N3 conditions. In all cases, intervals were shorter for the wake condition than the sleep stages.
BOLD standard deviation changes in clusters defined via AVA changes
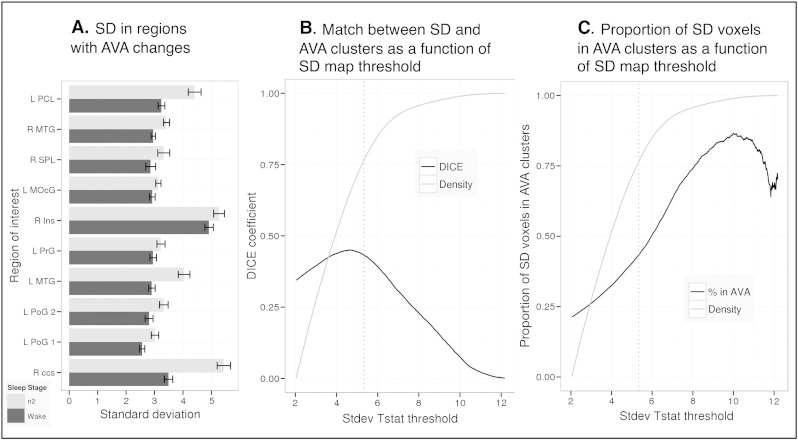
We analyzed the BOLD standard deviation (SD) within the spherical fROIs centered at the local maxima of the W vs. N2 contrast described in Table 1. We found that SD significantly increased from W to N2 in those regions defined by a significant decrease in AVA from W to N2 (see Fig. 5a, this relied on the same 33 participants as the AVA contrasts between the W and n2 conditions). This increase was significant at alpha level exceeding p < .005 in all but 3 fROIs (L middle occipital, p = .018, R insula, p = .049, and L precentral gyrus, p = .029).
Fig. 5.
Relation of AVA and SD sleep-related changes in W vs. N2. Panel A: Functional ROIs were defined from regions showing strong W > N2 patterns. In all these, SD was significantly higher during N2 than W (with exclusion of single region, all ps < .005). Panel B: The spatial match of sleep-related SD and AVA changes quantified via DICE coefficient. When thresholded at the same level (p < 05 single voxel uncorrected, Family-Wise-Error p < .05 using cluster extent constraint) the DICE coefficient was moderate (0.34); it increased slightly with stricter thresholding of the SD map, but remained below 0.5. Panel C: While the overlap between the maps was moderate, areas showing stronger sleep effects on SD were, proportionally, more likely to be found in AVA clusters. In Panels B and C, ‘density’ refers to the density of T values inside the initial SD map.
These findings demonstrate how changes in BOLD SD and BOLD AVA provide important but complementary information. Standard deviation increased in all fROIs. However, whereas in some fROIs this was accompanied by a loss of AVA signatures towards a chance value indicating a random asymmetry pattern, in two occipital regions AVA transformed from a significant floor mode to significant ceiling mode.
The relationship between the distribution of AVA and SD changes (W vs. N2) can be understood considering the whole-brain distribution of the effects: when thresholded at the same significance level (single voxel p < .05; cluster extent corrected at p < .05), clusters showing changes in AVA encompassed 9.8% of the combined WM + GM area, whereas clusters showing changes in SD encompassed 41% of this area. Thus, SD effects tended to be more widespread. Within the mask of regions showing changes in SD, 21% of the volume was associated with changes in AVA. Thus, it was clearly not the case that the two metrics target the same phenomenon. Within the mask of regions not showing changes in SD, only 2% of the volume was associated with changes in AVA, but we note that this latter mask likely includes low-signal-quality regions where it would be difficult to identify any systematic pattern across conditions.
When quantifying the spatial overlap of the AVA and SD effects (thresholded using cluster-based thresholding, p < .05 on cluster extent, p < .05 on single voxel uncorrected), we found a moderate overlap, as quantified by the DICE coefficient, D = .344. However, a stronger overlap was found when considering only voxels showing increasingly stronger SD effects. As shown in Fig. 5b, we recalculated the DICE coefficient while limiting the SD map to increasingly more stringent statistical thresholds, and found a maxima of overlap (D = 0.45) at a threshold of T = 4.68 (p < .0001; approximately 55% percentile of voxels' T value distribution within the SD mask). Thus, the maximal overlap was found when voxels showing less extreme SD effects (2.01 < T < 4.68) were ignored. However, increasing the threshold beyond T = 4.68 reduced the DICE coefficient (see Fig. 5b). Thus, it is not the case that the AVA patterns can be reduced to a set of areas showing a particularly strong SD effect. Finally, the strongest SD increases (from W to N2) did tend to localize in areas showing AVA effects. As seen in Fig. 5c, applying increased thresholds to the SD map tended to increase the relative proportion of SD voxels inside the AVA map.
Within the same ROIs (defined by the W vs. N2 AVA contrast) we also evaluated the impact of sleep on SD during the N1 and N3 sleep stages. For the contrast between W and N1 (using the 41 participants with data in both stages), only two regions showed a statistically significant difference (greater for N1: L paracentral lobule, p = .017; R calcerine, p < .0001). The results for contrast between W and N3 were similar to those found for the W vs. N2, with increased SD during N3.This increase was significant (p < 0.05) in all regions apart from three (R insula, p = .86; R superior parietal, p = .054; L precentral gyrus, p = 0.52).
