Summary
Homeostasis of short-lived blood cells is dependent on rapid proliferation of immature precursors. Using a conditional histone 2B-mCherry-labeling mouse model, we characterize hematopoietic stem cell (HSC) and progenitor proliferation dynamics in steady state and following several types of induced stress. HSC proliferation following HSC transplantation into lethally irradiated mice is fundamentally different not only from native hematopoiesis but also from other stress contexts. Whereas transplantation promoted sustained, long-term proliferation of HSCs, both cytokine-induced mobilization and acute depletion of selected blood cell lineages elicited very limited recruitment of HSCs to the proliferative pool. By coupling mCherry-based analysis of proliferation history with multiplex gene expression analyses on single cells, we have found that HSCs can be stratified into four distinct subtypes. These subtypes have distinct molecular signatures and differ significantly in their reconstitution potentials, showcasing the power of tracking proliferation history when resolving functional heterogeneity of HSCs.
Graphical Abstract

Highlights
-
•
Histone 2B-mCherry labeling enables cell division tracking in vivo
-
•
Stressors enforce proliferation of MPPs rather than HSCs
-
•
Transplantation enforces sustained HSC proliferation
-
•
Mitotic history allows for functional separation of candidate HSCs
Säwén et al. apply histone 2B-mCherry labeling to study proliferation dynamics in normal and perturbed hematopoiesis. Of several stressors evaluated, transplantation was unique in enforcing sustained stem cell proliferation. They also demonstrate the utility of histone 2B labeling to separate candidate stem cells into subpopulations with distinct molecular and functional characteristics.
Introduction
As most mature blood cells are short-lived, they are in need of continuous replacement to ensure a sufficient capacity of the hematopoietic system. Hematopoiesis is therefore characterized by vigorous proliferation, although magnitudes differ depending on the developmental stages at which defined progenitors reside (Passegué et al., 2005). Historically, it has been argued that hematopoietic stem cells (HSCs) are critically responsible for the maintenance of homeostasis within the hematopoietic system (Bryder et al., 2006), a presumption which is largely based on HSCs residing at the apex of the hematopoietic hierarchy, their multipotency, and their extensive longevity/self-renewal. Importantly, however, these features have been predominantly defined by transplantation experiments.
In clinical hematopoietic stem and progenitor cell (HSPC) transplantations, patients are commonly conditioned with myeloablative chemotherapy and/or irradiation before receiving a graft, with HSPCs to be used for transplantation typically harvested from donors following cytokine-induced mobilization. Challenges in assessing HSC quality and quantity in humans preclude assessment of how such therapeutic regimens influence HSC properties and functional potential both short- and long-term post-transplantation. This might be particularly relevant for the transplantation setting, in which HSCs are subjected to very high and arguably abnormal proliferation pressures that adult HSCs under physiological conditions are not exposed to.
Initial indications that proliferative status might be an important determinant for the functional capacity of HSC were obtained from transplantation studies in which bone marrow (BM) cells in active cell cycle, and enriched for HSC activity, displayed a diminished ability to rescue lethally irradiated hosts (Fleming et al., 1993). Later, more refined HSC enrichment strategies confirmed that adult HSCs are normally residing in the G0/G1 phase of the cell cycle (Cheshier et al., 1999, Morrison and Weissman, 1994, Morrison et al., 1997), with transplantation experiments revealing a sharp reduction in the reconstitution capacity of candidate and actively cycling HSCs (Glimm et al., 2000, Habibian et al., 1998, Nygren et al., 2006, Orschell-Traycoff et al., 2000). With this said, fetal liver HSCs, which are known to actively cycle, are nonetheless much more potent than adult HSCs in a transplantation setting (Jordan et al., 1995, Rebel et al., 1996a, Rebel et al., 1996b). In addition, convincing demonstrations that HSCs in active cell cycle can be reverted to a G0 state, with a robust regain in their reconstitution potential, are still lacking (Nygren et al., 2006). Therefore, when caught in active cell cycle, candidate HSCs might predominantly represent cells that have permanently lost their key HSC properties (Qiu et al., 2014). This might be particularly relevant for cell populations that cycle infrequently and where very few cycling cells can be obtained at a given moment in time. For such populations, it might be more feasible, or at least complementary, to study cell function from the perspective of their proliferative history (Foudi et al., 2009, Qiu et al., 2014, Wilson et al., 2008).
