Abstract
Background/Aims
The functional variant (rs56109847) in the 3′-untranslated regions (3′-UTR) of the serotonin receptor 3E (HTR3E) gene is associated with female diarrhea predominant irritable bowel syndrome (IBS-D) in British populations. However, the relationship of the polymorphism both to HTR3E expression in the intestine and to the occurrence of Chinese functional gastrointestinal disorders has yet to be examined.
Methods
Polymerase chain reaction amplification and restriction fragment length polymorphism analyses were employed to detect polymorphisms among Chinese Han women, particularly 107 patients with IBS-D, 99 patients with functional dyspepsia (FD), 115 patients with mixed IBS and 69 patients with IBS-D + FD. We also assessed microRNA-510 (miR-510) and HTR3E expression in human colonic mucosal tissues with immunohistochemistry and other methods. Dual-luciferase reporter assays were conducted to examine the binding ability of miR-510 and HTR3E 3′-UTR.
Results
Genotyping data showed the variant rs56109847 was significantly associated with IBS-D, but not with FD, mixed-IBS, or FD + IBS-D. HTR3E was abundantly expressed around the colonic mucosal glands but less expressed in the stroma. miR-510 expression decreased, whereas HTR3E expression increased in the colonic mucosal tissue of patients with IBS-D compared with those in controls. HTR3E expression was significantly higher in patients with the GA genotype than that in patients with the GG genotype. The single-nucleotide polymorphisms disrupted the binding site of miR-510 and significantly upregulated luciferase expression in HEK293 and HT-29 cells.
Conclusions
The single-nucleotide polymorphisms rs56109847 led to reduced microRNA binding and overexpression of the target gene in intestinal cells, thereby increasing IBS-D risk in the Chinese Han population. The decreased expression of miR-510 might contribute to IBS-D.
Keywords: Female, Functional gastrointestinal disorders, MicroRNA-510, Receptors, Serotonin, 5-HT3
Introduction
Functional dyspepsia (FD) and irritable bowel syndrome (IBS) are common functional gastrointestinal disorders (FGID) in the human population, particularly in females.1,2 The etiology of FGID is assumed to be associated with infection,3 alterations in immune system or visceral sensitivity,4 imbalance of the intestinal flora,5 and psychiatric factors.6,7 Recent studies on the familial aggregation of patients with FGID have indicated that hereditary factors may contribute to susceptibility to FGID.6,8,9
MicroRNAs (miRNAs) are a class of small noncoding RNAs that regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of the messenger RNA (mRNA) of the target gene.10 MicroRNAs are involved in the pathogenesis of FGID.11 Moreover, genomic variation within miRNA target sites may be important genetic sources that influence FGID risk in humans.12,13
Mechanistic studies of FGID showed that serotonin receptor 3 (5-HT3) plays an important role in the motor-sensory function of the gut.14 In all the subtypes of the 5-HT3 receptor, only the 5-HT3E subunit is exclusively expressed in gastrointestinal (GI) tissues, including colon and small intestine.15,16 This finding indicates that the 5-HT3E subunit may play a distinct and specific role in the formation and function of 5-HT3 receptors in the human GI tract. Previous studies in England indicated that the GA genotype of HTR3E may serve as a predisposing factor in patients with IBS.12 The high prevalence of overlap between FD and IBS has been consistently reported both locally and internationally17; this phenomenon demonstrated that both disorders share common pathophysiological disturbances.18 Female gender was also associated with FD overlapping IBS.19
In this study, we examined the association of the 5-HT3E polymorphism with susceptibility to FD, diarrhea predominant IBS (IBS-D), mixed IBS (IBS-M), and FD + IBS-D in a Chinese female Han population. We also assessed the mRNA and protein expression of miR-510 and 5-HT3E in the colonic mucosa of patients with IBS-D and controls to determine the association between the expression and the 5-HT3E polymorphism. We further examined the binding ability of miR-510 to the HTR3E 3′-UTR containing G or A alleles.
Materials and Methods
Study Population
In this study, 107 patients with IBS-D, 99 patients with FD, 115 patients with IBS-M, and 69 patients with IBS-D + FD were diagnosed by using the Rome III criteria. All patients were of the southern Chinese female Han population. The control group (n = 294) was age matched, and the subjects were healthy based on checkup examinations and had no history of GI diseases.
All the participants provided informed consent. Briefly, 10 mL of peripheral blood was collected. Each subject was asked to complete a questionnaire with data, including age, profession, and other demographic information. Due to reasons such as the patients’ wishes, age, and the high correlation between IBS-D and the single-nucleotide polymorphisms (SNP) rs56109847, we extracted a random 53 patients with IBS-D for biopsy.
Basic characteristics, such as age, profession, and place of residence of the patients and control subjects are presented in Table 1. Most of the patients were younger than 50 years, and were classified as people who lived in Yangzhou city. Over 50% of the patients underwent mental labor in their profession. Age, profession, place of residence, marital status, alcohol or smoking habits, and family history of bowel symptoms were not significantly different between the patients and control subjects (P > 0.05).
Table 1.