Functional connectivity among regions implicated in AVA activity during wakefulness
Functional connectivity of brain regions showing significant AVA during wakefulness
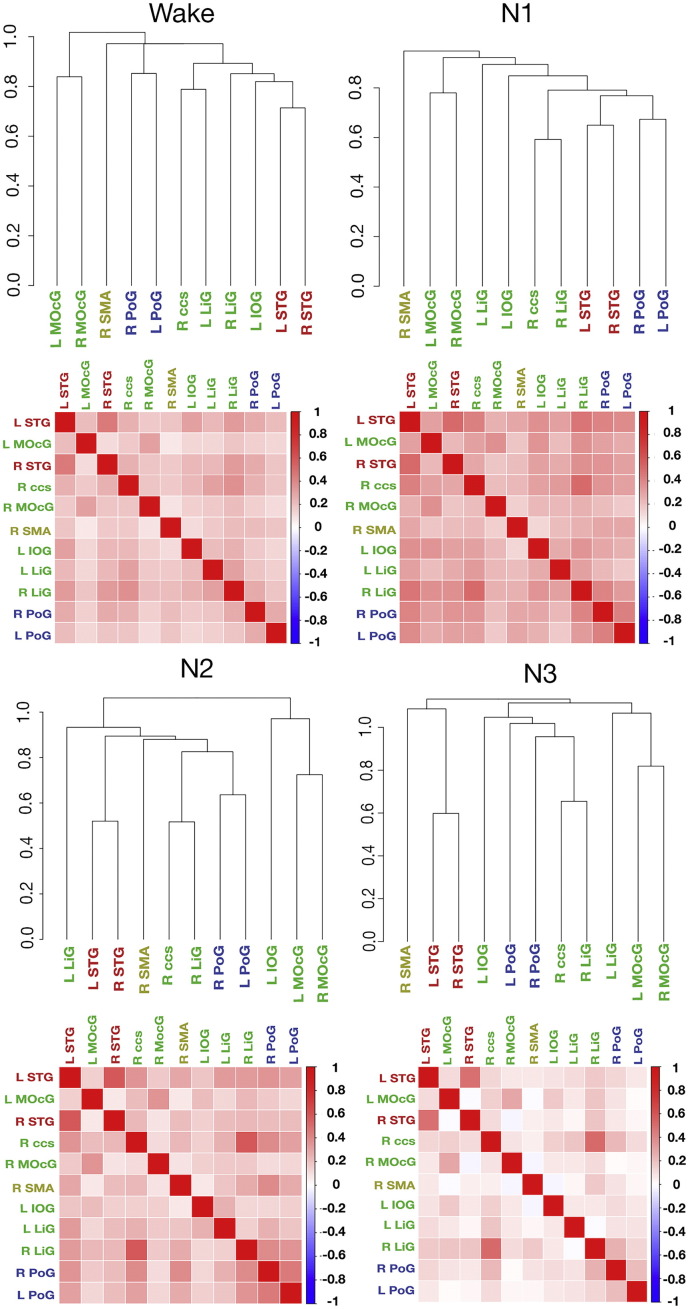
In this analysis we quantified the similarity of functional connectivity patterns in a network defined from brain regions showing significant AVA during wakefulness (11 activity peaks of W condition in Fig 1, see Supplementary Table 2 for locations). We used a hierarchical clustering solution (a dendrogram) to describe connectivity between regions showing significant AVA on the single-participant level. These analyses differ from quantifying functional connectivity between region pairs as they speak to the structure of connectivity between the set of regions as a whole, which can remain similar even given a change in connectivity strength per se, as we show below.
For the W, N1, N2 and N3 conditions, the consensus cluster analysis solution returned centroid-dendrograms that were similar to those we reported in prior work (Davis et al. 2014). As shown in Fig. 6, terminal leaves included STG bilaterally, with a separate cluster for middle occipital gyri, and a separate cluster for postcentral gyri. This supports our conclusion in prior work that AVA patterns in sensory systems are highly correlated within but not across systems. Finally, within each of the four conditions, the mean similarity of participants' dendrograms strongly exceeded that expected by chance (ps < .0001 for all conditions as indicated by permutation methods; see Methods).
Fig. 6.
Connectivity between regions with statistically significant AVA during wakefulness, shown for the W, N1, N2 and N3 conditions. We constructed functional ROIs around peak voxels showing AVA effects during wakefulness. For each condition, we characterized the connectivity of these regions using hierarchical clustering on the single-participant level, and then derived a consensus, centroid solution on the group level from the single-participant dendrograms. The resulting solutions were qualitatively similar across conditions (upper row). In contrast, the absolute correlations between time series decreased from wakefulness sleep.
Another approach to quantifying similarity in network structure is by evaluating the overall similarity of the group-level connectivity matrices across conditions (see Fig. 6). We treated each condition's connectivity matrix as a vector with 55 unique values, and examined the similarities of these values across conditions using Pearson's R as a similarity metric. (While most correlations were positive, a few were very slightly negative.) The pairwise similarities were as follows: W:N1 = 0.93, W:N2 = 0.71, W:N3 = 0.70, N1:N2 = 0.78, N1:N3 = 0.76, N2:N3 = 0.84. These considerable similarities, taken together with the overall similarity in dendrogram solutions across conditions do not suggest a qualitative difference in organization of functional connectivity within this network across wakefulness and sleep.
While network structures were quite similar across wakefulness and sleep, sleep was associated with a gradual and significant reduction in connectivity among regions (see Fig. 6). For the W vs. N1 contrast, 9 of the 55 region pairs showed statistically significant decreases, and none showed statistically significant increases (paired t-tests for each region pair, df = 41, p < .05 uncorrected; we used uncorrected tests as we aim to discover data patterns rather than specifically document which specific reductions are significant). The region showing the most consistent pattern of reduced connectivity was L. STG, for which 5 of its 10 connections showed statistically significant reduced correlations. For the W vs. N2 contrast, 14 of the 55 region pairs showed statistically significant decreases, and 4 showed statistically significant increases (paired t-tests for each region pair, df = 32, p < .05 uncorrected). The region showing the most consistent pattern of reduced connectivity was L. IOG, for which 7 of its 10 connections showed statistically significantly reduced correlations. The region pairs showing statistically significant increased correlations were {L STG, R STG}, {L STG, L LiG}, {R ccs, R LiG}, {R ccs, L PoG}. For the W vs. N3 contrast (paired t-tests for each region pair, df = 15, p < .05 uncorrected) we found statistically significant reductions in the connectivity strength of 26 of the 55 region pairs. A significantly higher correlation was found in one region pair, the {R ccs, R LiG}. Note that in all tests between conditions, we used matched-length BOLD time series (see Methods).
