ABSTRACT
The lack of vaccine and limited antiviral options against respiratory syncytial virus (RSV) highlights the need for novel therapeutic strategies. One alternative is to develop drugs that target host factors required for viral replication. Several microarray and proteomics studies had been published to identify possible host factors that are affected during RSV replication. In order to obtain a comprehensive understanding of RSV-host interaction, we integrated available proteome and transcriptome datasets and used it to construct a virus-host interaction network. Then, we interrogated the network to identify host factors that are targeted by the virus and we searched for drugs from the DrugBank database that interact with these host factors, which may have potential applications in repositioning for future treatment options of RSV infection.
KEYWORDS: Respiratory syncytial virus, human, Virus-host interactions, Microarray analysis, Proteomics, Protein-protein interaction network, Drug-drug target interaction network
Introduction
Respiratory syncytial virus (RSV) is classified in the Pneumovirus genus of the Paramyxoviridae family, which is an enveloped virus with a negative-sense single-stranded RNA genome of 15.2 kb in length that encodes for 10 subgenomic mRNAs and 11 proteins [1]. RSV is the leading cause of acute lower respiratory tract infection, which affects 33.8 million children younger than 5 years of age and responsible for 4 million hospital admissions and 200,000 deaths worldwide [2]. While most cases of RSV infections cause self-limited illness, about 3.4 million children worldwide develop severe symptoms including pneumonia or bronchiolitis and 99% of deaths occur in developing countries [3]. To date, there is no vaccine safe and effective against RSV, and the antiviral option available is limited. Currently, ribavirin is licensed for the treatment of RSV infection, which is a small molecule drug that acts as a nucleoside analog [1]. However, ribavirin is not recommended for the routine management of the disease due to its issues with delivery and safety [4]. Another option is palivizumab, which is a monoclonal antibody licensed for use as a prophylactic drug that targets the viral fusion (F) glycoprotein [5]. But it is recommended for high-risk individuals [1] and the price is prohibitive especially in developing countries. Several vaccine candidates are currently under clinical trials [6], but none of them are licensed. The limited option for RSV treatment and control underlines the need to find novel classes of drugs to minimize the global burden of RSV.
An alternative method in developing new drugs for viral infections is by identifying drugs that target host cellular factors needed for virus replication [7]. Due to its limited coding capacity, viruses must depend on host cellular factors to complete their replication cycle. In addition, viruses must escape from the host defense system in order to succeed in replication. Shedding light on the virus–host interaction allows the identification of host cellular networks that are utilized by the virus. These host factors required for viral replication may provide potential drug targets for RSV treatment. Several studies used microarray and proteomic methods to identify host factors required for RSV replication [8–23]. Excellent reviews on these host factors and their role in RSV disease and pathogenesis had been published [24,25]. Host factors for influenza virus [26–34], dengue virus [35,36], and HIV [37–39] that were identified by high-throughput transcriptomic and proteomic approaches have increased our understanding of the molecular mechanism of viral replication. An integrative proteome and transcriptome analysis on the RSV–host interaction will not only provide a comprehensive overview of these host factors but also suggest novel alternatives of these host factors as drug targets. An advantage of developing host factors as drug targets is the lower possibility of emergence of drug-resistant strains [7].
In order to reconstruct the RSV–host interaction network, this study analyzed the datasets of microarray and proteomics studies on RSV infection. Here, the overlap of host factors identified by microarray and proteomic methods was compared at both the levels of gene and protein identity and biological process. The virus–host interaction network was generated by integrating the transcriptome and proteome datasets. The host factors affected by RSV replication were combined with the DrugBank dataset to reconstruct the drug–host factor network with the aim to identify host factors that are targeted by US FDA-approved molecules which could be repositioned for RSV infection treatment.
Methods
Acquisition of microarray and proteomics datasets
Microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) [40]. Proteomics datasets were downloaded from the Proteomics Identifications database (PRIDE) (www.ebi.uk/pride/archive). List of host genes and proteins that were not deposited in the databases were obtained from the published papers (Table 1). The list was narrowed down by selecting host genes and proteins with two-fold and more change in abundance level and false discovery rate of less than 1% [41].
Table 1.
Summary of microarray and proteomic studies in identifying host factors affected during respiratory syncytial virus infection.
| Virus strain | Host system | Techniques | No. of host factors identified | Year | Reference |
|---|---|---|---|---|---|
| A2 | A549 | Microarray | 306 | 2001 | Zhang et al. [8] |
| Long | HeLa | Microarray | 380 | 2002 | Tian et al. [9] |
| A2 | BALB/c mice | Microarray | 411 | 2007 | Janssen et al. [10] |
| Long | A549 | Microarray | 377 | 2007 | Martinez et al. [11] |
| A2 | Various cell lines | Microarray | 184 | 2007 | Mayer et al. [12] |
| Circulating strain | Pediatric patients (whole blood) | Microarray | 395 | 2013 | Meijas et al. [13] |
| Circulating strain | Pediatric patients (PBMC) | Microarray | 293 | 2015 | Brand et al. [14] |
| A2 | A549 | 2DE; MALDI-TOF-MS | 19 | 2004 | Brasier et al. [15] |
| A2 | A549; Vero | 2D-DIGE; LC–MS/MS | 22 | 2012 | Hastie et al. [16] |
| A2 | A549 | 2D-DIGE; MALDI-TOF-MS | 34 | 2010 | Jamaluddin et al. [17] |
| A2 | A549 | 2D-DIGE; LC–MS/MS | 14 | 2010 | van Diepen et al. [18] |
| A2 | A549 (SILAC) | SDS-PAGE; LC–MS/MS | 431 | 2010 | Munday et al. [19] |
| Subgroup B | A549 (SILAC) | SDS-PAGE; LC–MS/MS | 112 | 2010 | Munday et al. [20] |
| A2 | HEp2 | In-solution IEF; LC–MS/MS | 69 | 2011 | Ternette et al. [21] |
| A2 | A549 | In-solution IEF; LC–MS/MS | 146 | 2014 | Dave et al. [22] |
| Recombinant vaccine (F,G) | BALB/c mice | SDS-PAGE; LC–MS/MS | 48 | 2015 | van Diepen et al. [23] |
PBMC: peripheral blood mononuclear cell; SILAC: stable isotope labeling with amino acids in cell culture; 2DE: two-dimensional gel electrophoresis; 2D-DIGE: two-dimensional differential gel electrophoresis; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; IEF: isoelectric focusing; MALDI-TOF-MS: matrix-assisted laser desorption ionization-time of flight-mass spectrometry; LC–MS/MS: liquid chromatography–tandem mass spectrometry
To merge and integrate the list of host factors, the gene probe identifiers or gene identifiers for microarray data and UniProt ID (www.uniprot.org) for proteomics data were converted to the unique official gene symbol curated by the human genome gene nomenclature committee (www.genenames.org).