Basic Characteristics of the Patients and Control Subjects
| IBS-D (n = 107) | FD (n = 99) | IBS-D + FD (n = 69) | Controls (n = 294) | χ2 | P-value | |
|---|---|---|---|---|---|---|
| Age (yr) | ||||||
| 18–30 | 40 (37.4) | 32 (32.3) | 25 (36.2) | 108 (36.7) | 5.932 | 0.748 |
| 31–40 | 29 (27.1) | 25 (25.3) | 19 (27.5) | 88 (29.9) | ||
| 41–50 | 20 (18.7) | 17 (17.2) | 10 (14.5) | 52 (17.7) | ||
| 51–71 | 18 (16.8) | 25 (25.3) | 15 (21.7) | 46 (15.6) | ||
| Profession | ||||||
| Mental labor | 74 (69.2) | 72 (72.7) | 53 (76.8) | 220 (74.8) | 1.721 | 0.632 |
| Manual labor | 33 (30.8) | 27 (27.3) | 16 (23.2) | 74 (25.2) | ||
| Place of residence | ||||||
| Yangzhou city | 104 (97.2) | 94 (94.9) | 66 (95.7) | 289 (98.3) | 4.185 | 0.221 |
| Other cities | 3 (2.8) | 5 (5.1) | 3 (4.3) | 5 (1.7) | ||
| Marital status | ||||||
| Yes | 61 (57.0) | 66 (66.7) | 38 (55.1) | 185 (62.9) | 3.484 | 0.323 |
| No | 46 (43.0) | 33 (33.3) | 31 (44.9) | 109 (37.1) | ||
| Alcohol or smoking habits | ||||||
| Yes | 10 (9.3) | 15 (15.2) | 11 (15.9) | 28 (9.5) | 4.297 | 0.231 |
| No | 97 (90.7) | 84 (84.8) | 58 (84.1) | 266 (90.5) | ||
| Family history of bowel symptoms | ||||||
| Yes | 14 (13.1) | 10 (10.1) | 10 (14.4) | 27 (9.2) | 2.416 | 0.491 |
| No | 93 (86.9) | 89 (89.9) | 59 (85.6) | 267 (90.8) | ||
IBS-D, diarrhea predominant irritable bowel syndrome; FD, functional dyspepsia.
Data are presented as n (%).
Ethics
This study was reviewed and approved by the Medical Ethics Committee of Yangzhou University prior to participant recruitment.
Genotyping
Genomic DNA was extracted from whole blood in buffy coat preparations through the method of Miller with minor modifications by using the TIANamp Blood DNA Midi Kit (TIANGEN, Beijing, China).20 Genotyping was performed using polymerase chain reaction (PCR) amplification and restriction fragment length polymorphism (RFLP) analyses. The reaction volume contained 100 ng of DNA, 10 μL of 2× Taq PCR Master Mix (Boerdi, Nanjing, China), and 5 pmol oligonucleotide primers flanking the polymorphic region (forward: 5′-CGTCATATGCCTCTGGAA-CA-3′ and reverse: 5′-ATAGGCGTGAACCACTGCAC-3′) in double-distilled water (ddH2O) to a final volume of 20 μL. PCR reactions were performed using a PTC-200 (MJ Research, Minnesota, USA) or a Mastercycler gradient thermal cycler (MJ Research) with the following protocols: incubation at 94°C for 2 minutes; 5 cycles of 94°C for 30 seconds, 68°C for 30 seconds, and 72°C for 30 seconds; 5 cycles of 94°C for 30 seconds, 64°C for 30 seconds, and 72°C for 30 seconds; 25 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; and a final extension step of 72°C for 5 minutes. A 3 μL aliquot of the PCR product was run on 1.5% agarose gel to visualize a 397-bp fragment. SNP were identified by restriction enzyme digestion. Briefly, 3 μL of the PCR product of HTR3E was digested with 0.5 μL of the restriction enzyme Hpy188III (New England Biolabs, Ipswich, MA, USA), according to the manufacturer’s recommendations. Two types of genotypes were determined through electrophoresis of the enzyme-digested product of HTR3E 3′UTR, GG (298 bp), and GA (344 and 298 bp). The enzyme-digested products were confirmed using nucleotide sequencing.
MicroRNA-510 and Serotonin Receptor 3E Expression in Colonic Mucosa
Preparation of tissue sections
Colonic mucosal tissues were obtained from 45 female patients with IBS-D and the GG genotype and 8 female patients with IBS-D and the GA genotype (21–63 years old). We collected colonic mucosal tissue samples from 53 healthy females (matched to the patient sample with respect to age and ethnicity) as controls.
Immunohistochemistry studies
Immunohistochemical staining was performed as below. Colonic mucosal tissues were immediately washed in cold phosphate-buffered saline (PBS, pH 7.4) and then fixed in 4% buffered paraformaldehyde at 4°C for 24 hours. The tissues were dehydrated, embedded in paraffin, and then sectioned sagittally at a thickness of 4 μm. Slides were incubated in boiled 0.01 mol/L citrate buffer (pH 7.2) for 10 minutes, cooled at room temperature, immersed in 3% H2O2 for 10 minutes, and washed in PBS (0.01 mol/ L, pH 7.2) 3 times. Subsequently, 10% normal goat serum was added for 20 minutes. The sections were incubated with primary anti-5-HT3E (1:1000; Sigma, Saint Louis, USA) at 4°C for 14–16 hours. PBS was used as a substitute for antibodies and as a negative control. The slides were then washed in PBS (0.01 mol/L, pH 7. 2) 3 times and then incubated with the anti-rabbit IgG-horseradish peroxidase (HRP) for 20 minutes at room temperature. The slides were stained with 3′3-diaminbezidine tetrahydrochloride. The presence of dark brown granules was considered as a positive result. Negative controls were prepared through incubation with equivalent concentrations of non-immune rabbit antiserum. Integrated optical density measurements of images were performed under the same illuminating setting for each sample. The images were captured using the JEDA801D capture software. Optical density was determined in the microscopic fields of the malformed portion of colonic mucosal tissues. Immunohistochemically stained slides were reviewed independently by 2 researchers.
