Abstract
Essential genes are defined by their requirement to sustain life in cells or whole organisms. The systematic identification of essential gene sets not only allows insights into the fundamental building blocks of life, but may also provide novel therapeutic targets in oncology. The discovery of essential genes has been tightly linked to the development and deployment of various screening technologies. Here, we describe how gene essentiality was addressed in different eukaryotic model organisms, covering a range of organisms from yeast to mouse. We describe how increasing knowledge of evolutionarily divergent genomes facilitate identification of gene essentiality across species. Finally, the impact of gene essentiality and synthetic lethality on cancer research and the clinical translation of screening results are highlighted.
Keywords: Cancer research, CRISPR/Cas9, essential genes, genetic interaction, RNAi, screening, synthetic lethality
Introduction
In the past decades, an increasing number of genomes have been completely sequenced (Adams, 2000; Hillier et al., 2008; Howe et al., 2013; Venter et al., 2001; Wood et al., 2002). With the increasing knowledge of the sequence composition of genomes, the next challenge has been to comprehensively analyze the function of encoded genes. Of particular interest have been genes that are essential for survival at the cellular or organismic level. Identifying the minimal set of genes necessary for sustaining life will allow better understanding of life itself and provide insight into the origin of diseases.
The search for essential genes has been extensively conducted in prokaryotic organisms, with the aim to identify novel targets for antibiotic therapy (Clatworthy et al., 2007) and critical building blocks for synthetic biology (Khalil & Collins, 2010). Due to the small size of prokaryotic genomes and their easy accessibility to genetic manipulation, essential genes have been identified for a broad panel of prokaryotic organisms (de Berardinis et al., 2008; Gerdes et al., 2003; Glass et al., 2006; Kobayashi et al., 2003). In eukaryotic organisms, multiple loss-of-function technologies have been developed to investigate gene functions, including chemical mutagenesis (Hrabé de Angelis et al., 2000), insertional mutagenesis (Bellen et al., 2004), RNAi technologies (Dietzl et al., 2007; Kamath et al., 2003) and CRISPR/Cas9 genome editing (Shalem et al., 2015; Wang et al., 2014). The effectiveness and also the limits of those screening technologies have determined the scope by which essential genes have been recovered.
In this review, we describe the development of screening technologies and their impact on discovery of essential genes for common eukaryotic model organisms. We illustrate how knowledge of gene essentiality contributes to understanding of human diseases and can be employed for anticancer therapy.
What is an essential gene?
Essential genes are defined as genes that are required for sustaining life (Juhas et al., 2011). The concept of gene essentiality and its limits was first discussed in 1963 (Gluecksohn-Waelsch, 1963). Presently, the general understanding of an essential gene is that it is required for survival and proliferation of single cell organisms. In multicellular organisms, loss of essential genes results in lethality during development or inability for reproduction. The estimated proportion of essential genes varies considerably between different species, and also among different publications (Table 1). Reasons for this discrepancy can be diverse and include differences in methods used to achieve loss-of-function, inability to perform genome-wide knockouts in many organisms and incomplete recovery of all phenotypes associated with gene essentiality. While there is a core set of essential genes that shows a stringently lethal phenotype upon loss, there is a larger group of genes on which survival depends on specific environmental conditions, in particular developmental stages or tissues. The impact of the environment on gene essentiality was extensively described for Saccharomyces cerevisiae, showing that under conditions other than rich in nutrients, the percentage and composition of essential genes varies (Giaever et al., 2002; Hillenmeyer et al., 2008). For instance, while 4769 homozygous deletion strains were considered non-essential in rich medium, only 205 strains (3% of the genome) were non-essential when growth was tested under multiple environmental conditions (Hillenmeyer et al., 2008). Furthermore, defects in genes related to the immune system can also lack any visible phenotypes under laboratory conditions, but quickly become essential upon challenge by infectious agents (Galiana-Arnoux et al., 2006; Gazit et al., 2006).
Table 1. Estimated proportion of essential genes in model organism.
| Saccharomyces cerevisiae | |
| 17–18.1% | (Giaever et al., 2002; Winzeler et al., 1999) |
| Caenorhabditis elegans | |
| 13.9% | (Johnsen & Baillie, 1991) |
| 1% | (Clark et al., 1988) |
| 8.5% | (Kamath et al., 2003) |
| Drosophila melanogaster | |
| 8-16.3% | (Bellen et al., 2004) |
| 30% | (Dietzl et al., 2007) |
| Danio rerio | |
| 5.4% | (Amsterdam et al., 2004) |
| 9.3% | (Haffter et al., 1996) |
| Mus musculus | |
| 13.3% | (Bradley et al., 2012) |
| Homo sapiens (core essential genes in a cancer cell line panel) | |
| 1.4% | (Hart et al., 2014) |
The percentage of essential genes is obtained by dividing the number of essential genes given in the indicated literature by the total number of protein coding genes in the respective genomes (retrieved from ENSEMBL database).
In multicellular organisms, gene essentiality can be restricted to specific tissues and developmental stages. For example, mice deficient in SLC2744/FATP4, a protein responsible for the cellular import of free fatty acids, die shortly after birth due to skin abnormalities (Herrmann et al., 2003). This lethal phenotype can be rescued by re-expression of the protein in the skin (Shim et al., 2009). In contrast, knockout of SLC2744/FATP4 in either adipose tissue or the intestine did not show any striking phenotype (Lenz et al., 2011; Shim et al., 2009). In another example, post-developmental knockdown of essential genes in Caenorhabditis elegans revealed a fraction of genes than can actually prolong life-span when their function is lost at a later developmental stage (Curran & Ruvkun, 2007).
Another concept that is closely related to gene essentiality is fitness. Fitness and fitness defects were originally used to describe changes of allele frequencies in population studies (Otto & Lenormand, 2002) (Figure 1). As opposed to gene essentiality, it is not measured on a single cell level, but is a population-level phenotype. Among others, it describes the exponential growth rate of a given population relative to its wild-type counterpart (Giaever et al., 2002; Hillenmeyer et al., 2008). Compared to essential genes, loss of genes that are associated with fitness defects can show only mild or no phenotypes within one generation. However, in a heterogeneous and dynamic population, selective pressure against fitness defects will result in the disappearance of individuals carrying the unfavorable trait within consecutive generations, as shown for C. elegans (Ramani et al., 2012). Thus, genes that cause fitness defects can also be considered as essential in the context of population dynamics. Two studies in C. elegans and S. cerevisiae have shown that genes previously considered to be dispensable are actually associated with fitness defects (Breslow et al., 2008; Kamath et al., 2003). In summary, while the definition of gene essentiality is seemingly straightforward, unambiguously classifying a gene as essential is difficult and remains highly dependent on the context by which its function is measured.
Figure 1.