To summarize, the structure of functional connectivity in the AVA network, as defined from regions of local maxima during wakefulness, appeared to largely maintain across sleep conditions. However, deeper sleep was associated with reduced correlation strength.
Functional connectivity of brain regions where AVA decreased from wakefulness to N2 sleep
We also examined the connectivity structure between a set of regions defined by the W vs. N2 contrast (Fig 2a and Table 1). The consensus clustering solution for regions defined in this network revealed a different sort of arrangement as the terminal leaves did not tend to cluster within sensory regions (see dendrograms in Supplementary Fig. 1), Among these regions, sleep was associated with decreased connectivity (see connectivity matrices in Supplementary Fig. 1). For the W vs. N1 contrast only 3 of the 45 region pairs showed statistically significant differences in connectivity. The L MTG–L POG2 pair showed a significant increase in connectivity strength, and the R SPL was revealed to have significantly decreased connectivity to the R ins and the L MOcg (paired t-test for each region, df = 40, p < .05 uncorrected). The W vs. N2 contrasts revealed 15 region pairs with statistically significant reduced connectivity (paired t-test for each region, df = 32, p < .05 uncorrected). For the W vs. N3 contrast, 27 of the 45 region pairs showed statistically significant reduced connectivity. No region pairs were significantly increased (paired t-tests for each region pair, df = 15, p < .05). Interestingly, an analysis of the correlation matrices constructed for this set of regions showed strong maintenance of structure across W, N1, N2 and N3. The similarity of the W and N1 correlation matrices was quite high (Pearson's R = 0.93), as was the similarity of the W and N2 correlation matrices (Pearson's R = 0.86), and the W and N3 correlation matrices (Pearson's R = 0.81).
Discussion
Human NREM sleep is accompanied by extensive cognitive and behavioral changes (e.g., Harsh et al. 1994) and an increased arousal threshold for the deepest sleep stages (e.g., Carskadon and Dement 2005). Surprisingly, the analysis of spontaneous brain activity obtained from fMRI and EEG recordings has failed to identify equally widespread changes in the structure of functional connectivity networks during NREM sleep (Larson-Prior et al., 2009, Larson-Prior et al., 2011, Horovitz et al., 2008, Horovitz et al., 2009, Boly et al., 2012, Brodbeck et al., 2012, Tagliazucchi et al., 2013a). In particular, coordinated patterns of intrinsic connectivity networks (ICN) are preserved and, while disconnections occur inside particular networks such as the DMN, all individual ICN are nevertheless identifiable in all stages of human NREM sleep (Tagliazucchi et al. 2013c). The maintenance of connectivity patterns is particularly noticeable in sensory systems (e.g., Horovitz et al., 2008, Larson-Prior et al., 2009). The maintenance of connectivity structure does not mean that the absolute connectivity strength is necessarily unaltered (see, e.g., Picchioni et al. 2014, for reduced thalamic connectivity). However, the preserved structure of sensory ICNs points at a potential for preserved function during sleep, perhaps suggesting (Horovitz et al. 2008, p. 677) that “BOLD fMRI resting state activity does not require conscious wakefulness, but rather, in most brain areas, persists during reduced levels of consciousness characteristic of light sleep.”
Resting state activity, however, takes many forms beyond functional connectivity patterns, and in the current study we aimed to apply a new method for studying sleep-related activity, based on our prior work that documented particular activity dynamics in sensory cortices. Specifically, we have previously shown (Davis et al. 2014) that adult wakefulness is dominated by AVA signatures in sensory cortices, and suggested that these dynamics may indicate preparedness for sensory input during calm wakefulness in the absence of any specific orienting task or exogenous stimulation.
Here we find that during N2 and N3 sleep (but not N1) there is no evidence for the sorts of floor-mode AVA patterns seen during wakefulness. Comparisons against chance/baseline showed a more limited AVA profile in N2 and N3, and direct comparisons of N2 to wakefulness documented multiple regions where AVA was significant during wakefulness but at chance during N2. For N2, these changes were found in both auditory and visual systems, whereas for N3 they were limited to visual systems. In both N2 and N3, the AVA patterns in visual cortex were statistically significant, but showed a different pattern (“ceiling mode”) as compared to wakefulness. The whole-brain contrast between wakefulness and N3 showed few differences, meaning there were limited regions where the effect size was large (on the single-voxel level). However, N3 was also associated with the weakest AVA patterns compared to baseline, and it could be that both these results reflect higher intrinsic noise in that condition. Thus, although the functional connectivity patterns of sensory systems maintain across wakefulness and sleep (Horovitz et al., 2008, Larson-Prior et al., 2009, Uehara et al., 2014), our work shows a very strong change in local dynamics in all three sensory systems.