Quantitative reverse-transcription polymerase chain reaction detection
Total RNA was isolated from colonic mucosal tissues by using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and subjected to reverse transcription with Takara PrimeScriptTM First-strand cDNA Synthesis kit (Takara Bio, Kusatsu, Japan) according to the manufacturer’s instructions. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed using an All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) on the 7500 Real-Time PCR system (Applied Biosystems, South San Francisco, CA, USA). The U6 small RNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were used as internal controls. All reactions were performed in triplicate. The primers for miR-510 and U6 were 5′-CTCAACTGGT-GTCGTGGAGTCGGCAATTCAGTTGAGGTGATTG-3′ and TACAT ACTGGTGTCGTGGAGTC; and 5′-CGCTTC-GGCAGCACATATAC-3′ and 5′-TTCACGAATTTGCGT-GTCAT-3′, respectively. The primers for 5-HT3E and GAPDH mRNAs included 5′-GTCCCCACCCAAGTCAACAT-3′ and 5′-AAATGTCTGGGAGCCACAGG-3′; and 5′-AAGGT-GAAGGTCGGAGTCAAC-3′, and 5′-GGGGTCATTGATG-GCAACAATA-3′, respectively.
Western blot analysis
Approximately 100 mg of tissues were minced into small pieces by using surgical blades and then sonicated in protein lysis buffer. Protein concentration was measured by using the Bradford method. The specimens were adjusted to the same protein concentration, aliquoted, and then stored at −80°C until further assay. Equal amounts of total proteins from tissues were separated on SDS-polyacryl-amide gels and then electro-transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The blots were incubated with anti-5-HT3E, an antibody produced in rabbit (1:1000; Sigma), overnight at 4°C. The samples were washed and incubated with goat anti-rabbit IgG-HRP secondary antibodies (1:10 000) for 1 hour at room temperature and detected using an enhanced chemiluminescence kit (BeyoECL Plus; Beyotime, ShangHai, China).
Role of MicroRNA-510 on the 3′ Untranslated Regions of the Serotonin Receptor 3E Wild Type and Mutant Messenger RNA
Based on the bioinformatics prediction and data in the literature, the SNP rs56109847 of the HTR3E 3′-UTR is located in the region of miRNA-510 recognition for binding. Variations in this region could affect miRNA-510 binding 5-HT3E mRNA and 5-HT3E. To test our hypothesis, we determined whether SNP could affect 5-HT3E expression by using the dual luciferase reporter assay.
Construction of pRL-TK–HTR3E 3′-UTR-c.*76G/A
The following procedures were performed to construct the pRL-TK–HTR3E 3′-UTR-c.*76G/A. The full-length HTR3E 3′-UTR fragment (Supplementary Figure A) was amplified from genomic DNA by using the forward primer 5′-ATTATCTAGAGCAGGTGCTCACCTGCCAACTT-3′(P1) and the reverse primer 5′-CATGTCTGCTCGAAGCGG CCGCCTCTGCAGAATTATTTATTGGG-3′(P2); the former contains a restriction site for Xba I (in bold), whereas the latter contains the reverse complementary sequence of the SV40 polyA 5′-downstream of the pRL-TK Renilla luciferase (Rluc) to yield HTR3E 3′-UTR. Second, the SV40 poly A between Xba I and BamH I of pRL-TK vector (Supplementary Figure B) was amplified using the forward primer 5′-CCCAATAAATAATTCTG CAGAGGCGGCCGCTTCGAGCAGACATG-3′(P3), with 22 bases of the downstream HTR3E 3′-UTR (in bold), and the reverse primer 5′-CGCGGATCCTTATCGATTTTACCACAT-3′(P4), with the restriction site for BamH I in bold, to generate the SV40 poly A fragment. The recombinant DNA fragment (Supplementary Figure C) was generated after PCR by using P1 and P4 primers from the mixture template of HTR3E 3′-UTR and SV40 poly A fragment. The recombinant PCR product was also digested with Xba I and BamH I and inserted with the corresponding restriction sites of the pRL-TK Rluc vector (Promega, Madison, WI, USA) to construct pRL-TK–HTR3E 3′-UTR-c.*76G/A vector.