Host factor overlap analysis
List of overlapping host factors identified by microarray and proteomics methods were determined and visualized using Venny v2.0, a venn diagram web resource (bioinfogp.cnb.csic.es/tools/venny/).
Gene ontology enrichment analysis
To assign enriched gene ontology (GO) terms of host factors, the DAVID Bioinformatics Resources v6.7 (https://david.ncifcrf.gov/home.jsp) was used to query the list of identified host factors that were common to two or more studies [42,43]. Biological processes that were identified with a confidence level of 95% were included in the analysis.
Generation of virus–host factor interaction network
To obtain a global host response during RSV infection, a virus–host interaction network was constructed by querying the list of host factors that are common to two or more studies to the human protein–protein interaction database (HIPPIE) (cbdm.mdc-berlin.de/tools/hippie/index.php) [44] and the virus–host interaction database, VirHostNet v2.0 (virhostnet.prabi.fr) [45]. Interactors of RSV proteins identified in recent studies are also included in the network [46–48]. In order to minimize false positives and to ensure meaningful interactions, proteins that are expressed only in blood and lung tissues were selected in the HIPPIE database [49]. The generated network was analyzed and visualized using Cytoscape software v3.2.1 (cytoscape.org) [50]. MCODE, a clustering algorithm-based application in Cytoscape, was used to identify a network hub, which contains highly connected proteins that may act as control points in the network (parameter: degree cutoff = 2, node score cutoff = 0.2, K-core = 2, max. depth = 100) [51]. BiNGO application of Cytoscape was used to annotate the biological processes of identified MCODE cluster [52].
Generation of drug–host factor network
The drug–host factor network was constructed by downloading the drug file from the DrugBank database v4.3 (www.drugbank.ca) [53] and matching the drug targets with host factors that are common to two or more studies. The drug–host factor network was analyzed and visualized using Cytoscape software [50]. A drug–host factor subnetwork was constructed by selecting only drugs that have interactions with host factors identified in MCODE clusters.
Results
Microarray studies for the identification of host factors during RSV infection
Several microarray studies had been performed to identify host responses in RSV infection both in vitro and in vivo (Table 1). Zhang et al. [8] and Martinez et al. [11] used laboratory-adapted strains, A2 and Long respectively, to determine the transcription profile of human lung epithelial cell line, A549 and identified several hundreds host genes with significant change in expression level in cells inoculated with RSV. Tian et al. used Long strain to infect a human cervical carcinoma cell line, HeLa, and identified 380 host genes that are affected during RSV replication [9]; Mayer et al. investigated the gene expression profile of several human bronchial epithelial cell lines and primary respiratory epithelial cells during infection with various viral and bacterial pathogens and found set of genes that are common to these pathogens as well as RSV-specific host genes (data accessible at National Center for Biotechnology Information (NCBI) GEO database: accession GSE6802) [12]. Janssen et al. used a mouse model to determine the transcription profiles of lungs and lymph nodes in BALB/c mice infected with A2 strain, which led to the identification of 411 differentially expressed host genes [10]. Mejias et al. (GEO accession GSE38900) [13] and Brand et al. (GEO accession GSE69606) [14] analyzed the transcriptional profiles of blood samples from children infected with RSV and identified host factors that are associated with disease severity.
Proteomics approaches for the identification of host factors during RSV infection
Proteomic studies in identifying host responses during RSV infection mostly used the A2 strain and the A549 cell line (Table 1). Brasier et al. employed two-dimensional gel electrophoresis (2DE) to separate host proteins and matrix-assisted laser desorption ionization–time of flight–mass spectrometry to determine the peptide sequence, which identified 19 proteins with significant changes in abundance level in virus-infected cells [15]. Jamaluddin et al. opted to use the two-dimensional differential gel electrophoresis (2D-DIGE) technique to separate the proteins and identified 34 significantly expressed proteins [17]. Van Diepen et al. [18] and Hastie et al. [16] used the same protein separation method, 2D-DIGE but utilized liquid chromatography–tandem mass spectrometry (LC–MS/MS) as the protein identification technique to detect 14 and 22 host proteins that are perturbed in RSV-infected cells. Munday et al. used stable isotope labeling with amino acids in cell culture (SILAC) reagent to label proteins from virus-infected and mock-infected cells, which allowed the identification of hundreds of cellular proteins in A2 strain-infected [19] and subgroup B strain-infected A549 cells (data accessible at PRIDE: accessions 13269, 13270) [20]. Both Ternette et al. [21] and Dave et al. [22] used a non-gel-based protein separation technique, in-solution isoelectric focusing (IEF), which identified 69 and 146 host proteins with significantly altered abundance level. Recently, van Diepen et al. used a mouse model and identified 48 host proteins that were significantly expressed during virus challenge in mice vaccinated with recombinant vaccinia virus displaying RSV fusion (F) and attachment (G) glycoproteins [23].
Integration of proteomics and transcriptomics to generate virus–host factor interaction network
The list of host factors that were affected during RSV infection was combined from seven microarray and nine proteomic studies. In order to merge the list, the microarray probe IDs, Entrez Gene name from NCBI, and UniProt accession numbers were converted to the unique official gene symbol. Using the OrthoDB (orthodb.org) [54], human orthologs of mouse genes were cross-referenced from the datasets of Janssen et al. [10] and van Diepen et al. [23] before converting to official gene symbol.
A total of 2853 host factors were obtained after combining the 2039 host genes identified from the seven microarray studies and 814 host proteins identified from the nine proteomic studies. Of these, 2676 were nonredundant in the list and 177 were common to both microarray and proteomics datasets (Figure 1). The overlaps were low among microarray studies with a maximum of 53 genes that are common between Tian and Martinez datasets (Table 2). Likewise, a low overlap was observed among proteomics studies with a maximum of 74 proteins common between the two proteomic studies of Munday et al., which investigated the host responses in A549 cells infected with subgroup A [19] and B [20] viruses.
Figure 1.