There are several potential explanations for these findings. One possibility is that during restful wakefulness, AVA reflects a cycling through the activity ‘repertoire’ of sensory regions (Deco et al. 2011). These might be related to maintaining a preparatory response set that could be implemented rapidly when observing or hearing particular stimuli, and it is this function that is reduced during sleep. Alternatively and specifically for somatomotor regions, the shift from positive AVA to a more neutral AVA signature may be related to a shift from a state of wakeful inhibition of sophisticated and volitional motor activity to the very different type of motor inhibition during sleep (Morales and Chase 1978). This could be related to the fact that motor actions initiated during sleep may be generated by different systems (Parrino et al. 2006). Additionally, activation in both auditory and visual cortex has been linked to generation of false percepts during wakeful rest (e.g., Hesselmann et al. 2010). The sleep-related reduction of AVA may indicate modulation of any or all of such processes, including, possibly, a different dynamic operation mode corresponding to active inhibition of sensory inputs during sleep, thus shifting towards a ceiling mode of activity in which spontaneous deactivations reflect sensory inhibition of environmental noise. Further work is needed to dissociate between such explanations, but there is some support for the presence of inhibitory mechanisms in prior neuroimaging work, documenting a massive decrease in the extent of BOLD activity to auditory stimulation during NREM sleep accompanied by deactivation in visual cortex (Czisch et al. 2002), as well as reduced (rather than increased) occipital responses during sleep (stimuli delivered via stroboscopic light through closed eyelids) (Born et al. 2002).
Another possibility is that the changes in AVA observed in sensory cortices during sleep are linked to disconnection from environmental stimuli, which is prominent through all stages of sleep1. It might be that during N1 sleep the sensory cortices might not yet have switched to bistable dynamics which manifest themselves during sleep (Pigorini et al., 2015, Compte et al., 2003, Sanchez-Vives and McCormick, 2000). It is possible that a bistable regime would manifest itself with low BOLD AVA, but this is an issue that should be investigated further. It is notable that while we observed no difference in AVA between the wake and the N1 stages, we did find a significant increase in the peak-to-peak interval in N1. This suggests that a change to the peak-to-peak timing might be an initial indicator of sleep onset, prior to changes in AVA dynamics proper.
This shift from a floor-mode AVA profile during wakefulness to a ceiling mode AVA profile in visual cortex was specific to deeper sleep stages and not observed in early N1 sleep. This result might seem counter-intuitive, since the protection against incoming stimuli in the progression from wakefulness to sleep, i.e., N1, is of critical importance. However, this does not take into account the delay between thalamo-cortical disconnection occurring at sleep onset (Spoormaker et al., 2010, Tagliazucchi and Laufs, 2014) and the “deactivation” of sensory regions (Magnin et al. 2010). This delay may account for residual conscious awareness after the transition to N1 sleep and possibly also for the preserved AVA during early sleep, during which incoming stimuli are blocked by the thalamic disconnection. During N2 sleep, thalamo-cortical communication is re-established (as shown by the cortical presence of sleep-spindles, of thalamic origin; De Gennaro and Ferrara 2003), but at this point the sensory cortices already react to stimulation with an inhibition of BOLD activity (i.e., ceiling mode) in order to raise the threshold for awakening.
Finally, we note that when interpreting these changes in AVA dynamics it is important to keep in mind that AVA is a BOLD signature, and that to better understand its origins, future work should study the underlying neural activity in these regions, potentially using EEG/MEG and source modeling approaches. The current work does however suggest that during wakefulness, AVA is a functionally related signature rather than simply reflecting very low-level processes that are independent of cognitive activity (e.g., Boly et al. 2009).
AVA, variance and connectivity offer complementary insights into descent to sleep
In addition to studying AVA changes during sleep, we also examined how sleep-induced changes in AVA relate to changes in three parameters typically used to characterize sleep-related changes: BOLD signal variance (SD of time series), the magnitude of connectivity between different sets of regions (as measured via pairwise correlations), and the structure of connectivity between these regions (as measured via dendrograms). This evaluation is important not only for understanding the relation between the measures, but also for determining whether AVA provides additional information above and beyond offered by other methods.
Increased BOLD signal variance has been reported in sensory cortices during NREM sleep (Horovitz et al., 2008, Tagliazucchi et al., 2013c), which could suggest a magnification of responses to sensory stimuli – either activation or deactivation – resulting in increased variance, or potentially be a result of non-neural factors such as changes in perfusion during sleep (Horovitz et al. 2008), or even changes in respirations patterns as these have shown to impact EEG patterns during sleep (Faes et al. 2015; but note that we partialed out heart rate variance effects to some extent). Horovitz et al. (2008) reported a gradual increase in standard deviation that correlated with an Inverse Index of Wakefulness. Our findings are in line with these conclusions in the regions we examined, as we found strong increases in standard deviation for N2 and N3, but more modest ones for N1. The findings of increased variance during sleep may appear to be at odds with the finding that sleep is associated with a more limited propagation of signals as reported in pivotal TMS work by Massimini et al. (2005). However the same study also reported a stronger local response during sleep, and the (local) variance of the BOLD signal may be a signature of such local responses rather than reverberation in an extended network. Interestingly, the N1 stage already showed very strong differences in the temporal dynamics of the BOLD response as measured by the peak-to-peak interval. Thus, it could be that a change in peak frequency precedes a strong change in variance, and this is an issue that can be explored in future work.
The observation of changes in AVA, in particular the shift towards a ceiling mode in occipital regions, is of a starkly different nature, as it relates to the variance in the peak/pit values of the signal only and not to the ratio of absolute activity between peaks and pits. This additional result points to a dynamical transformation inside a (in fact, more restricted) subset of sensory regions, which are those exhibiting highest AVA values during wakefulness, thus establishing a stronger link between AVA and sleep. While purely physiological changes can clearly impact magnitude of signal variance, it is difficult to explain how it could generate such subtle but robust changes in time-domain dynamics measured by AVA.
The relationship between the sets of brain regions that showed changes in SD and AVA during sleep was complex: there was no striking overlap between areas in which SD and AVA changed during sleep, though voxels showing strong SD effects tended to be found within AVA clusters. Again, it is essential to keep in mind that analytically, the two quantities are independent. SD can increase while AVA stays the same, as would be the case during any linear transformation of the time series, such as the one associated with an increase in gain or amplitude modulation. Thus, both analytically and practically the quantities communicate different information.