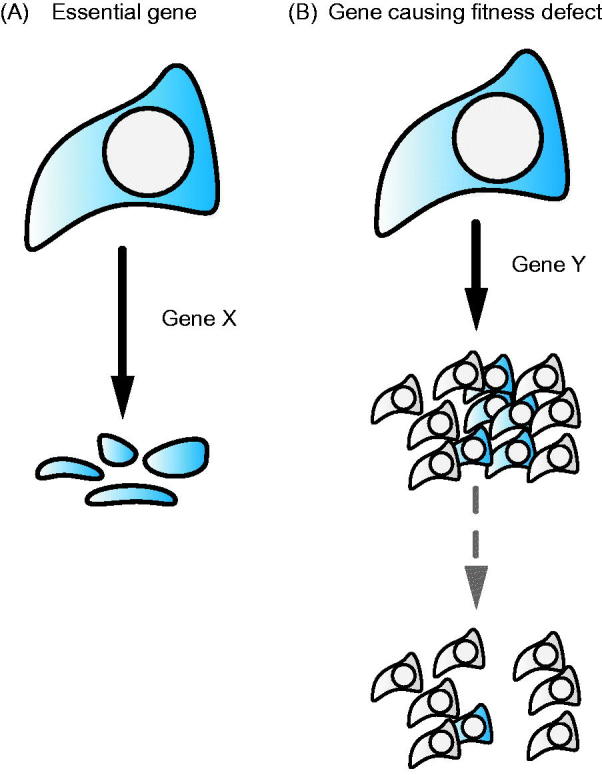
Model of essential genes and genes causing fitness defects. Loss-of-function of an essential gene X leads to cell death (A). In contrast, loss of a gene Y that is associated with a fitness defect leads to the gradual disappearance of the affected individuals from the population (B).
Discovery of gene essentiality in single cell organisms
Saccharomyces cerevisiae, or budding yeast, was one of the first eukaryotic organisms in which essential genes were studied by a systematic approach and on a genome-wide scale (Winzeler et al., 1999). Saccharomyces cerevisiae genes have many orthologs in common with other eukaryotes and the high rate of homologous recombination in S. cerevisiae enables its rapid genetic modification (Baudin et al., 1993). Thus, it has been a favorable model for studying gene function on a larger scale. In a first set of experiments by Winzeler et al. (1999), deletion strains for 2026 ORFs were created, of which 17% were found to be essential for growth and survival in rich medium. Using a competitive growth assay with a pool of homozygous deletions strains of non-essential genes, the authors additionally showed that 40% of the strains have fitness defects. The second large-scale deletion screen in S. cerevisiae already included 5916 genes (96% of all annotated ORFs), of which 18.7% turned out to be essential for growth in rich medium (Giaever et al., 2002). It was also observed that essential genes have more homologs in other organisms than their non-essential counterparts and that only 1% of essential genes had duplicates in the genome, as opposed to 8.5% of non-essential genes. While Winzeler et al. assessed phenotypes under two nutritional conditions (rich and low nutrients), Giaever et al. used five different conditions to demonstrate that gene essentiality and fitness both vary depending on the given environment. This observation was supported by another study in which a collection of ∼11 000 homo- or heterozygous deletion strains were tested against 726 different drugs or environmental stresses (Hillenmeyer et al., 2008). The authors observed that 97% of all mutants exhibited growth defects under at least one condition, and therefore suggest that nearly all genes are required under a specific environmental condition.
A main challenge for analyzing the function of essential genes in S. cerevisiae is the difficulty to generate hypomorphic mutants. Several methods have been developed to address this issue: essential genes can be shut off by inducible transcriptional repression (Mnaimneh et al., 2004), by heterozygous deletion (Deutschbauer et al., 2005) or by mRNA perturbation (DamP) (Breslow et al., 2008).
Recently, a genome scale collection of deletion mutations was generated for S. pombe (Kim et al., 2010). Saccharomyces cervisiae and S. pombe are distantly related and differ in many cellular functions (Wood et al., 2002), thus allowing for comparison and identification of genetic functions that are common to eukaryotes in general. In S. pombe, 4836 genes could be deleted, corresponding to 98.4% of all ORFs. Of those, 1260 or 26.1% were found to be essential. Similar to budding yeast, the proportion of single copy genes or singletons was higher among essential than non-essential genes. Gene sets of essential genes in both organisms were enriched for specific cellular processes (synthesis of DNA, RNA, lipids and proteins, transcriptional initiation, ribosome assembly).
Studies of essential genes in multicellular organisms
Compared to S. cerevisiae, the comprehensive study of essential genes in multicellular eukaryotic organism presents a greater technical challenge. Historically, most efforts to obtain genotype–phenotype interactions in multicellular organisms relied on forward genetic screening strategies using chemical or insertional mutagenesis. A major advance in functional genomics was introduced by the complete genome sequencing of model organisms and the development of RNAi technologies. The combination of both enabled the targeted knockdown of genes, allowing for reverse arrayed screens. Here, we describe how gene essentiality was explored in the three common model organisms Caenorhabditis elegans, Drosophila melanogaster and Danio rerio.
In C. elegans, the first screens that aimed at determining the number of essential genes relied on chemical mutagenesis with ethyl methanesulfonate (EMS) (Clark et al., 1988; Johnsen & Baillie, 1991). Based on the analysis of mutants in specific chromosomal regions including LGV(left) and unc-22 region, the authors estimated that the total number of essential genes in the C. elegans genome should range between 2850 and 3500 (Clark et al., 1988; Johnsen & Baillie, 1991). The first studies using RNAi in multicellular organisms were performed in C. elegans, by feeding animals with bacteria containing double stranded RNA or soaking animals in RNAi solution to achieve knockdowns (Fraser et al., 2000; Gönczy et al., 2000) (Figure 2A). Both screens started with the knockdown of genes on single chromosomes and steadily increased genomic coverage to genome scale (Kamath et al., 2003; Maeda et al., 2001; Sönnichsen et al., 2005). Roughly 800 genes were found to be critical for early embryonic development under laboratory conditions (Sönnichsen et al., 2005), which is only 4% of the C. elegans genome. The total number of essential genes was estimated to be around 1750 (Kamath et al., 2003). However, later studies indicate that the vast majority of non-essential genes show a measurable degree of fitness defect if measured over several generations, indicating that the number of essential genes might be underestimated (Ramani et al., 2012).