Recent studies have provided evidence that the contribution of HSCs to native hematopoiesis might be fundamentally different from that observed following transplantation (Busch et al., 2015, Sun et al., 2014). Experimental systems that allow for evaluation in steady state are therefore crucial to gain a thorough understanding of normal hematopoiesis. Recent adaptations and developments of histone 2B (H2B) fusion protein labeling systems (Foudi et al., 2009, Qiu et al., 2014, Wilson et al., 2008) have overcome many of the problems associated with earlier techniques to probe HSC proliferation in vivo (Cheshier et al., 1999, Kiel et al., 2007, Nygren and Bryder, 2008, Sudo et al., 2000, Takizawa et al., 2011) and allow for long-term evaluation of proliferation dynamics in a truly native setting (Foudi et al., 2009, Wilson et al., 2008). We therefore here made use of a doxycycline-inducible H2B-mCherry-labeling system (Egli et al., 2007) to investigate the proliferative responses of HSPCs following a range of pressures inflicted on the hematopoietic system, including transplantation, mobilization, and in vivo depletion of selected blood cell lineages.
Results
An Inducible H2B-mCherry System to Study Native Proliferation Dynamics within the Hematopoietic System
Our aim in this study was to explore the proliferation dynamics of HSPCs in steady state. For this purpose, we made use of a transgenic tet-ON mouse model that, upon doxycycline (DOX) administration, allows for expression of a H2B-mCherry (H2B-mCherry) fusion protein from the ubiquitously expressed Col1a1 locus (Col1a1-tetO-H2B-mCherry mice; Figure 1A; Egli et al., 2007). Following a pulse of H2B-mCherry induction, with concomitant incorporation of the fluorescent protein into nucleosomes, H2B-mCherry is divided between daughter cells upon proliferation (Figure 1B). As previously shown for H2B-GFP (Foudi et al., 2009), this permits measurement of H2B-mCherry fluorescence intensity to be correlated to proliferative history. We confirmed strong and inducible H2B-mCherry mRNA expression in homozygous Col1a1-tetO-H2B-mCherry mice after DOX administration, with minimal promiscuous expression 1 week after DOX withdrawal (Figure 1C).
Figure 1.
Description and Functional Validation of an Inducible H2B-mCherry Labeling Mouse Model
(A) Schematic representation of the Col1a1-tetO-H2B-mCherry model. Expression of the M2 reverse tetracycline transactivator (M2-rtTA) is driven by the Rosa26 promoter with H2B-mCherry positioned in the 3′ UTR of the Col1a1 gene under control of a tetracycline operon (TetOP). pA, polyadenylation signal; SA, splice acceptor.
(B) Schematic outline depicting the principle of H2B-mCherry labeling and label dilution using the Col1a1-tetO-H2B-mCherry system.
(C) H2B-mCherry mRNA expression relative to β-actin in HSCs and myeloid progenitors following a 7-day pulse and 7 days chase.
(D) Representative FACS histograms showing H2B-mCherry label in HSCs from heterozygous and homozygous Col1a1-tetO-H2B-mCherry mice fed for 1 week with “standard” (blue line; 200 mg/kg) or high (red filled histograms; 2 g/kg) DOX food pellets.
(E) H2B-mCherry label in progenitors downstream of HSCs following 1-week administration of high-concentration DOX followed by no or 7 days chase. Grey histograms depict mCherry expression in HSPCs from non-induced Col1a1-tetO-H2B-mCherry mice.
(F) Fgd5-ZsGreen expression and H2B-mCherry label retention in HSC, MPP, and GMLP subsets isolated from Col1a1-tetO-H2B-mCherry-Fgd5-ZsGreen mice following H2B-mCherry labeling and 4 weeks chase. Red dashed line indicates the boundary for ZsGreen positivity.