While the dynamics of activity in sensory cortices, as measured by AVA, changed with sleep, this was not the case for the structure of functional connectivity among these regions. We examined connectivity within these regions via dendrograms, and found that during wakefulness, these regions arranged according to a structure typified by strong clustering of regions within separate sensory systems, replicating our prior work (Davis et al. 2014). For W, N1 and N2 dendrograms, terminal nodes within a cluster always included regions linked to the same sensory functional system (e.g., lateral temporal cortex bilaterally, sensorimotor cortex bilaterally, and occipital cortex bilaterally). For N3, sensorimotor regions were not terminal nodes. It is possible that the constant scanner noise drove the consistent connectivity patterns during wake, N1 and N2 sleep for auditory regions (this is a general concern for any fMRI functional connectivity study). This concern could be evaluated via less noisy hemodynamic measures such as near-infrared spectroscopy, or by fMRI experiments with variable acoustic stimuli.
Additionally, we found that this structure of connectivity (among mainly sensory regions) maintained during sleep in a similar configuration to that found during wakefulness. This is very consistent with prior work mentioned earlier documenting maintained connectivity structure of sensory regions during sleep; see Horovitz et al. 2008 for visual seed region; Larson-Prior et al. 2009, for visual, auditory and somatomotor systems. Interestingly, this structure was maintained even though the absolute magnitude of pairwise time series correlations between these regions showed a systematic reduction with sleep. We found similar results when we defined seed regions via peaks in the W vs. N2 contrast. While spatial disintegration of networks would presumably be effective in insulating the sleeping brain from arousals and otherwise facilitating sleep (Boly et al., 2012, Tagliazucchi et al., 2013a), preservation of connectivity may serve to generate rapid responses to a sudden threat (Sadaghiani et al. 2010). In this apparent contradiction, the nature of our observation of preserved functional connectivity structure alongside drastically altered dynamics is manifest.
Interestingly, when focusing on the thalamus we found no significant changes in AVA patterns across wakefulness and sleep, in any thalamic subregion. This stands in contrast to the marked impact of sleep on thalamic functional connectivity (e.g., Picchioni et al. 2014). Similarly, there was no difference in AVA between wakefulness and N1, even though prior work (Tagliazucchi and Laufs 2014) has shown that N1 sleep is associated with an increase in cortico-cortico connectivity and a decrease in cortico-thalamic connectivity. However, it is important to note that it is possible that cortical changes in AVA are mediated by thalamic activity, without concomitant changes to AVA in the thalamus proper. That is, it could be that sleep's impact on sensory cortices is mediated indirectly by its effect on the thalamus and other subcortical structures with the consequence of reduced dynamics in the cortex. Future examinations of specific thalamic subregions across wake and sleep could dissociate between the stability of AVA and potential changes in connectivity (but studying connectivity of thalamic regions at high resolution would necessitate higher resolution scanning protocols). To summarize, we find that AVA dynamics, connectivity structure and correlation strength can vary independently and shed light on different aspects of sleep-related changes.
Alterations in the temporal dynamics of activity fluctuations are well-established features of EEG during sleep (AASM 2007) and fMRI is known to reflect neuroelectrical activity at specific frequencies (Laufs et al., 2003a, Laufs et al., 2003b, Mantini et al., 2007, Tagliazucchi et al., 2012b). In particular, it is known that the alpha frequency (between 8 Hz and 12 Hz) is present during wakefulness, is correlated with fMRI signals in sensory and visual (occipital) areas (Laufs et al., 2006, Ritter et al., 2009) and gives way to slower frequencies after sleep onset. The changes in AVA observed in sensory regions are consistent with the alterations in the dynamics of electrical activity, considering that similar amplitude asymmetries have been observed for the alpha wave in EEG recordings (Mazaheri and Jensen 2008).
Possible functional correlates of AVA
The sleep-related AVA changes we document may be related to different levels of conscious processing during wakefulness and sleep. Some higher level processing is maintained during sleep. Stimuli that fail to propagate to frontal and parietal regions and thus trigger a conscious percept (for instance, due to subliminal masking or due to the attentional blink) can still progress in the cortical hierarchy of sensory regions and in fact be processed to extract meaningful information, such as semantic information (Dehaene et al., 1998, Dehaene et al., 2006) or stop signals (van Gaal et al. 2009). However, the degree of information processing is diminished, for instance, lacking characteristic landmarks such as mismatch negativity (Loewy et al. 1996). In addition, the sleep state may involve unique modes of activity in visual cortex, including imagery or dreaming (see Born et al. 2002 for discussion), which could explain its unique mode of energy consumption during sleep. For instance, it has been shown it is possible to classify dream content from activity in visual cortex (Horikawa et al. 2013). This suggests that sleep unconsciousness is accompanied by unique activity regimen in the sensory cortices. AVA may reflect these differences in dynamical regimes across wakefulness and sleep, and the study of AVA in other brain states with even more restricted processing of sensory information (such as under anesthesia) should prove important to support this distinction. Future work examining AVA in relation to inter-individual differences in responding to sensory stimulation during sleep could address this issue by quantifying such differences in a quiet environment outside the scanner and then relating those observations to BOLD data as obtained and reported here.
While it is reasonable to postulate a “deactivation” of higher level, heteromodal cortical areas during sleep (i.e., the DMN and attention networks), the absence of AVA changes in these regions is most simply attributable to the fact that these regions do not show AVA during wakeful rest (see Fig. 1 and prior findings in Davis et al. 2014). Indeed, sleep-induced alterations in these higher level cortical areas appear to be more temporally extended (such as diminished temporal integration; Tagliazucchi et al. 2013c), instead of corresponding to changes in the variance of instantaneous “point-like” events (Tagliazucchi et al., 2012a, Davis et al., 2014). The relation between AVA and consciousness is an interesting topic for future work. At minimum, our findings suggest there might not be a monotonic relation between AVA and consciousness, as the differences between wakefulness and N3 appeared more limited than the differences between wakefulness and N2.