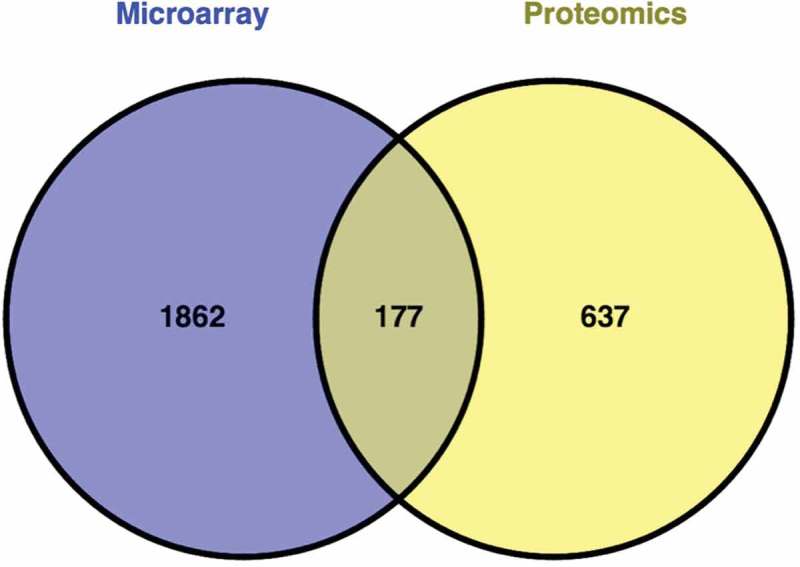
Comparison of host factors affected during respiratory syncytial virus infection identified by microarray and proteomics studies. A poor overlap was observed at the individual gene and protein level.
Table 2.
Overlap of host factors identified by microarray and proteomic studies.
| Microarray |
Proteomics |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang [8] | Tian [9] | Janssen [10] | Martinez [11] | Mayer [12] | Meijas [13] | Brand [14] | Brasier [15] | Hastie [16] | Jamaluddin [17] | Van Diepen [18] | Munday [19] | Munday [20] | Ternette [21] | Dave [22] | Van Diepen [23] | ||
| Microarray | Zhang [8] | 28 | 6 | 7 | 3 | 9 | 9 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | |
| Tian [9] | 28 | 37 | 53 | 29 | 23 | 12 | 1 | 4 | 2 | 0 | 10 | 6 | 4 | 36 | 5 | ||
| Janssen [10] | 6 | 37 | 44 | 23 | 37 | 17 | 1 | 10 | 2 | 0 | 14 | 8 | 2 | 45 | 16 | ||
| Martinez [11] | 7 | 53 | 44 | 20 | 29 | 15 | 2 | 5 | 1 | 2 | 25 | 12 | 6 | 53 | 9 | ||
| Mayer [12] | 3 | 29 | 23 | 20 | 5 | 3 | 0 | 0 | 0 | 0 | 4 | 3 | 0 | 8 | 0 | ||
| Meijas [13] | 9 | 23 | 37 | 29 | 5 | 34 | 0 | 4 | 1 | 0 | 10 | 6 | 4 | 27 | 7 | ||
| Brand [14] | 9 | 12 | 17 | 15 | 3 | 34 | 0 | 0 | 0 | 0 | 8 | 3 | 1 | 2 | 3 | ||
| Proteomics | Brasier [15] | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 4 | 1 | 9 | 4 | 0 | 2 | 0 | |
| Hastie [16] | 0 | 4 | 10 | 5 | 0 | 4 | 0 | 3 | 3 | 1 | 5 | 5 | 2 | 9 | 2 | ||
| Jamaluddin [17] | 0 | 2 | 2 | 1 | 0 | 1 | 0 | 4 | 3 | 0 | 14 | 6 | 1 | 2 | 0 | ||
| Van Diepen [18] | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 4 | 1 | 0 | 0 | 0 | ||
| Munday [19] | 3 | 10 | 14 | 25 | 4 | 10 | 8 | 9 | 5 | 14 | 4 | 74 | 4 | 14 | 4 | ||
| Munday [20] | 0 | 6 | 8 | 12 | 3 | 6 | 3 | 4 | 5 | 6 | 1 | 74 | 2 | 15 | 4 | ||
| Ternette [21] | 2 | 4 | 2 | 6 | 0 | 4 | 1 | 0 | 2 | 1 | 0 | 4 | 2 | 6 | 2 | ||
| Dave [22] | 0 | 36 | 45 | 53 | 8 | 27 | 2 | 2 | 9 | 2 | 0 | 14 | 15 | 6 | 13 | ||
| Van Diepen [23] | 0 | 5 | 16 | 9 | 0 | 7 | 3 | 0 | 2 | 0 | 0 | 4 | 4 | 2 | 13 | ||
The merged microarray and proteomics dataset of 2676 host factors was analyzed by DAVID for GO term enrichment in order to identify biological processes affected during RSV replication. The biological functions of these host factors include immune system process (p < 2.48 × 10−38), apoptosis (p < 3.83 × 10−19), RNA processing (p < 9.44 × 10−19), translation (p < 1.07 × 10−14), response to virus (p < 2.15 × 10−11), lymphocyte activation (p < 3.8 × 10−6), and regulation of NF-κB transcription (p < 6.05 × 10−5). At the level of biological process, these different host factors are shown to common cellular processes (Supplementary Table S1).
The merged list of 2676 host factors was narrowed down to 512, which included host factors identified in at least 2 datasets in order to increase the biological significance of the dataset. These host factors include IFIT3 and ISG15, which were common in eight studies; IFIT1, IFI35, MX1, and STA1, which were common in seven studies; IFIT2, which was common in six studies; CCL2, ENO1, IFI44, KLF4, PNPT1, RRBP1, SOP2, and WARS, which were common in 5 studies (Supplementary Table S2). Of the 512 host factors, 305 are upregulated and 109 host factors are downregulated during RSV infection. A subset of 98 host factors with discordant expression levels was observed when comparing between and among microarray and proteomics datasets. This subset of 512 host factors was selected for the construction of the RSV–host interaction network and was used to query for interactors using the HIPPIE database [44] and VirHostNet database [45]. The network consists of 1254 proteins and 1989 interactions (Figure 2). Network topology showed one large component of 1216 proteins, one group of 6 proteins (CAMPLG, DNM2, WDR48, USP21, PDIA4, PPIB), four groups of 3 proteins (GABPB1-TDRD7-TACC1, CDC42EP3-SEPT7-SEPT9, THBS1-MMP9-CXCL5, IL1R1-IL1RN-IL1R2), and seven groups of 2 proteins (NR4A1-PRDX4, CXorf40A-MT2A, IL16-KCNJ15, PSME3-ITPKB, IL2RG-IL15, CD81-IFITM1, PYCARD-AIM2). The rest of the proteins are not connected. The large component includes 10 viral proteins, which connect directly to 305 host proteins (Supplementary Table S3). Of these, 38 host factors were identified in the microarray and proteomics studies. Among the viral proteins, NS1 is connected to 219 host proteins while NS2, SH, G, and M2 are each connected to only one host protein.