Cell Culture, Transfection, and Dual Luciferase Reporter Assays
We obtained HEK293 and HT29 cell lines from the Shanghai Cell Bank, Chinese Academy of Sciences and which were then cultured in DMEM (HyClone, Logan, Utah, USA) supplemented with 10% fetal bovine serum (HyCloneUSA). For the luciferase assays, the cells were split into 24-well plates at approximately 5 × 105 cells per well prior to transfection. Subsequently, 2.5 μL of Lipofectamine 2000 (Invitrogen) per well was added with 400 ng (per well) of the Rluc recombinant plasmid pRL-TK–HTR3E 3′-UTR-c.*76A/G (constructed above) and 35 ng of the pGL3-control (firefly luciferase; Promega). The solution was then co-transfected into cultured cells with 500 ng (per well) of miR-NA-510 plasmid or negative control miRNA plasmid (synthesized by Genechem, Shanghai, China), according to the manufacturer’s instructions. After transfection for 48 hours, the dual-luciferase reporter assay system (Promega) was used and cell lysates were prepared according to the manufacturer’s instructions. The pRL-TK–HTR3E 3′-UTR-c.*76A/G and pGL3-control were transfected into cultured cells to investigate whether the SNP without miR-510 could affect HTR3E expression. Three procedures were performed for each transfection, and luciferase activity was measured at 3-fold.
Statistical Methods
The Statistical Program for Social Sciences, version 19.0 (SPSS, Chicago, IL, USA) was used for statistical analysis. Pearson’s χ2 test was performed to assess deviation from the Hardy-Weinberg equilibrium. Chi-square test was used to evaluate the basic characteristics of patients and control subjects, as well as differences in the frequency distributions of each allele and genotype of the miRSNPs between cases and controls. Group means were compared using the t test. A P-value < 0.05 was considered statistically significant, and all the statistical tests were two-sided.
Results
Significant Association of the Single-nucleotide Polymorphisms rs56109847 with Diarrhea Predominant Irritable Bowel Syndrome but Not Functional Dyspepsia or Functional Dyspepsia + Diarrhea Predominant Irritable Bowel Syndrome
For the HTR3E c.*76G>A variant, no deviation from Hardy-Weinberg equilibrium was detected in FD, IBS-D, and FD + IBS-D or healthy controls (P > 0.05). Genotyping data showed a significant association between female patients with IBS-D and the SNP rs56109847 in the HTR3E 3′-UTR (χ2 = 7.731, P = 0.005; Table 2). The frequency of the GA genotype relative to the GG genotype for the presence of FD alone, as well as the coexistence of FD and IBS-D, did not significantly differ among the groups.
Table 2.
Genotype and Allele Frequency of the Serotonin Receptor 3E 3′-Untranslated Regions Variant c._76G>A (rs56109847) by Female Diarrhea Predominant Irritable Bowel Syndrome and Healthy Control Groups
| Group | Genotype | Allele frequency | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| G/G | G/A | χ2 | P | G | A | χ2 | P | |
| IBS-D (nfemale = 107) | 90 (84) | 17 (16) | 7.731 | 0.005 | 197 (92) | 17 (8) | 7.357 | 0.007 |
| FD (nfemale = 99) | 90 (91) | 9 (9) | 0.567 | 0.451 | 189 (96) | 9 (4) | 0.546 | 0.460 |
| IBS-D + FD (nfemale = 69) | 61 (88) | 8 (12) | 1.802 | 0.179 | 130 (94) | 8 (6) | 1.730 | 0.188 |
| IBS-M (nfemale = 115) | 102 (89) | 13 (11) | 2.258 | 0.133 | 217 (94) | 13 (6) | 2.163 | 0.141 |
| Controls (nfemale = 294) | 274 (93) | 20 (7) | 568 (97) | 20 (3) | ||||
IBS-D, diarrhea predominant irritable bowel syndrome; FD, functional dyspepsia; IBS-M, mixed irritable bowel syndrome.
Data are presented as n (%).
Positive Expression Rate of Serotonin Receptor 3E in Patients with Diarrhea Predominant Irritable Bowel Syndrome and the GA Genotype Was Significantly Higher Compared with Patients with the GG Genotype, as Determined Using Immunohistochemistry Studies
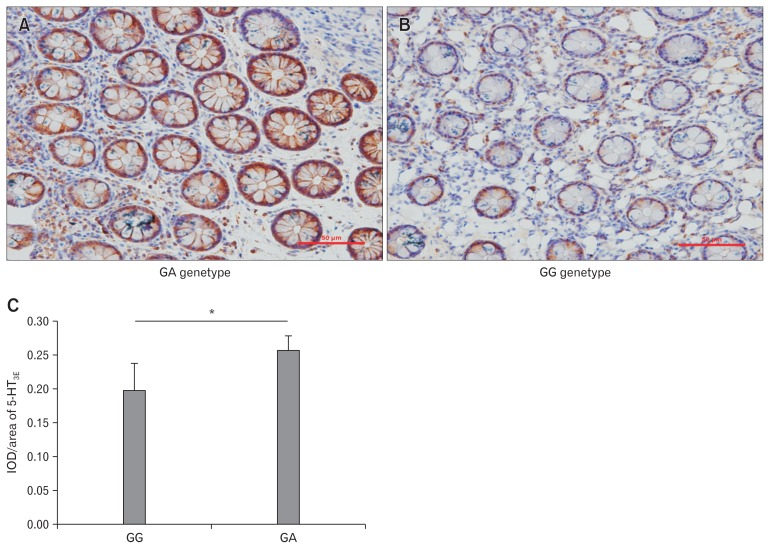
5-HT3E was abundantly expressed around the colonic mucosal glands with minimal expression in the stroma (Fig. 1A and 1B). 5-HT3E expression was significantly higher in patients with the GA genotype as compared with patients with the GG genotype (P < 0.05; Fig. 1C).
Figure 1.