Summary
Our findings lend support to the view that during sleep, sensory systems display a qualitatively different dynamical behavior as evidenced by the inhibition of particular BOLD dynamics in these regions. At the same time, the structure of connectivity between these regions remains stable. The findings also suggest that during wakefulness, AVA reflects a functional signature of information processing in sensory cortices, and as such is strongly reduced during sleep. The change in dynamics from wakefulness to sleep likely reflects different responses to environmental stimuli, thus protecting the brain against arousals in spite of well-preserved functional connectivity structure, which might endow the brain with rapid reactivity. The link between AVA and sensory information processing and preparedness is also highlighted by these findings, thus suggesting further research to elucidate the precise neural mechanisms underlying this association.
Acknowledgment
This project was supported by a European Research Council Starting Grant (ERC-STG #263318 NeuroInt) to U.H.
Footnotes
We thank an anonymous reviewer for this suggestion.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2015.12.034.
Appendix A. Supplementary data
Supplementary material
References
- AASM . 2007. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. [Google Scholar]
- Allen P.J., Polizzi G., Krakow K., Fish D.R., Lemieux L. Identification of EEG events in the MR scanner: the problem of pulse artifact and a method for its subtraction. Neuroimage. 1998;8:229–239. doi: 10.1006/nimg.1998.0361. [DOI] [PubMed] [Google Scholar]
- Atienza M., Cantero J.L., Escera C. Auditory information processing during human sleep as revealed by event-related brain potentials. Clin Neurophysiol. 2001;112:2031–2045. doi: 10.1016/s1388-2457(01)00650-2. [DOI] [PubMed] [Google Scholar]
- Birn R.M., Diamond J.B., Smith M.A., Bandettini P.A. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Boly M., Tshibanda L., Vanhaudenhuyse a., Noirhomme Q., Schnakers C., Ledoux D., Laureys S. Functional connectivity in the default network during resting state is preserved in a vegetative but not in a brain dead patient. Human Brain Mapping. 2009;30(8):2393–2400. doi: 10.1002/hbm.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boly M., Perlbarg V., Marrelec G., Schabus M., Laureys S., Doyon J., Pélégrini-Issac M., Maquet M., Benali H. Hierarchical clustering of brain activity during human nonrapid eye movement sleep. Proc Natl Acad Sci USA. 2012;15:5856–5861. doi: 10.1073/pnas.1111133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born A.P., Law I., Lund T.E., Rostrup E., Hanson L.G., Wildschiodtz G.…Paulson O.B. Cortical deactivation induced by visual stimulation in human slow-wave sleep. Neuroimage. 2002;17(3):1325–1335. doi: 10.1006/nimg.2002.1249. [DOI] [PubMed] [Google Scholar]
- Britz J., Van De Ville D., Michel C.M. BOLD correlates of EEG topography reveal rapid resting-state network dynamics. Neuroimage. 2010;52:1162–1170. doi: 10.1016/j.neuroimage.2010.02.052. [DOI] [PubMed] [Google Scholar]
- Brodbeck V., Kuhn A., von Wegner F., Morzelewski A., Tagliazucchi E., Borisov S., Michel C.M., Laufs H. EEG microstates of wakefulness and NREM sleep. Neuroimage. 2012;62:2129–2139. doi: 10.1016/j.neuroimage.2012.05.060. [DOI] [PubMed] [Google Scholar]
- Carskadon M.A., Dement W.E. Normal human sleep: an overview. In: Kryger M.H., Roth T., Dement W.C., editors. Principles and Practice of Sleep Medicine. fourth ed. Elsevier Saunders; Philadelphia: 2005. pp. 13–23. [Google Scholar]
- Chang C., Glover G.H. Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. Neuroimage. 2009;47:1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compte A., Sanchez-Vives M.V., McCormick D.A., Wang X.-J. Cellular and network mechanisms of slow oscillatory activity (<1 Hz) and wave propagations in a cortical network model. J. Neurophysiol. 2003;89:2707–2725. doi: 10.1152/jn.00845.2002. [DOI] [PubMed] [Google Scholar]
- Czisch M., Wetter T.C., Kaufmann C., Pollmächer T., Holsboer F., Auer D.P. Altered processing of acoustic stimuli during sleep: reduced auditory activation and visual deactivation detected by a combined fMRI/EEG study. Neuroimage. 2002;16:251–258. doi: 10.1006/nimg.2002.1071. [DOI] [PubMed] [Google Scholar]
- Davis B., Jovicich J., Iacovella V., Hasson U. Functional and developmental significance of amplitude variance asymmetry in the BOLD resting-state signal. Cereb Cortex. 2014;24(5):1332–1350. doi: 10.1093/cercor/bhs416. [DOI] [PubMed] [Google Scholar]
- De Gennaro L., Ferrara M. Sleep spindles: an overview. Sleep Med Rev. 2003;7(5):423–440. doi: 10.1053/smrv.2002.0252. [DOI] [PubMed] [Google Scholar]
- Deco G., Jirsa V.K., McIntosh A.R. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nature Reviews Neuroscience. 2011;12(1):43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Dehaene S., Naccache L., Le Clec'H G., Koechlin E., Mueller M., Dehaene-Lambertz G., van de Moortele P.F., Le Bihan D. Imaging unconscious semantic priming. Nature. 1998;395:597–600. doi: 10.1038/26967. [DOI] [PubMed] [Google Scholar]
- Dehaene S., Changeux J.P., Naccache L., Sackur J., Sergent C. Conscious, preconscious, and subliminal processing: a testable taxonomy. Trends Cogn Sci. 2006;10:204–211. doi: 10.1016/j.tics.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Dice L.R. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26:297–302. [Google Scholar]
- Faes L., Marinazzo D., Jurysta F., Nollo G. Linear and non-linear brain–heart and brain–brain interactions during sleep. Physiol Meas. 2015;36(4):683–698. doi: 10.1088/0967-3334/36/4/683. [DOI] [PubMed] [Google Scholar]