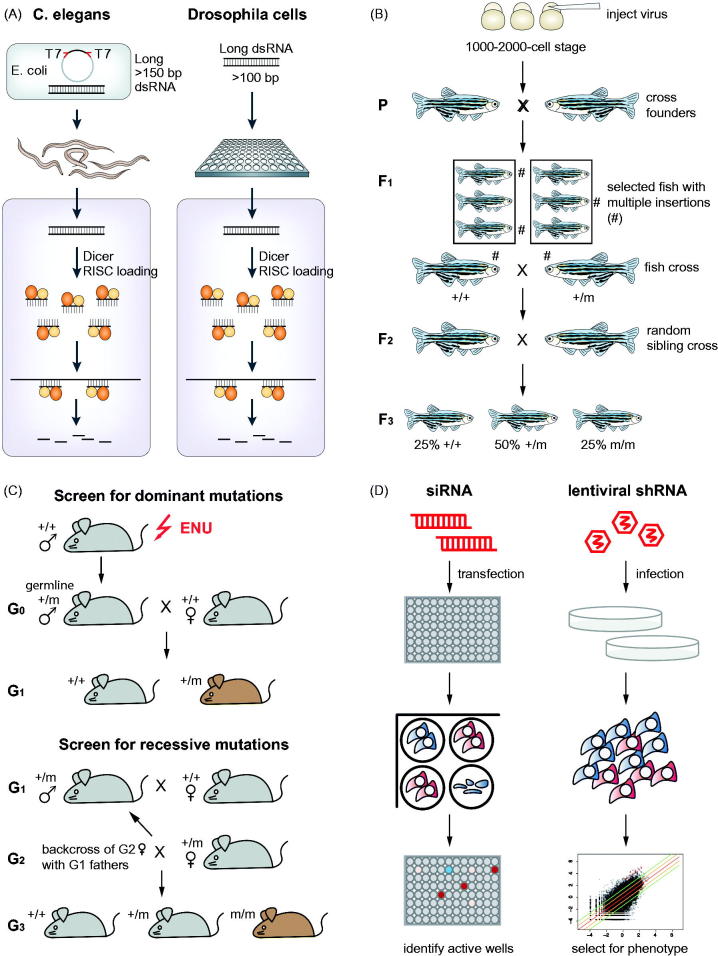
Figure 2.
Screening strategies in different model organisms. (A) Schematic overview of RNAi screening approaches in C. elegans by ingestion of E.coli and in Drosophila cells by bathing (modified from Boutros & Ahringer (2008)). Long double-stranded RNAs are introduced into the respective organisms and diced intracellularly into small-interfering RNAs (siRNAs). This results in many different siRNAs targeting a single transcript. (B) Outline of insertional mutagenesis screen in zebrafish (adapted by permission from Macmillan Publishers Ltd.: Nature Reviews Genetics (Patton & Zon, 2001), (c) 2002). Embryos are injected with a retrovirus at a 1000–2000-cell stage. These embryos are raised (= founder generation P), mated and the mutations transmitted to the F1 generation. Individual fish from the F1 generation with multiple insertions are selected and further crossed with each other to generate the F2 generation. Siblings of each F2 family are crossed with each other to generate homozygous mutations. (C) Overview of screening approaches using ENU induced mutagenesis in mouse. For dominant mutations, ENU mutagenized males carrying mutations in their germ lines are crossed with wild-type females (G0). Dominant mutations will be detected in the G1 generation. For recessive mutations, males of the G1 generation carrying mutations are crossed with wild type females. Then females of the resulting G2 generation are backcrossed with G1 males, thereby generating the G3 generation that potentially carries individuals with homozygous mutations. (D) Arrayed versus pooled loss-of-function screens in cancer cell lines. siRNAs are used for arrayed screens in a multi-well plates. Each well harbors a distinct gene knockdown. Candidate genes are detected by measuring signal levels (e.g. luminescence) of individual wells. In contrast, pooled loss-of-function screens rely on viral infection of cells with shRNA vectors. Each vector contains a barcode allowing identification of the specific shRNA. Pools of cells with different gene knockdowns are generated and cultured for several doubling times. Depletion of cells with specific gene knockdowns is detected by sequencing of barcodes and measuring their relative abundance at different time points.
The use of transposable elements for insertional mutagenesis has been a major tool for studying genotype–phenotype interactions in D. melanogaster. Using P-element transposons, essential genes were identified by screening mutants on individual chromosomes (Bourbon et al., 2002; Deak et al., 1997; Oh et al., 2003; Peter et al., 2002). The number of identified essential genes ranged from 130 to 850. The effort to generate and classify P-element insertions in every gene is systematically conducted by the Berkeley Drosophila Gene Project. However, achieving genomic saturation with P-elements is difficult and so far, only 40% of all drosophila genes have been successfully disrupted (Bellen et al., 2004). The percentage of lethal genes found by the Berkeley Drosophila Gene Project ranged between 8 and 16.3%, depending on the study included (Bellen et al., 2004). The first genome-scale knockdown screen with dsRNA in D. melanogaster was performed in cultured blood cells and identified >400 genes that show a strong reduction of viability upon knockdown, many of which lacked mutant alleles (Boutros et al., 2004) (Figure 2A). In 2007, a genome-wide transgenic RNAi fly library was published and found that roughly 30% of the fly lines showed a lethal phenotype (Dietzl et al., 2007).
In D. rerio, chemical mutagenesis with N-ethyl-N-nitrosourea (ENU) is an effective tool to introduce germline point mutations. Hence, several studies used ENU to generate large collections of mutants (Driever et al., 1996; Haffter et al., 1996). However, major general drawbacks of this method included laborious positional cloning to retrieve the underlying mutation and low genomic saturation that can be achieved (Justice, 1999). In the most extensive ENU screen in D. rerio, mutants could be assigned to 375 genes, covering only a fraction of the zebrafish genome (Haffter et al., 1996). Based on the results, the authors estimated that the percentage of lethal genes is roughly 2400, which is approximately 10% of the complete genome. However, only mutants with specific organ dysfunctions were selected for genotyping while mutants with multiple, non-viable malformations were not considered. Thus, the number of essential genes is most likely underestimated. Another forward genetic screening approach in D. rerio relied on the use of insertional mutagenesis (Patton & Zon, 2001) (Figure 2B). Two large-scale retroviral insertion screens were conducted in zebrafish, but only few essential genes could be retrieved (Gaiano et al., 1996; Golling et al., 2002). An insertional screen for embryonic and early larval development identified 315 essential genes, but only achieved a genomic saturation of 25% (Amsterdam et al., 2004). Based on these numbers, the authors estimated that ∼1,400 genes would be essential for embryonic development (Amsterdam et al., 2004). Of the genes identified, a high proportion had homologs in yeast (72%) and human (99%), indicating that essential genes are phylogenetically conserved.