To clarify to what extent HSPCs in Col1a1-tetO-H2B-mCherry mice are labeled by H2B-mCherry protein after DOX administration, we investigated the influence of genetic dose on label uptake in HSCs by comparing labeling in mice heterozygous or homozygous for the H2B-mCherry transgene (and rtTA). We also evaluated the influence of DOX concentration provided to animals. H2B-mCherry label in the HSC compartment was analyzed by flow cytometry 1 week following DOX administration (Figure 1D), revealing that HSCs in homozygous animals were highly and uniformly labeled, with a slightly higher label observed upon administration of more DOX (2 g/kg versus 200 mg/kg). The beneficial effect of higher DOX concentration on H2B-mCherry uptake in HSCs was more prominent in heterozygous animals (Figure 1D). Initial H2B-mCherry label in other primitive progenitor subsets was also analyzed (Figure 1E). Multipotent progenitors (MPPs) (also called short-term HSCs; Lineage−Sca1+c-kit+CD48−CD150− cells; Kiel et al., 2005, Kiel et al., 2008, Pronk et al., 2007) displayed a uniform initial label, strikingly similar to the initial label of HSCs, whereas granulocyte/macrophage/lymphoid progenitors (GMLPs) (Lineage−Sca1+c-kit+CD48+CD150−) and myeloid progenitors (MyP) (Lineage−Sca1−c-kit+CD150−) were more heterogeneously labeled. This likely reflects higher proliferation rates in these compartments (compared to HSCs and MPPs) during the labeling period, an interpretation supported by analyses of label dilution after 7 days chase (Figure 1E). Finally, to verify our results using an alternative approach for cell identification, we generated a double reporter strain by crossing H2B-mCherry mice with Fgd5-ZsGreen mice, which specifically mark the most-primitive HSPC compartment (Gazit et al., 2014). Following H2B-mCherry labeling and 4 weeks chase, the highest H2B-mCherry label retention was observed in HSC and MPP subsets, with highest Fgd5-ZsGreen expression and concomitant mCherry labeling observed in HSCs. By contrast, GMLPs were essentially negative for the Fgd5 reporter and retained less H2B-mCherry label (Figure 1F). The compatibility of the inducible H2B-mCherry mice with existing GFP reporter mouse lines illustrates an additional advantage of this particular model system.
The rapid labeling observed in homozygous animals compared to previously described H2B-labeling strategies (Foudi et al., 2009, Qiu et al., 2014, Wilson et al., 2008) should be advantageous, as less proliferation-associated label dilution during a shorter labeling period results in a more-uniform initial H2B-mCherry label. We therefore decided to use homozygous animals in combination with high-dose DOX for all subsequent experiments.
Long-Term H2B-mCherry Label Retention
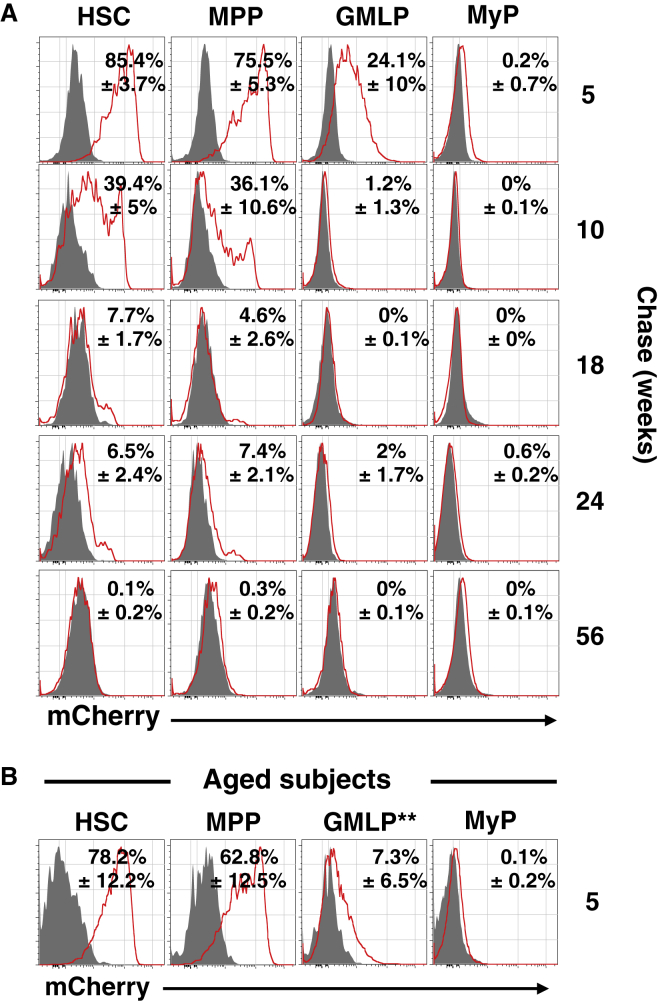
To analyze steady-state proliferation in HSPCs, cohorts of Col1a1-tetO-H2B-mCherry mice were administered DOX for 1 week, followed by analysis of label dilution in HSPCs after different periods of chase (Figure 2A). After 5 weeks chase, the majority of HSCs and MPPs retained high levels of H2B-mCherry label. By contrast, most GMLPs and MyPs had retained substantially less H2B-mCherry label. This confirmed a higher proliferation rate of MyPs in steady state (Nygren and Bryder, 2008), with GMLPs having an in vivo proliferation rate “intermediate” to that of HSCs/MPPs and MyPs. After 10 weeks chase, GMLPs and MyPs had lost all H2B-mCherry label, whereas a large fraction of MPPs and HSCs retained H2B-mCherry label. Following 18 and 24 weeks of chase, a small fraction of HSCs and MPPs still retained H2B-mCherry fluorescence, whereas none of the investigated progenitor subsets displayed any detectable label retention after 56 weeks of chase. To analyze steady-state proliferation in aged mice, a cohort of 16-month-old Col1a1-tetO-H2B-mCherry mice was administered DOX for 1 week followed by 5 weeks of chase (Figure 2B). Whereas no significant difference in proliferation of HSCs or MPPs could be observed as a consequence of age, GMLPs from old mice had lost significantly more H2B-mCherry label compared to GMLPs in young mice. Collectively, these results confirmed the quiescent nature of HSCs (Cheshier et al., 1999, Foudi et al., 2009, Wilson et al., 2008) both in young and aged mice and demonstrated that MPPs are as quiescent as HSCs in steady state. By contrast, aging associated with a higher steady-state proliferation rate of GMLPs.