Figure 2.

Respiratory syncytial virus (RSV)-host interaction network. The network contains 1,254 proteins (nodes) and 1,989 interactions (edges), which was constructed using HIPPIE and VirHostNet databases and visualized using Cytoscape. Yellow nodes represent RSV proteins, red nodes indicate upregulated host factors and blue nodes represent downregulated host factors during RSV infection. Gray nodes represent disconcordant expression level of host factors between and among transcriptome and proteome datasets. White nodes represent protein interactors identified from the databases.
MCODE analysis showed 10 clusters that were densely connected with a score of 2 or greater (Table 3). The highest ranked MCODE cluster had 8 proteins and 18 interactions that are involved in response to virus (p < 2.63 × 10−3). Other identified interaction hubs are involved in DNA replication (p < 1.64 × 10−5), SMAD protein phosphorylation (p < 6.15 × 10−6), ubiquitination (p < 5.9 × 10−7), interferon-mediated immunity (p < 2.23 × 10−5), RNA processing (p < 5.86 × 10−4), vesicle-mediated transport (p < 6.56x10−4), T-cell differentiation and JAK-STAT signaling (p < 3.1 × 10−7). Two interaction hubs contained viral proteins and its host interactors as members, which are involved in protein sumoylation (p < 4.31 × 10−5) and Rho protein signaling (p < 1.59 × 10−4).
Table 3.
MCODE predicted clusters in RSV–host interaction network.
| Cluster rank | Score | Proteins | Interactions | Process | p-value | Protein names |
|---|---|---|---|---|---|---|
| 1 | 5.14 | 8 | 18 | Response to virus | 2.63 × 10−3 | NCOR2,CREBBP,JUN,TSC22D,BCL3,EP300,FOS,RELA |
| 2 | 3.00 | 3 | 2 | DNA replication | 1.64 × 10−5 | CASP4,SMC3,SMC1A |
| 3 | 3.00 | 3 | 3 | SMAD protein phosphorylation | 6.15 × 10−6 | FKBP1A,ACVR1B,SMAD7 |
| 4 | 3.00 | 3 | 3 | Ubiquitination | 5.90 × 10−7 | PSMA2,PSMB8,PSMB9 |
| 5 | 3.00 | 3 | 3 | Interferon-mediated immunity | 2.23 × 10−5 | IFIT1,IFIT2,IFIT3 |
| 6 | 2.67 | 7 | 8 | Protein sumoylation | 4.31 × 10−6 | P,SUMO1,HNRNPF,SUMO2,HNRNPU,FAS,FLNA |
| 7 | 2.50 | 5 | 5 | RNA processing | 5.86 × 10−4 | HNRNPK,FUS,TOP1,YBX1,HIST1H1 C |
| 8 | 2.50 | 5 | 5 | Vesicle-mediated transport | 6.56 × 10−4 | KRT18,ANXA2,GRB2,YWHAZ,EPOR |
| 9 | 2.46 | 14 | 16 | T-cell differentiation, JAK-STAT signaling | 3.10 × 10−7 | STAT5A,HSPA8,PTPN1,PRKDC,SHC1,NR3C1,CRKL,TRIM28,APC,HSF1,ABL1,CTNNB1,JAK2,HSPA1B |
| 10 | 2.40 | 6 | 6 | Rho protein signaling | 1.59 × 10−4 | F,G,M,BCR,RAC1,RHOA |
JAK-STAT: Janus kinase-signal transducer and activator of transcription.
Exploration of drug–host factor interaction network for drug repositioning
The drug–host factor network was reconstructed in order to find alternative drugs that can inhibit RSV replication. Drugs that interact with the subset of 512 host factors were queried using the DrugBank database [53]. The network comprised 78 host factors and 177 drugs with 212 host factor–drug interactions (Figure 3). The topology of the network showed 2 major components that emerged from the drug and host factor interactions. The largest component contained 7 host factors and 56 drugs with 66 interactions and the second largest component contained 7 host factors and 34 drugs with 40 interactions. The connectivity of network was brought about by drugs with several targets, such as Tenecteplase connecting to five host factors (ANXA2, KRT8, PLAUR, SEPINB2, and SERPINE1) and Thalidomide targeting four host factors (FGFR2, ORM1, ORM2, and PTGS2). The presence of host factors being targeted by several drugs contributed also to the connectivity of the network, including Prostaglandin G/H synthase 2 (PTGS2) and DNA topoisomerase 2-alpha (TOP2A). PTGS2, which was upregulated during virus replication, are targeted by 49 different drugs while TOP2A, which was downregulated, are targeted by 24 different drugs (Supplementary Table S4). All 177 drugs in the network are FDA-approved with 22 biotech drug and 155 small molecule drug types. These drugs belong to different categories based as Anatomical Therapeutic Chemical classification, which include 18 anti-infectives, 10 respiratory system, and 37 anti-cancer drugs.
Figure 3.

Drug-host factor interaction network. This network contains 78 host factors that interact with 177 FDA-approved drugs, which were retrieved from the DrugBank database. These drugs can be repositioned for the treatment of RSV infection. Host factors are indicated by open circles with red color border representing upregulated and blue color border representing downregulated level of expression. FDA-approved small molecule drugs are represented by arrowheads and biotech drugs are represented by octagons. Fill colors of drugs represent the Anatomical Therapeutic Chemical (ATC) classification: Metabolism (cyan); Blood (red); Cardiovascular (pink); Dermatologicals (blue); Genito-urinary (light blue); Anti-infectives (green); Anti-neoplastics (orange); Musculo-skeletal (light purple); Nervous system (salmon pink); Anti-parasitic (yellow); Respiratory system (light green); Sensory organs (blue-green); and Various systems (white).