Essential genes in mouse
The mouse is the best studied mammalian model organism and identification of essential genes is of particular interest due to its close phylogenetic relationship to humans. Chemical mutagenesis with ENU has been the predominant screening tool to generate mouse mutants with novel phenotypes. ENU is a very powerful mutagen and predominantly creates single base mutations with the highest mutation rate in male spermatogonial stem cells (Balling, 2001; Russell et al., 1979). Chemical mutagenesis with ENU can cause both loss and gain of function mutations, and specific crossing strategies are required to obtain the desired mutations (Figure 2C). One of the first efforts to identify lethal genes in mouse used ENU to generate mutants and back-crossings to identify affected genomic loci, but without recovering the precise point mutation (Rinchik & Carpenter, 1993). A major drawback of ENU-based screens has been the laborious positional cloning necessary to identify underlying point mutations, which limited the rate by which novel mutants could be genotyped. In spite of this drawback, several large-scale ENU screens were initiated in the past to systematically generate, characterize and genotype novel mouse mutants (Hrabé de Angelis et al., 2000; Nolan et al., 2000a,b). While a large panel of phenotypic traits was documented for every mutant including fertility, other phenotypes of essential genes such as embryonic lethality were missed. Thus, only few essential genes were found in ENU screens. With the development of technologies for targeted gene disruption, many essential genes were identified by studying single gene functions in murine knockout models (Matsui et al., 1996; Varfolomeev et al., 1998).
Since the establishment of homologous recombination in embryonic stem cells as a tool for targeted gene deletion (Thomas & Capecchi, 1987), this technology has been further developed to enable generation of knockout cells on a larger scale (Skarnes et al., 2011). Consequently, large efforts aiming at systematically generating knockout mouse models for every gene were started (Bradley et al., 2012; White et al., 2013). A subset of mice mutants with knockouts of 472 secreted proteins have been screened for specific phenotypes, and 8% of those showed pre-weaning lethality (Tang et al., 2010). So far, roughly 3000 genes were identified to be essential upon knockout, which accounts for ∼13% of the murine genome (Georgi et al., 2013).
Bioinformatic resources
With the wealth of data available from both large loss-of-functions screens and genome sequencing projects, web-based depositories for genotype–phenotype interactions have been developed. The Online Essential Gene Database OGEE integrates results from large-scale screens from 16 prokaryotic and 8 eukaryotic organisms (Chen et al., 2012). The database offers annotations to each essential protein-coding gene, including corresponding expression profile, duplication status or involvement in embryonic development. Another repository is the Database of Essential Genes (DEG), which since its first publication in 2004 has been updated several times (Luo et al., 2013). The most recent release, DEG 11, includes essential genomic elements beyond protein-coding genes, such as promotors or non-coding RNAs. For common model organism such as M. musculus, C. elegans or D. melanogaster, community databases exist that systematically collect available phenotypes for every gene of the respective organisms (Blake et al., 2014; Dos Santos et al., 2015; Harris et al., 2014) and are therefore a useful resource for detailed information on essential genes. The growing amount of data also enables comparative genomics approaches to explore common features of essential genes across different species (Figure 3). For example, the propensity of genes to be lost in evolution was studied for a set of eukaryotic organisms (Krylov et al., 2003). Essential genes were found to be associated with a low propensity to be lost during evolution, to accumulate fewer substitutions in the protein sequence and to be highly expressed. Furthermore, essential/lethal genes are found to be highly connected in protein networks (Jeong et al., 2001) and have a high degree of phylogenetic retention (Gustafson et al., 2006). In contrast, non-essential genes were more frequently targeted by many transcriptional factors, indicating that they are more dynamically regulated (Yu et al., 2004). Based on functional data from yeast, essential genes were initially thought to be predominantly singletons. However, later studies could show that both singletons and duplicates are equally represented among essential genes in other organisms (Liang & Li, 2007; Liao & Zhang, 2007). Comparative genomics also allows dissecting characteristics of potentially essential genes in humans, which are not directly amenable to experimental studies. Analysis of the evolutionary and population genetics property of 2472 human orthologs of murine essential genes showed that they have less variants and are more frequently haploinsufficient (Georgi et al., 2013). Whether human orthologs of murine essential genes are more frequently associated with human diseases is under debate, as some studies found associations (Dickerson et al., 2011; Georgi et al., 2013) while others did not (Park et al., 2008). In summary, comparative genomic approaches help to understand the global structural features of essential genes and thus allow sequence-based prediction of essential genes in organisms without data from functional genomics experiments.
Figure 3.
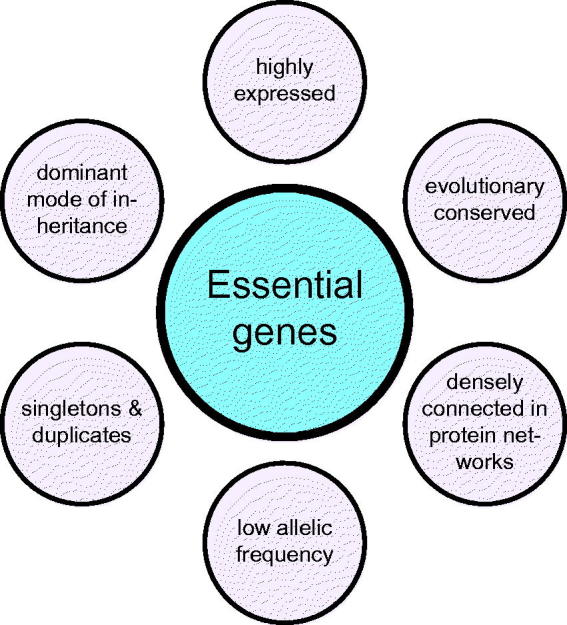
General characteristics of essential genes. General characteristics of essential genes found across different species by comparative genomics are presented.
Genetic interactions in model organisms
To systematically study the functions and interaction partners of genes on a larger scale, synthetic genetic arrays have been developed in S. cerevisiae as a powerful tool (Boone et al., 2007). In these experiments, every gene from a panel of query genes is deleted in combination with a gene from a second panel, resulting in large set of mutants with the loss of two genes. The growth behavior of all mutants is then individually measured and compared to each other. Three possible outcomes can result from a combinatorial loss-of-function: if the observed phenotype is the same as the single knockout, no genetic interaction is assumed. If the knockout of one gene can compensate for the loss of the other gene, then a positive or alleviating genetic interaction is present. If the knockout of one gene aggravates the phenotype caused by the loss of the second gene, a negative genetic interaction is found. If two genes share the same genetic interaction pattern across a large panel of genes, it can be assumed that they are functionally related.
Synthetic genetic arrays were first performed with deletion strains of non-essential genes and could uncover several novel genetic interactions (Tong et al., 2001,2004). Based on these results, large-scale screens were conducted in S. cerevisiae mutants that harbor a conditional repressed essential gene and the loss of a query gene (Davierwala et al., 2005). The effect of the genetic interaction between 575 essential genes and 30 query genes on growth behavior of mutants was analyzed. Similar to non-essential genes, essential genes also tend to share similar interaction partners if they are functionally related. However, the density of genetic interaction, i.e. the number of interaction partners was five times higher in essential genes, underlining that they are central hubs within the cellular network. Furthermore, the function of previously unknown essential genes could be assigned due to similarity of interaction partners, e.g. PGA1, which is required for specific functions of the endoplasmic reticulum.