Figure 2.
Long-Term but Finite H2B-mCherry Label Retention in HSCs
(A) Representative FACS histograms of H2B-mCherry label in HSPCs after indicated chase periods. Grey histograms depict mCherry expression in HSPCs from non-induced Col1a1-tetO-H2B-mCherry mice. Red histograms denote H2B-mCherry label in HSPCs pulsed with DOX for 1 week and analyzed following indicated periods of chase. Numbers indicate the mean % mCherry-positive cells ± SD over unlabeled controls of the indicated populations. (n = 7 at 5 weeks, 12 at 10 weeks, 3 at 18 weeks, 7 at 24 weeks, and 7 at 56 weeks).
(B) Representative histograms depicting H2B-mCherry label in HSPCs isolated from 17-month-old Col1a1-tetO-H2B-mCherry mice, pulsed with DOX for 1 week, followed by 5 weeks chase (n = 6). Asterisks indicate a significant difference when comparing the % of mCherry-positive cells over unlabeled controls in the indicated population isolated from young and old mice.
Transplanted HSCs Maintain Higher Proliferation Rates Long-Term Post-transplantation
The most commonly used assay to evaluate HSC function is by transplantation, a regimen that is also in routine clinical practice for various hematological conditions (Gratwohl et al., 2010). To investigate how HSC proliferation is affected by transplantation and whether the transplantation dose might influence these processes, we isolated BM cells from unlabeled Col1a1-tetO-H2B-mCherry mice and transplanted two doses of cells (3 × 105 and 1 × 107 cells) into lethally irradiated mice. As an indicator of hematopoietic recovery, we monitored peripheral blood values over time using automated cell counting. This revealed that white blood cell counts in all recipients were similar to pre-transplantation values 16–18 weeks after transplantation (data not shown). Upon hematopoietic recovery, we induced H2B-mCherry, followed by a 4-week chase period. Thereafter, recipients were sacrificed and H2B-mCherry label in donor HSCs analyzed and compared to age- and sex-matched non-transplanted control mice (Figures 3 and S1). Strikingly, the transplanted HSCs from both cell doses exhibited significant reductions in mCherry levels compared to the untransplanted controls, demonstrating that the transplanted HSCs maintained a higher proliferation rate even 4 months post-transplant. Interestingly, a trend toward greater H2B-mCherry label dilution was observed in the HSCs isolated from recipients transplanted with a lower cell dose, though this did not reach statistical significance. These data indicate that transplantation of HSCs over a wide cell dose range greatly influences the long-term proliferation dynamics of HSCs in the BM.
Figure 3.
The Long-Term Effect of Transplantation on Hematopoietic Stem and Progenitor Cell Proliferation
Two experiments where 3 × 105 or 1 × 107 unfractionated bone marrow (UBM) cells from Col1a1-tetO-H2B-mCherry mice were transplanted into lethally irradiated wild-type hosts were performed. After stable reconstitution of the blood system (16 and 18 weeks after transfer), all recipients were given DOX for 1 week followed by 4 weeks chase before analysis. Age- and sex-matched (to the donor cells) non-transplanted control mice were induced and analyzed simultaneously. Secondary transplantation was performed using 3 × 106 UBM cells together with previously transplanted competitor cells. Sixteen weeks post-secondary transplantation, DOX were given to recipients followed by 4 weeks chase and H2B-mCherry label retention analysis of donor HSCs. Displayed are mCherry MFI in HSCs normalized to the mean MFI in HSCs of controls for each experiment, with error bars denoting the mean ± SD. n = 13 controls, 12 primary recipients receiving 3 × 105 cells, 11 primary recipients of 1 × 107 cells, 4 secondary recipients from primary recipients receiving 1 × 107 cells, and 14 secondary recipients from primary recipients transplanted with 3 × 105 cells.