To identify the drugs that interact with host factors belonging to network hubs, a subnetwork was generated containing only host factors belonging to MCODE clusters (Table 3) and the drugs targeting those host factors. Of the 10 MCODE clusters identified in this study, 5 of these highly connected subnetworks contain 9 host factors that interact with 15 FDA-approved drugs (Figure 4).
Figure 4.

Drug-host factor interaction subnetwork. This subnetwork contains 9 host factors that interact with 15 FDA-approved drugs, which can be prioritized for repositioning as antiviral option for RSV infection. Host factors are indicated by open circles with red color border representing upregulated and blue color border representing downregulated level of expression. FDA-approved small molecule drugs are represented by arrowheads and biotech drugs are represented by octagons. Fill colors of drugs represent the Anatomical Therapeutic Chemical (ATC) classification: Blood (red); Cardiovascular (pink); Dermatologicals (blue); Anti-neoplastics (orange); and Anti-parasitic (yellow).
Expert commentary and 5-year view
The lack of vaccine and limited antiviral option for RSV emphasized the need to understand the molecular mechanism of pathogenesis of the virus. Recent advances in transcriptomic and proteomic technologies have increased our knowledge of host responses during RSV infection. However, a meta-analysis on the datasets generated from these high-throughput experiments has not yet been done. In this study, an integrated approach was performed on published microarray and proteomics datasets in order to obtain a more complete picture of virus–host interaction during RSV infection. This study also explored the possibility of identifying host factors as possible drug targets and repositioning of FDA-approved drugs for RSV treatment using drug–host factor network analysis.
Earlier microarray studies on host responses used cell lines infected with laboratory-adapted RSV strains, which gave a comprehensive picture of chemokines and cellular processes involved during virus infection [8,9,11,12]. However, these in vitro studies may not reflect the clinical outcome of the disease, such as severe pneumonia and bronchiolitis. A mouse model by Janssen et al. identified processes and pathways that were activated immediately after RSV infection, which can be applied in studying host genes associated with severe RSV infection in children [10]. Two recent microarray studies reported by Mejias et al. [13] in 2013 and Brand et al. [14] in 2015 shed light on the host responses associated with disease severity in hospitalized children with RSV infection.
Comparison of host genes identified by microarray experiments during RSV infection showed a poor overlap. A maximum overlap of only 53 genes that were significantly expressed during RSV infection was observed between Tian et al. [9] and Martinez et al. [11] datasets, which may be due to using the same RSV Long strain in their microarray experiments. Several factors may explain the low concordance of microarray datasets including differences in virus strain, host system, time-point of analysis, and microarray platform used in the experiments. The low concordance can be improved by following the Minimum Information about a Microarray Experiment standards and using the MicroArray Quality Control tools [55]. Concordance can be improved also by analyzing only genes common to different microarray platforms [56]. An alternative is to use RNA-seq technology, which offers higher reproducibility with less technical variation than microarray [57].
Earlier proteomics studies utilized 2DE techniques, which identified host responses such as heat shock, antioxidant, and stress response proteins during RSV replication [15–18]. However, gel-based protein separation technique is limited to detecting changes in highly abundant proteins and identified only less the 40 host proteins that are affected during RSV infection. Munday et al. overcame this limitation by employing a SILAC reagent that can differentially label proteins in RSV-infected and mock-infected cells and utilizing LC–MS/MS to identify several hundreds of host proteins [19,20]. An excellent review on the application of SILAC and quantitative proteomics to study virus–host interaction has been published [58]. Ternette et al. [21] and Dave et al. [22] used in-solution IEF, a non-gel-based technique to separate the proteins and LC–MS/MS for comprehensive identification of host proteins.
Most of the proteomics studies on host responses to RSV infection used in vitro system and so far, only one proteomic study by van Diepen et al. used a mouse model to investigate host responses during vaccination with chimeric vaccinia virus expressing RSV F and G proteins [23]. The study identified differential expression of seven host proteins in the lung tissues of RSV-challenged mice with characteristic lung inflammation and infiltration of white blood cells [23]. These host proteins can serve as molecular markers for RSV vaccine-induced enhanced disease, which can be used to evaluate the different vaccine candidates currently undergoing preclinical and clinical trials. To date, there are no proteomic studies on clinical samples from RSV-infected patients and we hope that proteomic studies will be performed in the future to complement the microarray studies of Mejias et al. [13] and Brand et al. [14].
Comparison of overlaps between microarray and proteomics datasets of host responses during RSV infection showed a small number of host factors that are common to both datasets. The changes in the expression of mRNA transcripts do not always correspond to the changes in the abundance of proteins, which may be due not entirely to technical differences but also to biological differences, such as posttranscriptional mechanisms controlling the rate of protein translation, different half-lives of mRNA and corresponding protein products, and differences in intracellular location of expressed proteins [59]. This discordant expression level of RNA transcripts and proteins is also observed in yeast cells [59], leukemia cells [60], mice [61], and psoriasis [62]. This low overlap highlights the need to integrate transcriptomic and proteomic approaches. Recent studies have combined both techniques in the investigation of influenza virus–host interaction. Shapira et al. [30] used a yeast two-hybrid system while Watanabe et al. [63] used co-immunoprecipitation technique to identify host proteins that interact with viral proteins and they both used genome-wide RNA interference (RNAi) screens to determine the transcriptional profile during influenza virus infection. An advantage of using loss-of-function method such as RNAi screening compared to standard DNA probe-based microarray technique is the ability to see the direct effect on virus replication of host gene knockdown. Hopefully, experiments that identify host factors, which interact directly to RSV proteins by yeast two-hybrid or co-immunoprecipitation techniques coupled with RNAi screens will be performed in the future.
While a poor overlap was observed at the individual gene and protein level, analysis at the level of biological process showed similar cellular functions shared by these factors during RSV replication. This indicates that different host genes and proteins identified in microarray and proteomics experiments contribute to the same biological processes. In addition, comparison of the results of GO term-enrichment analysis of highly connected subnetworks between the subset of 512 host factors that are common in at least two studies and the full dataset of 2676 host factors showed similar biological processes that are affected during RSV infection. This suggests robustness of the data against noise or stochastic fluctuations in gene expression or protein abundance.