Synthetic genetic arrays were also performed in cultured drosophila cells using combinatorial knockdown with siRNA and image-based analysis of morphological features of cells (Fischer et al., 2015; Horn et al., 2011). In the publication by Horn et al., 93 genes involved in MAPK, JNK and p38 signaling were knocked down in pairwise combinations, resulting in positive and negative genetic interactions. For example, single knockdown of Ras85D resulted in a reduced cell number, indicating that this gene is essential. However, the effect of Ras85D knockout could be compensated by the knockdown of a second gene, CG13197 (Horn et al., 2011). The same approach was performed on a larger scale by Fischer et al., by combined knockdown of a panel of 1367 genes involved in key cellular process (chromatin biology, cell cycle regulation, protein homeostasis) against a panel of 72 query genes. By grouping genes according to the pattern of their genetic interaction, genes could be assigned to known functional groups and directionality of genetic interaction could be inferred. Using this approach, novel links between the ERK signaling and chromatin remodeling could be discovered.
Essential genes in cancer cells
The identification of essential genes in tumor cells is of outstanding interest in cancer research, as they present potential targets for novel therapeutic interventions. The first evidence that cancer cells may depend on specific mutated genes for proliferation and survival was shown in Kras mutant colorectal cancer cells (Shirasawa et al., 1993). It was shown that the targeted deletion of mutant Kras resulted in a significant growth defect of cancer cells in nude mice. This observation was generalized under the concept of oncogene addiction (Weinstein & Joe, 2008), which proposes that cancer can become dependent on specific mutated genes (oncogenes). These oncogenes then take over an essential role within a specific pathway that is not found in normal cells.
Thus, in pursuit of those conditionally essential genes, many loss-of-function screens have been performed. The two main screening strategies that are used to identify candidate genes in cancer cells are arrayed screens using siRNA and pooled screens using shRNA (Figure 2D). A pilot study tested the effect of siRNA-mediated knockdown of 21 genes in transformed and non-transformed mammalian cells, measuring cell viability as outcome (Harborth et al., 2001). While the chosen siRNA library was small, the study highlighted that screening with siRNAs was feasible in mammalian cell lines. Three years later, a first large-scale RNAi screen using 5305 siRNAs was performed to identify genes that regulate cell division in HeLa cells (Kittler et al., 2004). Although not primarily focusing on essential genes, this study showed that knockdown of previously known essential genes such as ribosomal proteins or proteasome core units result in a lethal phenotype.
One of the first shRNA screens was performed in diffuse B-cell lymphoma using a retroviral library targeting 2500 genes, uncovering an essential role of NFkappaB pathway members for cell survival (Ngo et al., 2006).Two years later, several screens that used large pools of shRNAs to identify essential genes in cancer cells were published. The screen by Silva et al. used shRNA pools of different scales to knockdown genes in five breast cancer cell lines (Silva et al., 2008). Among the identified essential genes were several cell cycle regulators and components of the protein translation machinery. The sensitivity of cells towards knockdown of essential genes varied between cell lines and this observation could be confirmed using drugs with the same targets. Similar findings were obtained from another study that used 8204 shRNAs targeting 2924 genes in four cell lines (Schlabach et al., 2008). The number of depleted genes varied significantly between cell lines, from as low as 2.5% of the gene panel to 23.8%. Another shRNA screen used the TRC library developed by the RNAi consortium (Moffat et al., 2006), which contains 170 000 shRNAs targeting 17 200 human genes, to screen for essential genes in 12 cancer cell lines (Luo et al., 2008). Two sets of essential genes could be discriminated: one set of global essential genes that was found in all cell lines and enriched for cellular processes including mRNA processing, translation and proteasomal degradation. In addition, another set of essential genes was identified that was specific to selected cell lines and often included oncogenes. One example of such a lineage-specific essential gene is IRF4, which was identified to be essential in multiple myeloma (Shaffer et al., 2008). IRF4 is not genetically altered itself, indicating that essential genes do not necessarily correlate with mutation status in cancer. Another example is Brd4, which was found to be essential in a genetically defined model of acute myeloid leukemia and is a regulator of Myc (Zuber et al., 2011).
Due to the promising findings of the first screens, larger efforts were conducted to identify global and lineage-specific essential genes across larger sets of cell lines. The largest of such efforts to date, termed Project Achilles, assessed gene essentiality for 11 000 genes across 216 human cancer cell lines by pooled lentiviral shRNA screens (Cowley et al., 2014). In ovarian cancer, 54 lineage-specific essential genes were identified (Cheung et al., 2011), among which PAX8 was further validated and found to be also amplified in ovarian cancer. Another large-scale screen used pooled shRNAs to target 16 000 genes, but focused on a large panel of cell lines for a few selected tumor entities (Marcotte et al., 2012). A total of 72 different breast, pancreatic and ovarian cancer cell lines were screened and a large set of essential genes was detected which overlaps with previous shRNA screens, indicating that the method is robust (Koh et al., 2012). In total, a core set of 291 genes were discovered to be essential across many cell lines (Hart et al., 2014).
Synthetic lethality in cancer
A concept of genetic interaction that has major implication for cancer therapy is synthetic lethality, with the potential to target the selected loss of tumor suppressor genes or addiction to oncogenes in cancer cells (Chan & Giaccia, 2011; Kaelin, 2005). Briefly, synthetic lethality occurs in cells that survive with an altered gene function in either gene A or gene B, but does not survive if the function of gene A and B are both altered (Figure 4). Within the framework of gene essentiality, synthetic lethality can also be understood as an essential functional relationship between two genes.
Figure 4.
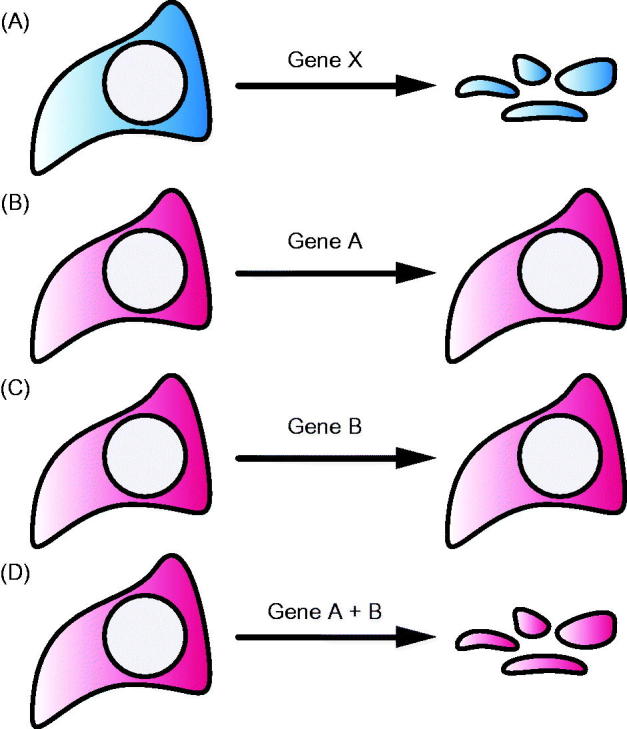
Concept of synthetic lethality. Loss-of-function of the essential gene x leads to immediate cell death (A), while loss-of-function of either gene A or gene B does not have a phenotypic effect. In contrast, combined loss-of-function of gene A and B results in a synthetic lethal interaction.