See also Figure S1.
Serial transplantation is routinely used to evaluate HSC self-renewal. To assess how such procedures impact HSC turnover rates, we transplanted BM from a cohort of primary recipients into secondary recipients, followed by H2B-mCherry labeling and chase after stable blood cell reconstitution was reached. Analysis of HSCs from these secondary recipients revealed an even greater increase in HSC proliferation rates than we had observed in the primary recipients (Figures 3 and S1). No detectable differences in HSC proliferation rates as a consequence of the initial transplantation dose were observed.
Mobilization Depletes Multipotent Progenitors from the Bone Marrow and Increases Proliferation of Hematopoietic Stem and Progenitor Cells
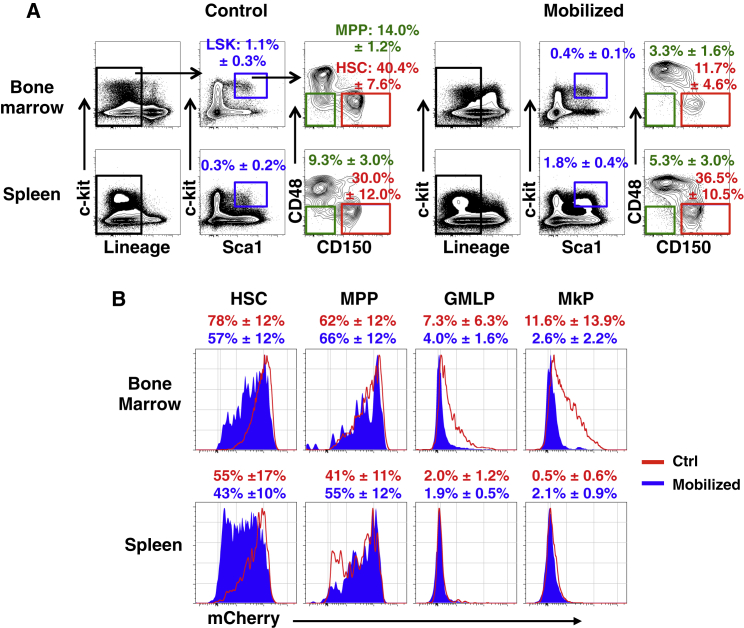
As induced mobilization of HSPCs has been suggested to be interlinked with proliferation (Morrison et al., 1997), we next set out to investigate this question using the Col1a1-tetO-H2B-mCherry system. We chose to use a mobilization protocol where an initial injection of cyclophosphamide (Cy) is combined with multiple injections of granulocyte-colony stimulating factor (G-CSF), which mimics protocols regularly used to mobilize hematopoietic progenitors in humans before transplantation (Milone et al., 2003). Previously induced Col1a1-tetO-H2B-mCherry mice were injected with Cy and then once per day for 5 days with G-CSF. The day after the last G-CSF injection, we analyzed HSPCs in spleen and BM. As expected, both the frequency and absolute numbers of the Lineage−Sca1+ckit+ HSPCs were reduced in the BM of mobilized mice (Figure 4A; data not shown), which was accompanied by a 6-fold increase in the frequency of these cells in spleen (Figure 4A). Somewhat surprisingly, mobilizations caused a selective and almost complete depletion of MPPs from the BM (Figure 4A). H2B-mCherry label retention in mobilized mice revealed enhanced proliferation in BM megakaryocyte progenitors (MkP) and GMLPs. For HSCs, mobilization moderately increased proliferation in the BM, which was even more pronounced for splenic HSCs (Figure 4B). Finally, comparing H2B-mCherry label retention in HSPCs from spleen and BM in control (non-mobilized) mice revealed that splenic HSPCs, and in particular the more mature GMLP and MkP subsets, had undergone more proliferation than their BM counterparts in steady state.
Figure 4.
HSPC Mobilization by Cy and G-CSF Depletes Multipotent Progenitors and Induces Proliferation in Bone Marrow and Splenic HSPCs
Labeled Col1a1-tetO-H2B-mCherry mice were chased for 4 weeks before Cy/G-CSF mobilization. One day after the last G-CSF injection, mice were analyzed and compared to age- and sex-matched non-mobilized animals.
(A) (Top) FACS plots from BM obtained from control and mobilized mice, showing depletion of LSKCD150−CD48− (MPP) cells following mobilization. Numbers depict the mean % of Lineage−Sca1+ckit+ (LSK) cells ± SD (blue box) and the frequency of MPP (green box) and HSC (red box) subsets of LSK cells. (Bottom) Similar analysis as in top panels but conducted on spleen cells is shown.