- Forman S.D., Cohen J.D., Fitzgerald M., Eddy W.F., Mintun M.A., Noll D.C. Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): use of a cluster-size threshold. Magn Reson Med. 1995;33:636–647. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- Glover G.H., Li T.Q., Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Harsh J., Voss U., Hull J., Schrepfer S., Badia P. ERP and behavioral changes during the wake/sleep transition. Psychophysiology. 1994;31:244–252. doi: 10.1111/j.1469-8986.1994.tb02213.x. [DOI] [PubMed] [Google Scholar]
- Hesselmann G., Sadaghiani S., Friston K.J., Kleinschmidt A. Predictive coding or evidence accumulation? False inference and neuronal fluctuations. PLoS One. 2010;5(3) doi: 10.1371/journal.pone.0009926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa T., Tamaki M., Miyawaki Y., Kamitani Y. Neural decoding of visual imagery during sleep. Science. 2013;340(6132):639–642. doi: 10.1126/science.1234330. [DOI] [PubMed] [Google Scholar]
- Horovitz S.G., Fukunaga M., de Zwart J.A., van Gelderen P., Fulton S.C., Balkin T.J., Duyn J.H. Low frequency BOLD fluctuations during resting wakefulness and light sleep: a simultaneous EEG–fMRI study. Hum Brain Mapp. 2008;29:671–682. doi: 10.1002/hbm.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz S.G., Braun A.R., Carr W.S., Picchioni D., Balkin T.J., Fukunaga M., Duyn J.H. Decoupling of the brain's default mode network during deep sleep. Proc Natl Acad Sci USA. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez F., Etienne P.G.M. 2006. Pastecs: package for analysis of space-time ecological series. R package version, pp. 1–3. [Google Scholar]
- Issa E.B., Wang X. Altered neural responses to sounds in primate primary auditory cortex during slow-wave sleep. J. Neurosci. 2011;31(8):2965–2973. doi: 10.1523/JNEUROSCI.4920-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke K., von Wegner F., Morzelewski A., Borisov S., Maischein M., Steinmetz H., Laufs H. To wake or not to wake? The two-sided nature of the human K-complex. Neuroimage. 2012;59(2):1631–1638. doi: 10.1016/j.neuroimage.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Krauth B., Poveda, Jeanmonod, Morel, Székely A mean three-dimensional atlas of the human thalamus: generation from multiple histological data. NeuroImage. 2010;49(3):2053–-2062. doi: 10.1016/j.neuroimage.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Larson-Prior L.J., Zempel J.M., Nolan T.S., Prior F.W., Snyder A.Z., Raichle M.E. Cortical network functional connectivity in the descent to sleep. Proc Natl Acad Sci USA. 2009;106:4489–4494. doi: 10.1073/pnas.0900924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior L.J., Power J.D., Vincent J.L., Nolan T.S., Coalson R.S., Zempel J., Snyder A.Z. Modulation of the brain's functional network architecture in the transition from wake to sleep. Prog Brain Res. 2011;193:277–294. doi: 10.1016/B978-0-444-53839-0.00018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H., Krakow K., Sterzer P., Eger E., Beyerle A., Salek-Haddadi A., Kleinschmidt A. Electroencephalographic signatures of attentional and cognitive default modes in spontaneous brain activity fluctuations at rest. Proc Natl Acad Sci USA. 2003;100:11053–11058. doi: 10.1073/pnas.1831638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H., Kleinschmidt A., Beyerle A., Eger E., Salek-Haddadi A., Preibisch C., Krakow K. EEG-correlated fMRI of human alpha activity. Neuroimage. 2003;19:1463–1476. doi: 10.1016/s1053-8119(03)00286-6. [DOI] [PubMed] [Google Scholar]
- Laufs H., Holt J.L., Elfont R., Krams M., Paul J.S., Krakow K., Kleinschmidt A. Where the BOLD signal goes when alpha EEG leaves. Neuroimage. 2006;31:1408–1418. doi: 10.1016/j.neuroimage.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Laufs H., Daunizeau J., Carmichael D.W., Kleinschmidt A. Recent advances in recording electrophysiological data simultaneously with magnetic resonance imaging. Neuroimage. 2008;40:515–528. doi: 10.1016/j.neuroimage.2007.11.039. [DOI] [PubMed] [Google Scholar]
- Loewy D.H., Campbell K.B., Bastien C. The mismatch negativity to frequency deviant stimuli during natural sleep. Electroencephalogr. Clin. Neurophysiol. 1996;98:493–501. doi: 10.1016/0013-4694(96)95553-4. [DOI] [PubMed] [Google Scholar]
- Mantini D., Perrucci M.G., Del Gratta C., Romani G.L., Corbetta M. Electrophysiological signatures of resting state networks in the human brain. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13170–13175. doi: 10.1073/pnas.0700668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaheri A., Jensen O. Asymmetric amplitude modulations of brain oscillations generate slow evoked responses. J. Neurosci. 2008;28:7781–7787. doi: 10.1523/JNEUROSCI.1631-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin M., Rey M., Bastuji H., Guillemant P., Mauguière F., Garcia-Larrea L. Thalamic deactivation at sleep onset precedes that of the cerebral cortex in humans. Proc Natl Acad Sci USA. 2010;107:3829–3833. doi: 10.1073/pnas.0909710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimini M., Ferrarelli F., Huber R., Esser S.K., Singh H., Tononi G. Breakdown of cortical effective connectivity during sleep. Science. 2005;309(5744):2228–2232. doi: 10.1126/science.1117256. [DOI] [PubMed] [Google Scholar]
- Morales F.R., Chase M.H. Intracellular recording of lumbar motoneuron membrane potential during sleep and wakefulness. Exp Neurol. 1978;62(3):821–827. doi: 10.1016/0014-4886(78)90289-3. [DOI] [PubMed] [Google Scholar]