Since most oncogenes or tumor suppressor genes are not directly amenable to pharmacological therapy, there is an urge to identify genes that become essential due to their functional interaction with oncogenes or tumor suppressors (Garber, 2002; Kaelin, 2005). In addition, genes that gain essentiality when specific cellular pathways are blocked by anticancer drugs are also of particular interest for combinatorial drug treatment. Knowing those genes would considerably enlarge the repertoire of cancer therapy and allow more selective killing of cancer cells.
A first proof-of-principle experiment in a mammalian cell model used a hypoxanthine–guanine phosphoribosyl transferase (HPRT1) deficient cell line that expresses HPRT1 and a GFP reporter on an episomal plasmid (Simons et al., 2001). HPRT1 is non-essential under normal conditions and the episomal plasmid is consequently lost. However, when the biosynthetic pathway leading to guanine monophosphate production is perturbed by specific inhibitors, HPRT1 becomes essential and only cells that were able to retain the expression plasmid survived.
Subsequently, several studies exploited the concept of synthetic lethality to identify genes with chemosensitizing potential. For example, an arrayed siRNA screen was performed by Whitehurst et al. with the small lung cancer cell line NCI-H1155, measuring viability of gene knockdowns in the presence of sub-lethal concentrations of paclitaxel (Whitehurst et al., 2007). The authors identified 87 genes that render the lung cancer cells sensitive to treatment with this microtubule inhibitor. In another arrayed siRNA screen, breast cancer cells were treated with a PARP-inhibitor and a number of kinases were identified to act synthetically lethal with the inhibitor (Turner et al., 2008). Using the same approach, synthetic lethal interactions were found for a multiplicity of drugs, including inhibitors of PLK1 (Liu-Sullivan et al., 2011; van der Meer et al., 2014), DNA-PK (Dietlein et al., 2014), ATR (Mohni et al., 2014) or EGFR (Astsaturov et al., 2010). A different approach making use of synthetic lethality aims at identifying genes that are essential in a specific genetic background, i.e. the presence of a gain or loss of function mutation. Targeting a panel of genes, either with RNAi or selective drugs, would reveal candidates that act synthetically lethal with the respective mutation. One of the first studies based on this concept screened 23 550 compounds to identify drugs that selectively kill cells transformed by different combinations of oncogenes, but not their isogenic non-transformed counterparts (Dolma et al., 2003). The authors found that specific combinations of oncogenes increased topoisomerase expression, rendering cells sensitive to topoisomerase inhibitors. A further early study aimed at identifying kinases that are required by clear renal cancers lacking the von Hippel-Lindau tumor suppressor (Bommi-Reddy et al., 2008). A small shRNA screen was performed and revealed several kinases that act synthetically lethal, of which some could be confirmed using drugs.
Later, several genome-scale RNAi screens have been performed using isogenic cell lines, which either have a RAS mutation (Schlabach et al., 2008) or loss of TP53 (Krastev et al., 2011) background. In another screen, Vizeacoumar et al. used genome-scale shRNA libraries to systematically identify negative genetic interactions across five isogenic cell lines with loss of function of major tumor suppressors (Vizeacoumar et al., 2013). An alternative approach is to compare genetic interactions in multiple cell lines harboring the desired mutational background to a panel of cells without this background. Two hallmark papers used this approach to identify synthetic lethal interactions with mutated KRAS using a large set of cell lines (Barbie et al., 2009; Scholl et al., 2009), hereby identifying TBK1 and STK33 as negative interactors. TBK1 activates anti-apoptotic signals via NfKappaB (Barbie et al., 2009), while STK33 suppresses mitochondrial apoptosis through S6K1 and BAD (Scholl et al., 2009). However, different results were obtained with pharmacological inhibitors, indicating that the interaction might be independent of the kinase activity of STK33 (Babij et al., 2011; Luo et al., 2012; Weïwer et al., 2012).
With the wealth of data from functional screens and mutation data from sequencing projects at hands, integrated approaches to identify synthetic lethal interactions were also possible. For example, RNAi profiles of cancer cells were compared to genome-wide copy number abberations, hereby identifying 56 genes for which reduction in growth only occurred if cells also harbored a copy number loss of the respective gene (Nijhawan et al., 2012). Another study combined results from a Wnt pathway activity readout with data on lethal phenotypes identified by pooled shRNA screens for a set of 85 cell lines (Rosenbluh et al., 2012). The authors found out that cancer with high active Wnt levels rely on YAP1 that forms a complex with TBX5 and mediates expression of anti-apoptotic genes BCL2L1 and BIRC5.
Although there is a constantly growing amount of data from structural and functional studies, only few identified genes have been exploited as drug targets in a clinical setting. Initial successes by targeting lineage specific essential genes (BCR-Abl fusion protein in chronic myeloid leukemia by imatinib mesylate (Kantarjian et al., 2002)) have raised high hopes for a more efficient cancer therapy. However, while the repertoire for targeted therapy is constantly enlarging and improving overall survival rates of many cancers, the initial success of BCR-Abl imatinib remains an exception rather than the rule. Potential reasons for this development include inherent limitations of cell lines as a cancer model (Wilding & Bodmer, 2014), tumor heterogeneity (Gillies et al., 2012) and the involvement of the tumor microenvironment (Straussman et al., 2012).
Clinical translation of synthetic lethality has been (in part) successful in two cases: PARP inhibitors in BRCA mutated breast cancer and the combination of retinoid acid and arsenic trioxide for treating promyelocytic leukemia. In 2005, two parallel publications described that in breast cancer cells with deficient BRCA1 and BRCA2, the Poly(ADP-ribose) polymerase PARP1 takes over an essential function in repairing DNA lesions (Bryant et al., 2005; Farmer et al., 2005). Pharmacological inhibition of PARP1 was highly effective in eradicating BRCA1 and BRCA2 deficient cancer cells. With a strong biological rationale behind, PARP inhibitors quickly went to phase I (Fong et al., 2009) and phase II (Audeh et al., 2010; Tutt et al., 2010) trials for treatment of BRCA1/2 mutated breast and ovarian cancer, with promising results regarding response rate and clinical benefit. Several consecutive trials, including phase 3 trials, have been started, but the results so far are mixed and recent findings suggest that the significance of BRCA germline mutations in determining therapy response needs to be reassessed (Scott et al., 2015).