Changes in the expression and abundance levels of interferon-stimulated genes and their protein products were detected commonly in both microarray and proteomics datasets. Interferon-induced protein with tetracopeptide repeats 1, 2, and 3 (IFIT1, IFIT2, and IFIT3), which were detected commonly in both platforms, were upregulated during RSV replication. These proteins form a complex that has an antiviral activity with IFIT1 recognizing the 5′-triphosphate of viral RNA and IFIT3 mediating interferon-induced antiproliferative responses [21,64]. Myxovirus resistance 1 protein (MX1) was detected also in seven studies and possesses antiviral activity by acting on IFIT-sequestered viral RNA [64]. In influenza virus-infected cells, MX1 inhibits ribonucleoprotein complex assembly by binding to viral NP and PB2 proteins, which in turn prevents PB2–NP interaction [65]. Further studies are needed to elucidate the fate of IFIT-sequestered viral RNA during RSV replication.
In order for the virus to utilize the host cellular machinery, it must target preferentially host proteins that are well connected in the virus–host interaction network allowing the virus to rewire the network to favor viral replication [66]. Indeed, 2 of the 10 MCODE clusters identified in this study contain viral proteins. One MCODE cluster has the RSV phosphoprotein (P) that interacts with host proteins involved in the sumoylation process, which is a posttranslational modification process of adding a small ubiquitin-like modifier (SUMO) to target proteins. The P protein of parainfluenza virus was found to be sumoylated with SUMO1 and acts as cofactor of viral RNA-dependent RNA polymerase during virus replication [67]. The other MCODE cluster contains viral proteins F, G, and M and host proteins associated with Rho protein signaling, which may play an important role in initiating cell-to-cell fusion resulting in syncytium formation and filamentous virion morphology [68]. Host factors that are involved in cell fusion may be promising drug targets for RSV antiviral drug development.
Host factors localized in network hubs that are being targeted by the virus can be explored also as potential drug targets by searching for drugs that interact with the same host factors, which can be repositioned for RSV treatment. This strategy has been utilized in identifying drug targets for influenza [69]. In this study, 5 of the 10 MCODE clusters contain host factors that interact with FDA-approved drugs (Figure 4). The first MCODE cluster had host factors involved in response to virus with drugs Nadroparin and Irbesartan connecting to FOS and JUN, respectively. During RSV infection, FOS was found to be upregulated in the microarray studies of Martinez et al. [11] and Mayer et al. [12] while JUN was also upregulated in the microarray studies of Tian et al. [9], Martinez et al. [11], Mayer et al. [12], and in the proteomics study of Dave et al. [22]. RSV-infected cells treated with an agonist of peroxisome-proliferator-activated receptor-γ resulted in the reduced binding activity of transcription factor, AP-1, which is composed of FOS and JUN and this prevented the release of proinflammatory cytokines [70]. Thus, inhibition of FOS and JUN by Nadroparin and Irbesartan may have potential applications as anti-inflammatory options for RSV infection.
The second subnetwork contained host factors that participate in SMAD protein phosphorylation, which contains the host factor FK506 binding protein 1A (FKBP1A) that connects with three immunomodulatory agents (Pimecrolimus, Tacrolimus, and Sirolimus). In RSV-infected A549 cells, FKBP1A expression is suppressed, which is a glucocorticoid receptor-regulated gene and may explain why glucocorticoid treatment is ineffective in children with RSV-induced bronchiolitis and wheezing [71]. Developing drugs that could abrogate the suppression or enhance the expression of FKBP1A may prevent the severe form of RSV infection.
The third subnetwork contained host proteins involved in ubiquitination with the drug Carfilzomib connecting to two host factors, proteasome subunit beta type-8 (PSMB8) and type-9 (PSMB9). PSMB8 was upregulated both in microarray study of Janssen et al. [10] and in the proteomics study of Dave et al. [22]. Similarly, PSMB9 was upregulated in the two studies and was detected also in the Martinez et al. dataset [11]. Confirmatory experiments are needed to see if Carfilzomib, which is an antineoplastic agent and proteasome inhibitor [72], has therapeutic potential against RSV.
The fourth cluster had members that participate in RNA processing with host factor DNA topoisomerase 1 (TOP1) connecting to four drugs. The abundance level of TOP1 decreased in both proteomics studies by Munday et al. [19] and Dave et al. [22]. Since the expression level of this protein is low during infection, molecules and drugs that increase the abundance level or stabilize the protein may have an effect on RSV replication.
The fifth cluster had host factors involved in vesicle-mediated transport with host factors keratin 18 (KRT18) and annexin (ANXA2) connecting to the drug Tenecteplase, and erythropoietin receptor (EPOR) connecting to four other drugs. KRT18 and ANXA2 were both detected in four proteomics studies [15,17,19,20], while EPOR was downregulated in two microarray studies [8,9]. The four drugs (Darbepoetin alfa, Peginesatide, Epoetin alfa, and Epoetin zeta) are agonists to the host factor EPOR, which facilitates the activation of Jak-STAT signaling pathway [53]. This signaling pathway is targeted by the virus through STAT2 suppression by RSV NS1 protein, which leads to reduction of expression of antiviral host factors [73]. Thus, further studies are needed to test the antiviral activity of these four drugs that target EPOR.
The virus–host interaction network generated in this study is still far from complete. There is still a big gap in the knowledge of RSV proteins that interact directly with host proteins. Hopefully, more experiments using yeast two-hybrid system, co-immunoprecipitation experiments as employed by Oliveira et al. [74], or microfluidics screens as used by Kipper et al. [46] will be performed in future studies in order to systematically identify all primary targets of RSV proteins. Then, the obtained results can be combined with functional genomic approaches such as genome-wide RNAi screens or CRISPR-cas9 screens, which is a genome editing system that can generate gene knock-outs and knock-in of host factors at the DNA level [75,76]. The results of functional genomic studies can then be integrated with proteomics in order to have a more complete picture of RSV–host interactions. We also hope that proteomics studies using clinical samples will be investigated in the future to complement the published microarray studies with the goal of validating the association of host proteins with RSV disease severity and its potential application in the validation of RSV vaccine trials. We also hope that in addition to developing antivirals that directly target the virus, host factors as novel drug targets would be included in the pipeline of RSV drug development. Thus, integrative approach will not only increase our understanding of the complexity of RSV–host factor interaction but also provide new insights on potential antiviral targets.