- Morgan V.L., Gore J.C., Szaflarski J.P. Temporal clustering analysis: what does it tell us about the resting state of the brain? Med. Sci. Monit. 2008;14:345–352. [PMC free article] [PubMed] [Google Scholar]
- Murphy K., Birn R.M., Handwerker D.A., Jones T.B., Bandettini P.A. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashida T., Yabe H., Sato Y., Hiruma T., Sutoh T., Shinozaki N., Kaneko S. Automatic auditory information processing in sleep. Sleep. 2000;23:821–828. [PubMed] [Google Scholar]
- Parrino L., Halasz P., Tassinari C.A., Terzano M.G. CAP, epilepsy and motor events during sleep: the unifying role of arousal. Sleep Med Rev. 2006;10(4):267–285. doi: 10.1016/j.smrv.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Picchioni D., Pixa M.L., Fukunaga M., Carr W.S., Horovitz S.G., Braun A.R., Duyn J.H. Decreased connectivity between the thalamus and the neocortex during human nonrapid eye movement sleep. Sleep. 2014;37(2):387–397. doi: 10.5665/sleep.3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigorini A., Sarasso S., Proserpio P., Szymanski C., Arnulfo G., Casarotto S., Fecchio M., Rosanova M., Mariotti M., Lo Russo G., Palva J.M., Nobili L., Massimini M. Bistability breaks-off deterministic responses to intracortical stimulation during non-REM sleep. Neuroimage. 2015;112:105–113. doi: 10.1016/j.neuroimage.2015.02.056. [DOI] [PubMed] [Google Scholar]
- R Core Team (2012). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, (URL http://www.R-project.org/)
- Ritter P., Moosmann M., Villringer A. Rolandic alpha and beta EEG rhythms' strengths are inversely related to fMRI-BOLD signal in primary somatosensory and motor cortex. Hum. Brain. Mapp. 2009;30:1168–1187. doi: 10.1002/hbm.20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sämann P.G., Wehrle R., Hoehn D., Spoormaker V.I., Peters H., Tully C., Holsboer F., Czisch M. Development of the brain's default mode network from wakefulness to slow wave sleep. Cereb. Cortex. 2011;21:2082–2093. doi: 10.1093/cercor/bhq295. [DOI] [PubMed] [Google Scholar]
- Schölvinck M.L., Maier A., Frank Q.Y., Duyn J.H., Leopold D.A. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci USA. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoormaker V.I., Schröter M.S., Gleiser P.M., Andrade K.C., Dresler M., Wehrle R., Sämann P.G., Czisch M. Development of a large-scale functional brain network during human non-rapid eye movement sleep. J. Neurosci. 2010;30:11379–11387. doi: 10.1523/JNEUROSCI.2015-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani S., Hesselmann G., Friston K.J., Kleinschmidt A. The relation of ongoing brain activity, evoked neural responses, and cognition. Front. Syst. Neurosci. 2010;4 doi: 10.3389/fnsys.2010.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vives M.V., McCormick D.A. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat Neurosci. 2000;3:1027–1034. doi: 10.1038/79848. [DOI] [PubMed] [Google Scholar]
- Skipper J.I., Goldin-Meadow S., Nusbaum H.C., Small S.L. Gestures orchestrate brain networks for language understanding. Curr Biol. 2009;19(8):661–667. doi: 10.1016/j.cub.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Balenzuela P., Fraiman D., Chialvo D.R. Criticality in large-scale brain fMRI dynamics unveiled by a novel point process analysis. Front. Physiol. 2012;3 doi: 10.3389/fphys.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., von Wegner F., Morzelewski A., Borisov S., Jahnke K., Laufs H. Automatic sleep staging using fMRI functional connectivity data. Neuroimage. 2012;63(1):63–72. doi: 10.1016/j.neuroimage.2012.06.036. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E., Von Wegner F., Morzelewski A., Brodbeck V., Borisov S., Jahnke K., Laufs H. Large-scale brain functional modularity is reflected in slow electroencephalographic rhythms across the human non-rapid eye movement sleep cycle. Neuroimage. 2013;70:327–339. doi: 10.1016/j.neuroimage.2012.12.073. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E., Von Wegner F., Morzelewski A., Brodbeck V., Laufs H. Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Front. Hum. Neurosci. 2012;6 doi: 10.3389/fnhum.2012.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Behrens M., Laufs H. Sleep neuroimaging and models of consciousness. Front Psychol. 2013;4 doi: 10.3389/fpsyg.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Von Wegner F., Morzelewski A., Brodbeck V., Jahnke K., Laufs H. Breakdown of long-range temporal dependence in default mode and attention networks during deep sleep. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15419–15424. doi: 10.1073/pnas.1312848110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Laufs H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron. 2014;82(3):695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Uehara T., Yamasaki T., Okamoto T., Koike T., Kan S., Miyauchi S.…Tobimatsu S. Efficiency of a “small-world” brain network depends on consciousness level: a resting-state FMRI study. Cereb. Cortex. 2014;24(6):1529–1539. doi: 10.1093/cercor/bht004. [DOI] [PubMed] [Google Scholar]
- van Gaal S., Ridderinkhof K.R., van den Wildenberg W.P., Lamme V.A. Dissociating consciousness from inhibitory control: evidence for unconsciously triggered response inhibition in the stop-signal task. J. Exp. Psychol. 2009;35:1129. doi: 10.1037/a0013551. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Brady M., Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Greve D.N., Fischl B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage. 2009;48:63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material