The combination of all-trans retinoic acid (ATRA) and arsenic trioxide in treatment of promyelocytic leukemia (PML) is another example for clinical translation of synthetic lethality. The standard treatment of PML has been a combination of retinoic acid and anthracyclines, with overall high success rates. However, several studies show that ATRA and arsenic trioxide can strongly synergize to eradicate PML in in vitro and in vivo models (Lallemand-Breitenbach et al., 1999; Shao et al., 1998). Both compounds bind at different moieties of the PML-RARA fusion protein, thereby synergistically accelerating its degradation. A phase 3 clinical trial showed that combination of both substances is most likely superior to standard therapy and associated with less hematological toxicity, but higher hepatic toxicity (Lo-Coco et al., 2013). In summary, while systematic screens in cancer cell lines have yielded a wealth of data on essential genes and synthetic lethality, translating these findings into novel clinical therapy still remains a major challenge.
Novel methods for discovery of essential genes
RNAi has been the main workhorse for targeted identification of novel gene functions for almost a decade, with constant refinement of design and application. However, there are also limits and disadvantages inherent to RNAi technology. These include off-target effects (Ma et al., 2006), toxicity and incompleteness of generating knockdown for selected genes (Boutros & Ahringer, 2008). Knockout efficiency is also dependent on biological sources of variability, such as AGO2 expression levels (Hart et al., 2014). Additionally, RNAi exclusively targets the mRNA of transcribed genomic regions, creating only loss-of-function. This however precludes analysis of non-coding genomic regions and gain-of-function phenotypes.
Recently, the development of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 system of S. pyogenes into a genome editing tool (Cong et al., 2013; Mali et al., 2013) has opened new avenues for functional genomics. In brief, a synthetic small guide RNA targets the modified Cas9 endonuclease to a complementary sequence in the genome, where it introduces a double strand break. In most cases, these breaks are repaired by non-homologs end joining, which is error-prone and frequently results in loss-of-function deletion or insertions. CRISPR/Cas9 can be used in a wide range of species with high efficiency (Friedland et al., 2013; Gratz et al., 2013; Hwang et al., 2013), making it an universal tool.
The simplicity of the CRISPR/Cas9 technology allows high-throughput screening in a similar scale as pooled shRNAs. CRISPR/Cas9 screens were performed in cancer cells and murine embryonic stem cells to identify resistance mechanisms towards toxins or drugs (Koike-Yusa et al., 2014; Shalem et al., 2014; Zhou et al., 2014). In addition, screens have been conducted to identify essential genes in haploid and diploid leukemia cells (Wang et al., 2014) and colorectal cancer cells (Hart et al., 2015). Interestingly, identified hits between shRNA and CRISPR screens were partly non-overlapping, indicating that complete loss of function may result in different phenotypes (Hart et al., 2015; Shalem et al., 2014). In addition, the number of essential genes identified by a CRISPR/Cas9 screen was higher than by a shRNA screen in HCT116 (Hart et al., 2015,2014). It is thus anticipated that large-scale loss of function screens will be repeated using CRISPR/Cas9 technology. In addition to loss-of-function of protein coding genes, CRISPR/Cas9 also allows targeting and functional characterization of long non-coding RNAs and non-transcribed regions (Ho et al., 2015; Kearns et al., 2015; Yin et al., 2015). Moreover, modifications of the CRISPR/Cas system for transcriptional activation of genes will in the future enable identification of gene essentiality and genetic interaction with gain-of-function CRISPR/Cas9 libraries (Gilbert et al., 2014; Konermann et al., 2014). CRISPR/Cas9 also enables reconstruction of point mutations frequently found in cancer (Antal et al., 2015), thereby allowing screening for synthetic lethality in very specific genetic backgrounds. In face of all those possibilities opened up by novel screening methods, technical standardizations of screening procedures and bioinformatics analysis pipelines are essential to obtain comparable results across different screens.
Acknowledgements
We thank Marco Breinig and Christian Scheeder for careful reading of the manuscript and critical comments.
Declaration of interest
Work in the lab of M.B. is in part supported by an ERC Advanced Grant. T.Z. is supported by a fellowship from the DKFZ International Postdoc program.
References
- Adams MD. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal CE, Hudson AM, Kang E, et al. Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell. 2015;160:489–502. doi: 10.1016/j.cell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astsaturov I, Ratushny V, Sukhanova A, et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal. 2010;3:ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet (London, England) 2010;376:245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Babij C, Zhang Y, Kurzeja RJ, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer. Res. 2011;71:5818–26. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- Balling R. ENU mutagenesis: analyzing gene function in mice. Annu Rev Genomics Hum Genet. 2001;2:463–92. doi: 10.1146/annurev.genom.2.1.463. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, et al. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–30. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–81. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Eppig JT, et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–17. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommi-Reddy A, Almeciga I, Sawyer J, et al. Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in. A pilot synthetic lethal screen . Proc Natl Acad Sci USA. 2008;105:16484–9. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–49. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Bourbon HM, Gonzy-Treboul G, Peronnet F, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–66. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–5. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–6. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 2008;5:711–18. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–17. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov. 2011;10:351–64. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Minguez P, Lercher MJ, Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40:D901–6. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108:12372–7. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DV, Rogalski TM, Donati LM, Baillie DL. The unc-22(IV) region of Caenorhabditis elegans: genetic analysis of lethal mutations. Genetics. 1988;119:345–53. doi: 10.1093/genetics/119.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley GS, Weir BA, Vazquez F, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davierwala AP, Haynes J, Li Z, et al. The synthetic genetic interaction spectrum of essential genes. Nat Genet. 2005;37:1147–52. doi: 10.1038/ng1640. [DOI] [PubMed] [Google Scholar]
- de Berardinis V, Vallenet D, Castelli V, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak P, Omar MM, Saunders R, et al. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: correlation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics. 1997;147:1697–722. doi: 10.1093/genetics/147.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–25. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson JE, Zhu A, Robertson DL, Hentges KE. Defining the role of essential genes in human disease. PLoS One. 2011;6:e27368. doi: 10.1371/journal.pone.0027368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietlein F, Thelen L, Jokic M, et al. A functional cancer genomics screen identifies a druggable synthetic lethal interaction between MSH3 and PRKDC. Cancer Discov. 2014;4:592–605. doi: 10.1158/2159-8290.CD-13-0907. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–96. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- Dos Santos G, Schroeder AJ, Goodman JL, et al. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–7. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fischer B, Sandmann T, Horn T, et al. A map of directional genetic interactions in a metazoan cell. Elife. 2015;4 doi: 10.7554/eLife.05464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–30. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–3. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N, Amsterdam A, Kawakami K, et al. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–32. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, et al. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–7. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Garber K. Synthetic lethality: killing cancer with cancer. J Natl Cancer Inst. 2002;94:1666–8. doi: 10.1093/jnci/94.22.1666. [DOI] [PubMed] [Google Scholar]
- Gazit R, Gruda R, Elboim M, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–23. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- Georgi B, Voight BF, Bućan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–84. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12:487–93. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, et al. Essential genes of a minimal bacterium. Proc Natl Acad Sci USA. 2006;103:425–30. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluecksohn-Waelsch S. Lethal genes and analysis of differentiation. Science. 1963;142:1269–76. doi: 10.1126/science.142.3597.1269. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–40. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Echeverri C, Oegema K, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–6. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–35. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Snitkin ES, Parker SCJ, et al. Towards the identification of essential genes using targeted genome sequencing and comparative analysis. BMC Genomics. 2006;7:265. doi: 10.1186/1471-2164-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, et al. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–65. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–93. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, et al. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, et al. Systematic discovery and classification of human cell line essential genes, bioRxiv. Cold Spring Harbor Labs J. 2015 doi: 10.1101/015412. [DOI] [Google Scholar]