Expression of serotonin receptor 3E (5-HT3E) in colonic mucosa of female diarrhea predominant irritable bowel syndrome (IBS-D) patients with GA and GG genotypes detected by immunohistochemistry. (A) Expression of 5-HT3E in colonic mucosa of female IBS-D patients with GA genotype as detected using immunohistochemistry (magnification, ×100). (B) Expression of 5-HT3E in colonic mucosa of female IBS-D patients with GG genotype as detected using immunohistochemistry (magnification, ×100). (C) Positive expression rate of 5-HT3E in patients with GA genotype was significantly higher than that in patients with GG genotype as revealed by immunohistochemistry studies (*P < 0.05). IOD, integrated optical density.
Serotonin Receptor 3E Protein Expression in Colonic Mucosal Tissues in Patients with Diarrhea Predominant Irritable Bowel Syndrome and the GA Genotype Was Significantly Higher than that in Serotonin Receptor 3E GG Genotype Patients
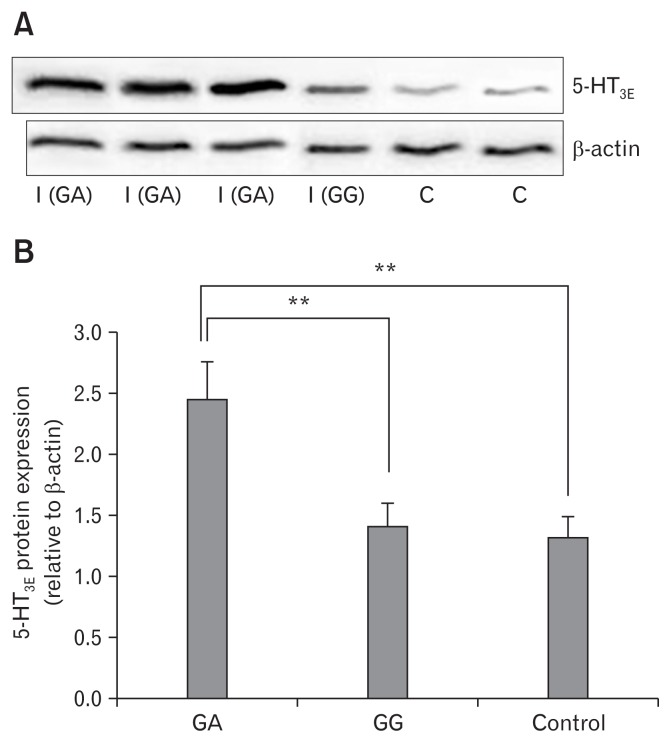
We detected 5-HT3E protein expression in colonic mucosal tissues in 53 IBS-D patients and 53 normal controls by using Western blot analysis. 5-HT3E protein expression in the colonic mucosal tissues of patients with IBS-D patients was higher compared with that in normal colonic mucosal tissues (P < 0.01; Fig. 2A). These results suggest that 5-HT3E may play an important role in the occurrence of IBS-D, particularly in female IBS-D. We detected 5-HT3E protein expression in colonic mucosal tissues in 45 patients with IBS-D, and the GG genotype and in 8 patients with IBS-D and the GA genotype by using the same method. 5-HT3E protein expression in the colonic mucosal tissues of patients with the GA genotype was significantly higher (P < 0.01; Fig. 2B).
Figure 2.
Expression of the serotonin receptor 3E (5-HT3E) protein in colonic mucosal tissues of female diarrhea predominant irritable bowel syndrome (IBS-D) patients and normal controls. (A) The expression of 5-HT3E protein in colonic mucosal tissue of female IBS-D patients and normal controls, as detected using Western blot analysis, I (GA) and I (GG) refer to IBS-D patients with the GA genotype and patients with the GG genotype, respectively; C refers to normal control. (B) The 5-HT3E protein expression in colonic mucosal tissues in patients with GA genotype were significantly higher than in patients with GG genotype and normal controls (**P < 0.01).
Results of Quantitative Reverse-transcription Polymerase Chain Reaction Detection
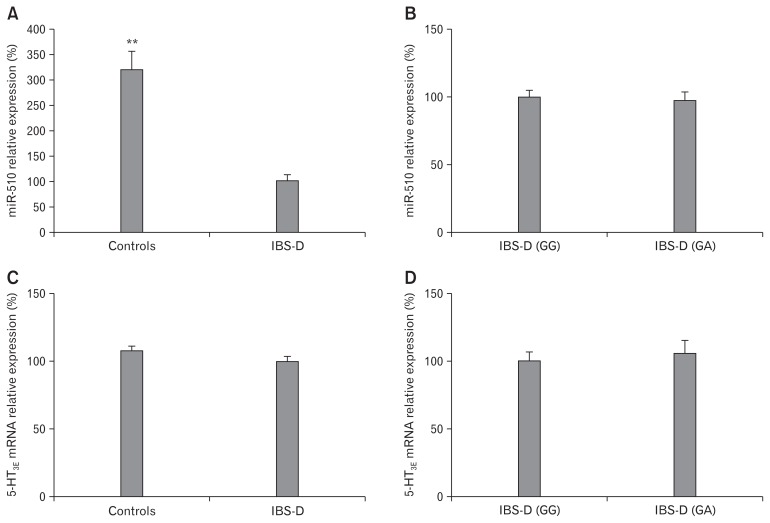
We detected miR-510 expression in the colonic mucosal tissues of 53 patients with IBS-D and 53 normal controls by using qRT-PCR. MicroRNA-510 expression was lower in the colonic mucosal tissues of patients with IBS-D as compared with normal controls (Fig. 3A). However, miR-510 expression in colonic mucosal tissues was not significantly different between patients with IBS-D and the GA genotype and patients with the GG genotype (Fig. 3B). The expression of 5-HT3E mRNA in tissue samples was also not significantly different between patients with IBS-D and normal controls (Fig. 3C) or between patients with IBS-D with the GA and GG genotypes (Fig. 3D) (P > 0.05).