Supplementary Material
Financial & competing interests disclosure
This work was supported by a grant-in-aid from the Japan Initiative for Global Research Network on Infectious Diseases (JGRID) from the Japan Agency for Medical and Research and Development (AMED), the Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science (JSPS), the Science and Technology Research Partnership for Sustainable Development (SATREPS) from AMED and Japan International Cooperation Agency (JICA).The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Key issues.
Respiratory syncytial virus (RSV) is the leading cause of viral acute respiratory infections in children.
There is no vaccine available and antiviral option against RSV is limited.
Understanding of virus–host interaction is important in developing vaccines and antiviral drugs.
Current evidence from microarray and proteomic studies has identified hundreds of host factors that are affected during RSV replication.
Comparison of the host factors identified by these high-throughput techniques showed a poor overlap.
Integration of transcriptome and proteome datasets provides a more global approach in constructing virus–host interaction network.
Virus–host interaction network generated in this study was interrogated to identify host factors affected during RSV infection and explored as potential drug targets.
Drugs that target RSV–host factors can be repositioned for therapeutic application in RSV infection.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
* One of the earliest studies to identify host genes during RSV infection in vitro using microarray technology.
* This study used mouse model to identify host genes affected during RSV infection.
** This study used microarray to identify host genes that are associated with severe pneumonia in children infected with RSV. The use of whole blood RNA transcriptional profiling has future applications in differentiating patients with mild or severe form of the disease.
** This study used microarray to identify host genes that are associated with severe pneumonia in children infected with RSV. The use of whole blood RNA transcriptional profiling has future applications in differentiating patients with mild or severe form of the disease.
* One of the earliest studies to identify host proteins during RSV infection in vitro using proteomics.
* This study used SILAC reagent to differentially label proteins from virus-infected cells and control. This technique coupled with LC-MS/MS increased the throughput and sensitivity in identifying and quantifying host proteins during RSV infection.
** This study used a chimeric vaccinia virus-RSV vaccine to infect mice and identified host proteins associated with pathophysiology in lungs, similar to the fatal cases involving formalin-inactivated RSV vaccine in the 1960s. Results of this study can be applied for evaluation purposes of next-generation RSV vaccines.
** This study used meta-analysis of datasets from genome-wide RNAi screens and identified host factors essential for influenza virus replication. The authors queried the DrugBank database for drugs that interact with the essential host factors.
- Knipe DM, Howley PM. Fields virology. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013. [Google Scholar]
- Legand A, Briand S, Shindo N. Addressing the public health burden of respiratory viruses: the battle against respiratory viruses (BRaVe) initiative. Future Virol. 2013;8(10):953–968. [Google Scholar]
- Nair H, Nokes DJ, Gessner BD. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez WJ, Bui RH, Connor JD. Environmental exposure of primary care personnel to ribavirin aerosol when supervising treatment of infants with respiratory syncytial virus infections. Antimicrob Agents Chemother. 1987;31(7):1143–1146. doi: 10.1128/aac.31.7.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler JA, Van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol. 1989;63(7):2941–2950. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron R. WHO consultation on respiratory syncytial virus (RSV) vaccine development. Geneva: World Health Organization; 2015. [Oct 30;]. Overview of the RSV vaccine field.http://www.who.int/immunization/research/meetings_workshop/rsv_vaccine_development/en/index1.html cited. Available from. [Google Scholar]
- Ludwig S. Disruption of virus-host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections. Biol Chem. 2011;392(10):837–847. doi: 10.1515/BC.2011.121. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Luxon BA, Casola A. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J Virol. 2001;75(19):9044–9058. doi: 10.1128/JVI.75.19.9044-9058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Zhang Y, Luxon BA. Identification of NF-kappaB-dependent gene networks in respiratory syncytial virus-infected cells. J Virol. 2002;76(13):6800–6814. doi: 10.1128/JVI.76.13.6800-6814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Pennings J, Hodemaekers H. Host transcription profiles upon primary respiratory syncytial virus infection. J Virol. 2007;81(11):5958–5967. doi: 10.1128/JVI.02220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Lombardia L, Garcia-Barreno B. Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells. J Gen Virol. 2007;88(Pt 2):570–581. doi: 10.1099/vir.0.82187-0. [DOI] [PubMed] [Google Scholar]
- Mayer AK, Muehmer M, Mages J. Differential recognition of TLR-dependent microbial ligands in human bronchial epithelial cells. J Immunol. 2007;178(5):3134–3142. doi: 10.4049/jimmunol.178.5.3134. [DOI] [PubMed] [Google Scholar]
- Mejias A, Dimo B, Suarez NM. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10(11):e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand HK, Ahout IM, de Ridder D. Olfactomedin 4 Serves as a Marker for Disease Severity in Pediatric Respiratory Syncytial Virus (RSV) Infection. PLoS One. 2015;10(7):e0131927. doi: 10.1371/journal.pone.0131927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier AR, Spratt H, Wu Z. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J Virol. 2004;78(21):11461–11476. doi: 10.1128/JVI.78.21.11461-11476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie ML, Headlam MJ, Patel NB. The human respiratory syncytial virus nonstructural protein 1 regulates type I and type II interferon pathways. Mol Cell Proteomics. 2012;11(5):108–127. doi: 10.1074/mcp.M111.015909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaluddin M, Wiktorowicz JE, Soman KV. Role of peroxiredoxin 1 and peroxiredoxin 4 in protection of respiratory syncytial virus-induced cysteinyl oxidation of nuclear cytoskeletal proteins. J Virol. 2010;84(18):9533–9545. doi: 10.1128/JVI.01005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diepen A, Brand HK, Sama I. Quantitative proteome profiling of respiratory virus-infected lung epithelial cells. J Proteomics. 2010;73(9):1680–1693. doi: 10.1016/j.jprot.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Munday DC, Emmott E, Surtees R. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol Cell Proteomics. 2010;9(11):2438–2459. doi: 10.1074/mcp.M110.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday DC, Hiscox JA, Barr JN. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus subgroup B using SILAC coupled to LC-MS/MS. Proteomics. 2010;10(23):4320–4334. doi: 10.1002/pmic.201000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternette N, Wright C, Kramer HB. Label-free quantitative proteomics reveals regulation of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) and 5’-3’-exoribonuclease 2 (XRN2) during respiratory syncytial virus infection. Virol J. 2011;8(1):442. doi: 10.1186/1743-422X-8-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave KA, Norris EL, Bukreyev AA. A comprehensive proteomic view of responses of A549 type II alveolar epithelial cells to human respiratory syncytial virus infection. Mol Cell Proteomics. 2014;13(12):3250–3269. doi: 10.1074/mcp.M114.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diepen A, Brand HK, De Waal L. Host proteome correlates of vaccine-mediated enhanced disease in a mouse model of respiratory syncytial virus infection. J Virol. 2015;89(9):5022–5031. doi: 10.1128/JVI.03630-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Graham BS. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol. 2008;82(5):2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp RA, Mejias A, Ramilo O. Host gene expression and respiratory syncytial virus infection. Curr Top Microbiol Immunol. 2013;372:193–209. doi: 10.1007/978-3-642-38919-1_10. [DOI] [PubMed] [Google Scholar]