- Herrmann T, van der Hoeven F, Grone HJ, et al. Mice with targeted disruption of the fatty acid transport protein 4 (Fatp 4, Slc27a4) gene show features of lethal restrictive dermopathy. J Cell Biol. 2003;161:1105–15. doi: 10.1083/jcb.200207080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–5. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Marth GT, Quinlan AR, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–8. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- Ho TT, Zhou N, Huang J, et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015;43:e17. doi: 10.1093/nar/gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat Methods. 2011;8:341–6. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabé de Angelis MH, Flaswinkel H, Fuchs H, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–7. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–2. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Johnsen RC, Baillie DL. Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans. Genetics. 1991;129:735–52. doi: 10.1093/genetics/129.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Eberl L, Glass JI. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21:562–8. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Justice MJ. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8:1955–63. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–52. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–3. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–79. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–23. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–40. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–83. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh JLY, Brown KR, Sayad A, et al. COLT-Cancer: functional genetic screening resource for essential genes in human cancer cell lines. Nucleic Acids Res. 2012;40:D957–63. doi: 10.1093/nar/gkr959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, et al. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krastev DB, Slabicki M, Paszkowski-Rogacz M, et al. A systematic RNAi synthetic interaction screen reveals a link between p53 and snoRNP assembly. Nat Cell Biol. 2011;13:809–18. doi: 10.1038/ncb2264. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–35. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Guillemin MC, Janin A, et al. Retinoic acid and arsenic synergize to eradicate leukemic cells in a mouse model of acute promyelocytic leukemia. J Exp Med. 1999;189:1043–52. doi: 10.1084/jem.189.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz LS, Marx J, Chamulitrat W, et al. Adipocyte-specific inactivation of Acyl-CoA synthetase fatty acid transport protein 4 (Fatp4) in mice causes adipose hypertrophy and alterations in metabolism of complex lipids under high fat diet. J Biol Chem. 2011;286:35578–87. doi: 10.1074/jbc.M111.226530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–8. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–81. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Liu-Sullivan N, Zhang J, Bakleh A, et al. Pooled shRNA screen for sensitizers to inhibition of the mitotic regulator polo-like kinase (PLK1). Oncotarget. 2011;2:1254–64. doi: 10.18632/oncotarget.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco F, Avvisati G, Vignetti M, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369:111–21. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Lin Y, Gao F, et al. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2013;42:D574–80. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Masson K, Jaffe JD, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc Natl Acad Sci USA. 2012;109:2860–5. doi: 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–63. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–6. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Brown KR, Suarez F, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–89. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Oshima M, Oshima H, et al. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev Biol. 1996;178:179–85. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–10. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Zack TI, Ren Y, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–54. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet. 2000a;25:440–3. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Vizor L, et al. Implementation of a large-scale ENU mutagenesis program: towards increasing the mouse mutant resource. Mamm Genome. 2000b;11:500–6. doi: 10.1007/s003350010096. [DOI] [PubMed] [Google Scholar]
- Oh SW, Kingsley T, Shin H, et al. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics. 2003;163:195–201. doi: 10.1093/genetics/163.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–61. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- Park D, Park J, Park SG, et al. Analysis of human disease genes in the context of gene essentiality. Genomics. 2008;92:414–18. doi: 10.1016/j.ygeno.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2:956–66. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Peter A, Schöttler P, Werner M, et al. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 2002;3:34–8. doi: 10.1093/embo-reports/kvf012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Chuluunbaatar T, Verster AJ, et al. The majority of animal genes are required for wild-type fitness. Cell. 2012;148:792–802. doi: 10.1016/j.cell.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Rinchik EM, Carpenter DA. N-ethyl-N-nitrosourea-induced prenatally lethal mutations define at least two complementation groups within the embryonic ectoderm development (EED) locus in mouse Chromosome 7. Mamm Genome. 1993;4:349–53. doi: 10.1007/BF00360583. [DOI] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, et al. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–73. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WL, Kelly EM, Hunsicker PR, et al. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci USA. 1979;76:5818–19. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–4. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Fröhling S, Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-Ribose) polymerase inhibitors: recent advances and future development. J Clin Oncol. 2015;33:1397–406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NCT, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Fanelli M, Ferrara FF, et al. Arsenic trioxide as an inducer of apoptosis and loss of PML/RAR alpha protein in acute promyelocytic leukemia cells. J Natl Cancer Inst. 1998;90:124–33. doi: 10.1093/jnci/90.2.124. [DOI] [PubMed] [Google Scholar]
- Shim J, Moulson CL, Newberry EP, et al. Fatty acid transport protein 4 is dispensable for intestinal lipid absorption in mice. J Lipid Res. 2009;50:491–500. doi: 10.1194/jlr.M800400-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–8. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–20. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A, Dafni N, Dotan I, et al. Establishment of a chemical synthetic lethality screen in cultured human cells. Genome Res. 2001;11:266–73. doi: 10.1101/gr.154201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Li L, Tang J, et al. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010;28:749–55. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–12. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, Iorns E, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–77. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet (London, England) 2010;376:235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- van der Meer R, Song HY, Park SH, et al. RNAi screen identifies a synthetic lethal interaction between PIM1 overexpression and PLK1 inhibition. Clin Cancer Res. 2014;20:3211–21. doi: 10.1158/1078-0432.CCR-13-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev EE, Schuchmann M, Luria V, et al. Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–76. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vizeacoumar FJ, Arnold R, Vizeacoumar FS, et al. A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Mol Syst Biol. 2013;9:696. doi: 10.1038/msb.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–80; discussion 3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- Weïwer M, Spoonamore J, Wei J, et al. A potent and selective quinoxalinone-based STK33 inhibitor does not show synthetic lethality in KRAS-dependent cells. ACS Med Chem Lett. 2012;3:1034–8. doi: 10.1021/ml300246r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–64. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst AW, Bodemann BO, Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–19. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–84. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–6. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–80. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015;16:504–16. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Yu H, Greenbaum D, Xin Lu H, et al. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–31. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–91. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]