Figure 3.
Relative expression of microRNA-510 (miR-510) in colonic mucosal tissues of diarrhea predominant irritable bowel syndrome (IBS-D) patients and controls. (A) The relative expression of miR-510 in colonic mucosal tissues of IBS-D patients (n = 53) or normal controls (n = 53) was determined using quantitative reverse-transcription polymerase chain reaction (qRT-PCR). (B) The relative expression of miR-510 in 45 IBS-D tissue with the GG genotype and 8 IBS-D tissue samples with the GA genotype. (C) Relative expression of serotonin receptor 3E (5-HT3E) mRNA in colonic mucosal tissues of IBS-D patients (n = 53) or normal controls (n = 53). (D) Relative expression of 5-HT3E mRNA in 45 IBS-D tissue with GG genotype and 8 IBS-D tissue samples with the GA genotype. The expression level of miR-510 was normalized against U6 small nuclear RNA. The expression level of 5-HT3E mRNA was normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. ** P < 0.01.
Serotonin Receptor 3E rs56109847 Disrupts the Binding Site for MicroRNA-510 and Significantly Upregulates Luciferase Expression in HEK293 and HT-29 Cells
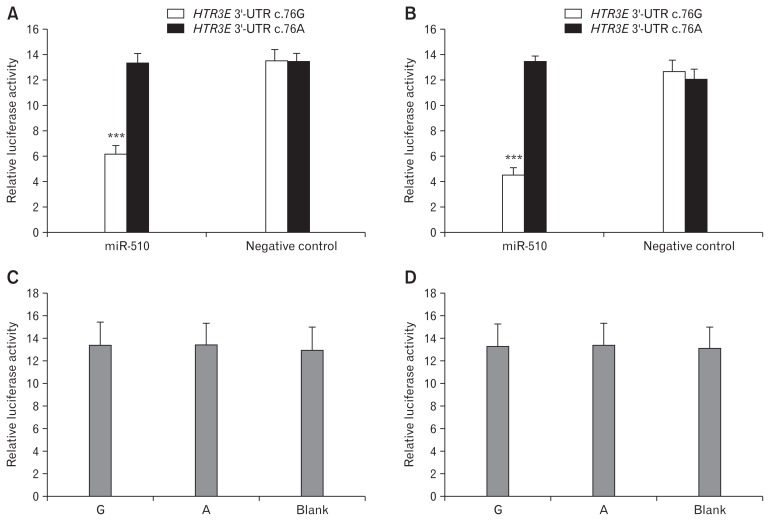
The HTR3E 3′-UTR variant c.*76G>A affected the binding of miR-510 to the HTR3E 3′-UTR, and the c.*76A variant constructs co-transfected with miR-510 showed significantly higher luciferase expression than the c.*76G allele constructs in HEK293 (Fig. 4A) and HT-29 (Fig. 4B) cells (P < 0.001). In both cell lines, luciferase activity was not significantly different when co-transfected with the negative controls (P > 0.05). When transfected with constructs containing the SNP (G/A) without miR-510 or negative control, no difference in luciferase activity was detected in both HEK293 and HT-29 cells (P > 0.05; Fig. 4C and Fig. 4D).
Figure 4.
Serotonin receptor 3E (HTR3E) variant rs56109847 significantly reduces binding and inhibitory effects of miR-510 in HEK293 and HT29 cells. (A) The HTR3E variant rs56109847 significantly reduces the binding and inhibitory effects of microRNA 510 (miR-510) in HEK293 cells (HTR3E 3′-UTR-c.*76G versus c.*76A, HTR3E 3′-UTR-c.*76G transfected with miR-510 versus with negative control). (B) The HTR3E variant rs56109847 significantly reduces the binding and inhibitory effects of miR-510 in HT29 cells (HTR3E 3′-UTR-c.*76G versus c.*76A, HTR3E 3′-UTR-c.*76G transfected with miR-510 versus with negative control). (C) No significant difference in the relative luciferase activity among pRL-TK–HTR3E 3′-UTR- c.*76G, pRL-TK–HTR3E 3′-UTR- c.*76A and blank EK293) (P > 0.05). (D) No significant difference in the relative luciferase activity among pRL-TK–HTR3E 3′-UTR-G, pRL-TK–HTR3E 3′-UTR-A and blank (HT29) (P > 0.05). Renilla luciferase (pRL-TK) activity was normalized against firefly luciferase (pGL3 control). Values are expressed as the mean ± SEM for 3 transfections, each measuring 3-fold. ***P < 0.001.