(B) H2B-mCherry label retention in BM and spleen from mobilized and control animals were compared in the indicated stem and progenitor populations. Red and blue histograms depict H2B-mCherry label in the indicated population from a representative control and from a mobilized animal, respectively. Numbers depict % mCherry-positive ± SD over unlabeled controls. A total of six control and seven mobilized animals were analyzed.
The Influence of Acute Depletion of Selected Subsets of Mature Cells on Hematopoietic Stem and Progenitor Cell Proliferation
We next decided to evaluate proliferation dynamics of HSPCs following selected depletion and subsequent recovery of mature blood cells, a more defined type of hematopoietic stress. We administered antibodies in vivo to deplete B cells (Keren et al., 2011) or Gr1+ myeloid cells (Tepper et al., 1992). Analysis of peripheral blood cells 3 days after depletion showed a nearly complete elimination of cells of the respective lineages (Figures 5A and 5C), confirming the efficacy of the depletion strategies. Next, B cells or Gr1+ cells were depleted from induced Col1a1-tetO-H2B-mCherry mice followed by H2B-mCherry label retention analysis. Despite the highly efficient B cell depletion, we could not observe any changes in proliferation of HSCs, MPPs, or GMLPs 14 days post-depletion (Figure 5B), suggesting that more mature progenitors are responsible for the rebound of the B cell lineage after its acute depletion. Gr1+ depletion slightly increased proliferation of GMLPs during the first 10 days after depletion, whereas HSCs and MPPs were unaffected (Figure 5D). We noted that increased proliferation had occurred in MPPs 5 weeks post-Gr1+ depletion, suggesting that a cellular feedback induced by Gr1+ depletion had led to increased proliferation of a subset of MPPs. However, this did not extend to the HSC compartment (Figure 5D). These results established that myeloid cell replenishment recruits more immature progenitors to proliferate, compared to B cell replenishment, although HSC proliferation was for the most part unaffected by either type of depletion.
Figure 5.
The Effects of Mature Cell Depletion on Hematopoietic Stem and Progenitor Cell Proliferation
(A) FACS plots depicting peripheral blood CD19+ cells before and 3 days after in vivo antibody mediated depletion of B cells. Numbers depict the mean % CD19+ cells of all white blood cells ± SD (n = 3).
(B) Representative FACS histograms of H2B-mCherry label 2 weeks after B cell depletion.
(C) FACS plots depicting peripheral blood Gr1+ cells before and 3 days after in vivo antibody-mediated Gr1+ cell depletion. Arrows indicate the gating hierarchy. Numbers depict the mean % of cells within the gates ± SD (n = 4).
(D) Representative FACS histograms of H2B-mCherry label retention 10 days and 5 weeks after Gr1+ cell depletion (two experiments were performed; n = 8 at 10 days; n = 11 at 5 weeks).
Proliferation History Reveals Unique Molecular Characteristics of HSCs
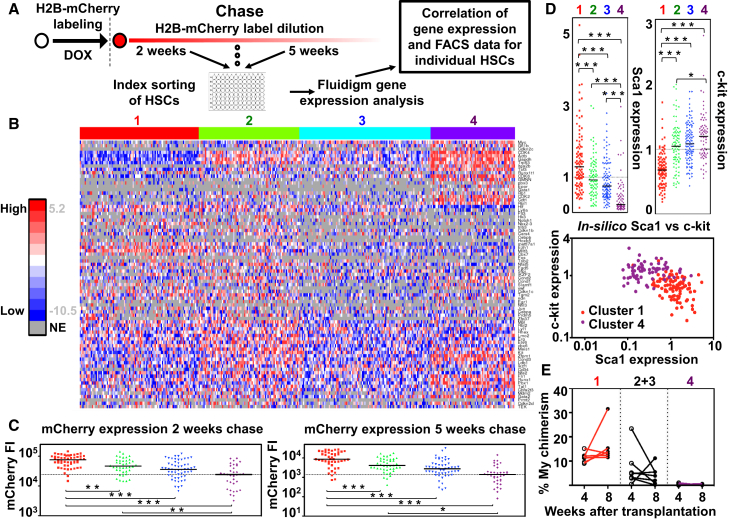
Whereas the initial H2B-mCherry label is uniform in HSCs (Figure 1D), HSCs retain different levels of label following chase, which presumably could reflect differences in the molecular properties of individual HSCs. To investigate this, we decided to study how proliferation history relates to gene expression in individual HSCs. We therefore next performed indexed fluorescence-activated cell sorting (FACS) of HSCs from labeled Col1a1-tetO-H2B-mCherry mice after 2 or 5 weeks chase (experimental outline described in Figure 6A), a strategy that facilitates evaluation of the proliferative history of each index-sorted HSC. Sorted HSCs were next subjected to gene expression analysis (Table S1) of a panel of genes including transcription factors, cytokine receptors, surface markers, and cell-cycle-related genes obtained from previous studies (Moignard et al., 2013, Qiu et al., 2014, Seita et al., 2012).