- Brass AL, Huang I-C, Benita Y. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454(7206):890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463(7282):818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- König R, Stertz S, Zhou Y. Human host factors required for influenza virus replication. Nature. 2010;463(7282):813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SD, Gat-Viks I, Shum BO. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs KM, Berard A, Xu W. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J Virol. 2010;84(20):10888–10906. doi: 10.1128/JVI.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove BK, Surtees R, Bean TJ. A quantitative proteomic analysis of lung epithelial (A549) cells infected with 2009 pandemic influenza A virus using stable isotope labelling with amino acids in cell culture. Proteomics. 2012;12(9):1431–1436. doi: 10.1002/pmic.201100470. [DOI] [PubMed] [Google Scholar]
- Kroeker AL, Ezzati P, Halayko AJ. Response of primary human airway epithelial cells to influenza infection: a quantitative proteomic study. J Proteome Res. 2012;11(8):4132–4146. doi: 10.1021/pr300239r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapat C, Saito R, Suzuki H. Quantitative phosphoproteomic analysis of host responses in human lung epithelial (A549) cells during influenza virus infection. Virus Res. 2014;179:53–63. doi: 10.1016/j.virusres.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA. Discovery of insect and human dengue virus host factors. Nature. 2009;458(7241):1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu HC, Hannemann H, Heesom KJ. High-throughput quantitative proteomic analysis of dengue virus type 2 infected A549 cells. PLoS One. 2014;9(3):e93305. doi: 10.1371/journal.pone.0093305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero CA, Datta PK, Sen S. HIV-1 Vpr modulates macrophage metabolic pathways: a SILAC-based quantitative analysis. PLoS One. 2013;8(7):e68376. doi: 10.1371/journal.pone.0068376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Huang D, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang D, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer MH, Fontaine JF, Vinayagam A. HIPPIE: integrating protein interaction networks with experiment based quality scores. PLoS One. 2012;7(2):e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand T, Delmotte S, Navratil V. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res. 2015;43(Database issue):D583–587. doi: 10.1093/nar/gku1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipper S, Hamad S, Caly L. New host factors important for respiratory syncytial virus (RSV) replication revealed by a novel microfluidics screen for interactors of matrix (M) protein. Mol Cell Proteomics. 2015;14(3):532–543. doi: 10.1074/mcp.M114.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jain N, Limpanawat S. Interaction between human BAP31 and respiratory syncytial virus small hydrophobic (SH) protein. Virology. 2015;482:105–110. doi: 10.1016/j.virol.2015.03.034. [DOI] [PubMed] [Google Scholar]
- Munday DC, Wu W, Smith N. Interactome analysis of the human respiratory syncytial virus RNA polymerase complex identifies protein chaperones as important cofactors that promote L-protein stability and RNA synthesis. J Virol. 2015;89(2):917–930. doi: 10.1128/JVI.01783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer MH, Lopes TJ, Mah N. Adding protein context to the human protein-protein interaction network to reveal meaningful interactions. PLoS Comput Biol. 2013;9(1):e1002860. doi: 10.1371/journal.pcbi.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Tegenfeldt F, Petty TJ. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43(Database issue):D250–256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki ES. The end of the microarray Tower of Babel: will universal standards lead the way? J Biomol Tech: JBT. 2006;17(3):200–206. [PMC free article] [PubMed] [Google Scholar]
- Petersen D, Chandramouli GV, Geoghegan J. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6(1) doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday DC, Surtees R, Emmott E. Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics. 2012;12(4–5):666–672. doi: 10.1002/pmic.201100488. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Jensen R, Gendeh G. Proteome and transcriptome analysis of retinoic acid-induced differentiation of human acute promyelocytic leukemia cells, NB4. J Proteome Res. 2004;3(3):627–635. doi: 10.1021/pr049976r. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA.et alComparative analysis of proteome and transcriptome variation in mouse PLoS Genet 201176e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Remmer HA, Sarkar MK. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015;7(1):86. doi: 10.1186/s13073-015-0208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kawakami E, Shoemaker JE. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 2014;16(6):795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle C-A. IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat Immunol. 2011;12(7):624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Verhelst J, Parthoens E, Schepens B. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol. 2012;86(24):13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A. 2007;104(18):7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Xu P, He B. Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5. J Virol. 2011;85(19):10261–10268. doi: 10.1128/JVI.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower TL, Pastey MK, Peeples ME. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J Virol. 2005;79(9):5326–5336. doi: 10.1128/JVI.79.9.5326-5336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chassey B, Meyniel-Schicklin L, Aublin-Gex A. Genetic screens for the control of influenza virus replication: from meta-analysis to drug discovery. Mol Biosyst. 2012;8(4):1297–1303. doi: 10.1039/c2mb05416g. [DOI] [PubMed] [Google Scholar]
- Arnold R, Konig W. Peroxisome-proliferator-activated receptor-gamma agonists inhibit the release of proinflammatory cytokines from RSV-infected epithelial cells. Virology. 2006;346(2):427–439. doi: 10.1016/j.virol.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Hinzey A, Alexander J, Corry J. Respiratory syncytial virus represses glucocorticoid receptor-mediated gene activation. Endocrinology. 2011;152(2):483–494. doi: 10.1210/en.2010-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DJ, Chen Q, Voorhees PM. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110(9):3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villenave R, Broadbent L, Douglas I. Induction and antagonism of antiviral responses in respiratory syncytial virus-infected pediatric airway epithelium. J Virol. 2015 doi: 10.1128/JVI.02119-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AP, Simabuco FM, Tamura RE. Human respiratory syncytial virus N, P and M protein interactions in HEK-293T cells. Virus Res. 2013;177(1):108–112. doi: 10.1016/j.virusres.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109(39):E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