Discussion
In this study the SNP rs56109847 was identified to be linked with the diarrhea phenotype of IBS (IBS-D) in the South Chinese Han female population, and the functional variant of HTR3E 3′-UTR could inhibit the binding of miR-510 to HTR3E 3′-UTR. The 5-HT3E protein expression level in human colonic mucosal tissues was shown to be enhanced in colonic tissues of IBS-D patients. The 5-HT3E protein expression level was significantly higher in IBS-D patients with the GA genotype than patients with the GG genotype. The polymorphic allele resulted in high expression of 5-HT3E in colonic cell lines. This indicated that microRNAs which target on HTR3E 3′-UTR could be potentially used for IBS-D therapy.
High prevalence of overlap between FD and IBS-D has been consistently displayed both locally and internationally.17 The SNP rs56109847 has been also shown in significant association with female IBS-D in the United Kingdum population.12 However, we failed to demonstrate an association between HTR3E polymorphism and occurrence of FD that overlapped with IBS-D or IBS-M. Furthermore, there was no significant association of the SNP rs56109847 with FD alone or 2 subtypes of FD, which is called epigastric pain syndrome and postprandial distress syndrome. This indicates that pathogenic mechanisms of IBS-D may be different from other functional gastrointestinal disorders.
The 5-HT3 receptor consists of 5 subunits, including 5-HT3A, 5-HT3B, 5-HT3C, 5-HT3D, and 5-HT3E.15,21,22 Only 5-HT3E is exclusively expressed in GI tissues, both colons and small intestines.15 Although the occurrence of IBS-D is associated with multiple genetic polymorphisms, 5-HT3E may play an important and specific role in the GI tract. SNPs residing in miRNA-binding sites affect expression of target mRNA and contribute to susceptibility of a lot of human diseases, such as cancer, kidney disease, and cognitive disorders.23–26 The enhanced level of gastrointestinal miR-29a expression increased intestinal permeability via down-regulation of target genes in patients with IBS-D.27,28 Here the 5-HT3E rs56109847 could directly inhibit the binding of miR-510 to HTR3E 3′-UTR in HEK293 and HT-29 cells confirmed by luciferase assays. This confirmed that the SNP (rs56109847) of the non-coding region of HTR3E affected the binding of mircoRNA, thus affecting the activity of the GI tract.
5-HT3E is abundantly expressed in colonic mucosal glands, but with less expression level in the stroma. The 5-HT3E protein expression level was significantly higher in IBS-D patients with the GA genotype than in patients with the GG genotype. However, there was no significant difference of miR-510 and 5-HT3E mRNA expression in colonic mucosal tissues in neither the GA nor GG genotype of IBS-D patients. This demonstrates that less binding of miR-510 to the 5-HT3E mRNA contributes to high expression of 5-HT3E protein in the GA genotype. In addition, it is still not clear why miR-510 was down-regulated in colonic mucosal tissues of IBS-D patients compared with controls, which may be associated with local chronic inflammation.
In conclusion, our studies confirm that the SNP rs56109847 is a susceptibility genetic polymorphism for the development of IBS-D in the South Chinese Han female population, and provide insights into the regulation of miR-510 on 5-HT3E expression of colonic tissues in patients with gastrointestinal disorders. The molecular mechanism that underlies the association of HTR3E SNP rs56109847 with IBS-D is also elucidated. Down-regulated expression of miR-510 in colonic mucosa would be a marker of the increased risk for developing IBS-D.
Supplementary Materials
Note: To access the supplementary figure mentioned in this article, visit the online version of Journal of Neurogastroenterology and Motility at http://www.jnmjournal.org/, and at http://dx.doi.org/10.5056/jnm15138.
Acknowlegements
Editorial support was provided by School of Medicine, Yangzhou University. The first author thanks all Yang-zhou women who participated in this study and the research staff Yaoyao Li, Zhenfeng Hao. The first author thanks Weijuan Gong especially for her support in the modification process.
Footnotes
Financial support: This study was supported by funding obtained from Jiangsu province, P.R. China (Grant No. 14KJD330004).
Conflicts of interest: None.
Author contributions: Ping Bo, Weijuan Gong, and Xiang-ming Li designed the research study; Yu Zhang, Yaoyao Li, and Zhenfeng Hao performed the experiment; Yu Zhang and Xiang-ming Li analyzed the data; and Yu Zhang drafted the manuscript.