Figure 6.
Single-Cell Gene Expression Analysis Combined with Index Sorting of HSCs Identifies Groups of HSCs with Different Proliferation Histories and Reconstitution Potential
(A) Experimental approach.
(B) Heatmap depicting the gene expression of 80 genes (rows) in 355 single HSCs (columns) from labeled Col1a1-tetO-H2B-mCherry mice after random forest clustering.
(C) Fluorescence intensity (FI) for mCherry in individual HSCs grouping to the clusters identified in (B). HSCs that were isolated after 2 (left) or 5 (right) weeks chase are shown in separate diagrams.
(D) (Top) Normalized expression of c-kit and Sca1 in individual HSCs of the identified clusters. (Bottom) In silico plot of Sca1 versus c-kit of candidate HSCs in clusters 1 and 4 is shown.
(E) One hundred HSCs (CD45.1+), sorted using Sca1 and c-kit gates based on the FACS index data from HSCs in the indicated cluster, were transplanted competitively into lethally irradiated WT recipients (CD45.2+). PB was analyzed for myeloid contribution at 4 and 8 weeks after transplantation (n = 6 for recipients of cluster 1 HSCs, 7 for recipients of cluster 2+3 HSCs, and 7 for recipients of cluster 4 HSCs). Cells in diagrams are grouped and colored according to which cluster they grouped to. Black lines indicate the median expression for cells in the indicated clusters.
See also Table S1.
To establish how the gene expression patterns obtained related to proliferative history of individual HSCs, we first performed random forest clustering using expression data from genes expressed in more than 20% of all candidate HSCs (Figure 6B). This allowed us to identify four clusters of cells. We next probed the H2B-mCherry index data from individual HSCs to the cluster they grouped with, which revealed a clear difference in the proliferative history between HSCs belonging to the different clusters (Figure 6C) both after 2 and 5 weeks chase. Investigating the data further for potential correlations to other surface markers used to sort HSCs, we could furthermore observe clear and differential expression of both Sca1 and c-kit between the different clusters (Figure 6D, top). HSCs with a more restricted proliferative history displayed higher Sca1 expression and lower c-kit expression when compared to HSCs located in clusters that had proliferated more extensively (Figure 6D, bottom). Finally, to functionally evaluate the HSC activity of cells within the identified clusters, we undertook transplantation experiments of cells prospectively isolated based on their differential Sca1 and c-kit expression. This revealed that Sca1highc-kitlow candidate HSCs (cluster 1) presented with a more robust reconstitution potential compared to cluster 2 and 3 cells. Reconstitution from cluster 2 and 3 cells also appeared to decrease rapidly over time (Figure 6E). Strikingly, HSCs belonging to cluster 4 were almost completely devoid of reconstitution potential.
Taken together, these data established that proliferation history can be an effective marker for further separation of candidate HSCs with distinct molecular and functional features and, as such, illuminates the clear correlation between HSC differentiation and proliferation.
Discussion
With the introduction of transgenic H2B-labeling models in hematopoietic research, functional comparisons between HSCs with differential proliferative histories have revealed that HSCs that harbor the highest reconstitution capacity after transplantation are found within the most slowly cycling cells (Foudi et al., 2009, Wilson et al., 2008). Although convincing, we noted a lack of several experimental details relating to parameters that influence on H2B-fusion protein expression but also dilution kinetics, as rather limited numbers of mice were analyzed in previous studies (Foudi et al., 2009).