References
- 1.Futagami S, Yamawaki H, Shimpuku M, et al. Impact of coexisting irritable bowel syndrome and non-erosive reflux disease on postprandial abdominal fullness and sleep disorders in functional dyspepsia. J Nippon Med Sch. 2013;80:362–370. doi: 10.1272/jnms.80.362. [DOI] [PubMed] [Google Scholar]
- 2.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 3.Tack J, Piessevaux H, Coulie B, Caenepeel P, Janssens J. Role of impaired gastric accommodation to a meal in functional dyspepsia. Gastroenterology. 1998;115:1346–1352. doi: 10.1016/S0016-5085(98)70012-5. [DOI] [PubMed] [Google Scholar]
- 4.Holtmann G, Goebell H, Jockenhoevel F, Talley NJ. Altered vagal and intestinal mechanosensory function in chronic unexplained dyspepsia. Gut. 1998;42:501–506. doi: 10.1136/gut.42.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nourrisson C, Scanzi J, Pereira B, et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS One. 2014;9:e111868. doi: 10.1371/journal.pone.0111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Z, Tang H. Decreased neuroplasticity may play a role in irritable bowel syndrome: implication from the comorbidity of depression and irritable bowel syndrome. J Neurogastroenterol Motil. 2015;21:298–299. doi: 10.5056/jnm14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arisawa T, Tahara T, Shiroeda H, et al. Genetic polymorphisms of SCN10A are associated with functional dyspepsia in Japanese subjects. J Gastroenterol. 2013;48:73–80. doi: 10.1007/s00535-012-0602-3. [DOI] [PubMed] [Google Scholar]
- 8.Bengtson MB, Rønning T, Vatn MH, Harris JR. Irritable bowel syndrome in twins: genes and environment. Gut. 2006;55:1754–1759. doi: 10.1136/gut.2006.097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Nie Y, Li Y, Zhang L. Association of cannabinoid type 1 receptor and fatty acid amide hydrolase genetic polymorphisms in Chinese patients with irritable bowel syndrome. J Gastroenterol Hepatol. 2014;29:1186–1191. doi: 10.1111/jgh.12513. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fourie NH, Peace RM, Abey SK, et al. Elevated circulating miR-150 and miR-342-3p in patients with irritable bowel syndrome. Exp Mol Pathol. 2014;96:422–425. doi: 10.1016/j.yexmp.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapeller J, Houghton LA, Mönnikes H, et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–2977. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- 13.Arisawa T, Tahara T, Fukuyama T, et al. Genetic polymorphism of pri-microRNA 325, targeting SLC6A4 3′-UTR, is closely associated with the risk of functional dyspepsia in Japan. J Gastroenterol. 2012;47:1091–1098. doi: 10.1007/s00535-012-0576-1. [DOI] [PubMed] [Google Scholar]
- 14.Gershon MD. Nerves, reflexes, and the enteric nervous system: pathogenesis of the irritable bowel syndrome. J Clin Gastroenterol. 2005;39(suppl 3):S184–S193. doi: 10.1097/01.mcg.0000156403.37240.30. [DOI] [PubMed] [Google Scholar]
- 15.Niesler B, Frank B, Kapeller J, Rappold GA. Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E. Gene. 2003;310:101–111. doi: 10.1016/S0378-1119(03)00503-1. [DOI] [PubMed] [Google Scholar]
- 16.Karnovsky AM, Gotow LF, McKinley DD, et al. A cluster of novel serotonin receptor 3-like genes on human chromosome 3. Gene. 2003;319:137–148. doi: 10.1016/S0378-1119(03)00803-5. [DOI] [PubMed] [Google Scholar]
- 17.Si JM, Wang LJ, Chen SJ, Sun LM, Dai N. Irritable bowel syndrome consulters in Zhejiang province: the symptoms pattern, predominant bowel habit subgroups and quality of life. World J Gastroenterol. 2004;10:1059–1064. doi: 10.3748/wjg.v10.i7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gwee KA, Chua AS. Functional dyspepsia and irritable bowel syndrome, are they different entities and does it matter? World J Gastroenterol. 2006;12:2708–2712. doi: 10.3748/wjg.v12.i17.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara Y, Kubo M, Kohata Y, et al. Cigarette smoking and its association with overlapping gastroesophageal reflux disease, functional dyspepsia, or irritable bowel syndrome. Intern Med. 2011;50:2443–2447. doi: 10.2169/internalmedicine.50.6012. [DOI] [PubMed] [Google Scholar]
- 20.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies PA, Pistis M, Hanna MC, et al. The 5-HT3B subunit is a major determinant of serotonin-receptor function. Nature. 1999;397:359–363. doi: 10.1038/16941. [DOI] [PubMed] [Google Scholar]
- 22.Miyake A, Mochizuki S, Takemoto Y, Akuzawa S. Molecular cloning of human 5-hydroxytryptamine3 receptor: heterogeneity in distribution and function among species. Mol Pharmacol. 1995;48:407–416. [PubMed] [Google Scholar]
- 23.Feng N, Xu B, Tao J, et al. A miR-125b binding site polymorphism in bone morphogenetic protein membrane receptor type IB gene and prostate cancer risk in China. Mol Biol Rep. 2012;39:369–373. doi: 10.1007/s11033-011-0747-9. [DOI] [PubMed] [Google Scholar]
- 24.Yang P, Tang R, Zhu J, et al. A functional variant at miR-24 binding site in B7-H2 alters susceptibility to gastric cancer in a Chinese Han population. Mol Immunol. 2013;56:98–103. doi: 10.1016/j.molimm.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Papagregoriou G, Erguler K, Dweep H, et al. A miR-1207-5p binding site polymorphism abolishes regulation of HBEGF and is associated with disease severity in CFHR5 nephropathy. PLoS One. 2012;7:e31021. doi: 10.1371/journal.pone.0031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Zhang R, Li M, et al. A functional MiR-124 binding-site polymorphism in IQGAP1 affects human cognitive performance. PLoS One. 2014;9:e107065. doi: 10.1371/journal.pone.0107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Q, Souba WW, Croce CM, Verne GN. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59:775–784. doi: 10.1136/gut.2009.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q, Costinean S, Croce CM, et al. MicroRNA 29 targets nuclear factor-κB–repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology. 2015;148:158–169. doi: 10.1053/j.gastro.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: To access the supplementary figure mentioned in this article, visit the online version of Journal of Neurogastroenterology and Motility at http://www.jnmjournal.org/, and at http://dx.doi.org/10.5056/jnm15138.