Using an inducible H2B-mCherry labeling system, we demonstrate that all HSCs can be highly labeled after only 1 week of induction and that the Tet ON system used here (analogous to the work of Foudi et al.) is particularly appropriate, given the strict requirement of DOX for transgene expression. In this regard, Tet OFF systems should at least in theory be more vulnerable to experimental artifacts, as they by contrast require continuous DOX intake during extensive time for continuous repression of the transgene. The very short labeling period, compared to the turnover in the HSC compartment, excludes dependence on proliferation for labeling and reaffirms that H2B labeling to a large extent is driven by histone exchange (Kimura and Cook, 2001) that involves the transiently expressed H2B-mCherry. In addition to the obvious benefits of reduced experimental times and costs, swift labeling is advantageous because proliferation during the “pulse” period can be minimized, thereby inducing a more-homogeneous initial label. This was clearly illustrated by comparing the initial label intensity in highly proliferative progenitors to the slowly cycling HSCs, which revealed that initial label is less homogenous in progenitors compared to HSCs due to the proliferation-associated label dilution during the pulse period. We did note a minor degree of spontaneous H2B label dilution during extended chase periods, which might limit very long-term analysis of deeply quiescent populations and potentially might obscure resolution between cells that, similarly to HSCs and MPPs here, differ only slightly in proliferative history. We find it reasonable that the histone exchange responsible for H2B-fusion protein incorporation should also influence on label retention.
In steady state, we observed that HSCs and, perhaps more surprisingly, MPPs divided slowly compared to other more-mature hematopoietic progenitors, which was similar regardless of age. By contrast, the proliferation of GMLPs was increased in aged animals. According to the standard model of blood formation, MPPs are progeny of HSCs, but they cannot give rise to HSCs themselves. Therefore, in a ubiquitous labeling system such as the one used here, proliferating MPPs identified during the chase period could be descendants from either HSCs or a consequence of self-replication. Their label retention is therefore influenced both by the proliferation rate of HSCs and that of MPP, which might further obscure differences in proliferation between these closely related cell types. Although we noted a trend toward less proliferation in HSCs compared to MPPs after 5 weeks chase, this was not as pronounced after longer periods of chase, likely because considerations of percentage label positive cells over a control might yield unclear results when there are only subtle differences in proliferation between populations. Population homogeneity is another potential factor, which might explain why the long-term proliferation rates of MPPs and HSCs are more similar in our experiments than in another report, which used a more refined subfractionation of the MPP compartment (Oguro et al., 2013). Slow proliferation of HSCs and MPPs is consistent with a recent study (Busch et al., 2015) in which GMLPs and more mature progenitors were shown to be important amplifiers in mature blood cell production, with less contribution from HSCs. This agrees also with the framework presented in another recent study of native hematopoiesis (Sun et al., 2014), in which multipotent (GMLPs) and lineage-restricted progenitors were proposed to drive hematopoiesis, with only minor contribution from HSCs. Our studies therefore consolidate the recent understanding that HSC behavior after transplantation is fundamentally different from HSC behavior in more unperturbed settings. More directly, we demonstrate that the extraordinary pressure put on HSCs after transplantation alters their proliferative behavior even long-term and upon restoration of normal hematopoiesis. By contrast, hematopoietic stress caused by depletion of specific blood lineages (B cells and granulocytes) exerted little proliferative pressure on HSCs, with the brunt of such stress being shouldered by more mature MPPs and/or lineage-restricted progenitors, depending on the lineage depleted. Myeloid cell depletion induced proliferation of progenitors immediately downstream of HSCs, whereas no effect on proliferation of these immature progenitors was observed after lymphoid cell depletion. In sharp contrast, systemic mobilization resulted in increases in HSC proliferation, which however was far less than that observed for downstream progenitors. In fact, enforced mobilization resulted in a nearly complete depletion of MPPs, highlighting a particularly prominent role of these cells in emergence hematopoiesis. Our interpretation that MPPs are transiently depleted following mobilization assumes stable expression of the CD48 marker. Whereas this interpretation is consistent with stable expression of CD48 and CD150 in HSPCs throughout ontogeny, across mouse strains, and a stable lack of CD48 in HSCs after transplantation and mobilization (Kiel et al., 2005, Kim et al., 2006, Yilmaz et al., 2006), we can however at this point not fully exclude that the stability of CD48 expression might be different in MPPs.
Following our investigations on HSC proliferation, we set out to investigate to what extent proliferation history and gene expression patterns could be utilized to reveal potential heterogeneity of candidate HSCs. For this, we chose an approach where we combined indexed single-cell flow cytometric cell sorting and multiplexed qRT-PCR. This approach allowed successful clustering based on gene expression patterns, which could be correlated to the proliferation history of the candidate HSCs. This approach also aided us in correlating cell surface marker expression to the gene-expression patterns in individual HSCs. Consistent with previous reports, differential c-kit surface expression was noted among candidate HSCs (Grinenko et al., 2014, Shin et al., 2014), with c-kitlow cells displaying a more restricted proliferative history. Reciprocally, and in line with a recent report (Wilson et al., 2015), we also noted differences in levels of Sca1 among the separate clusters. Sca1low HSCs had proliferated substantially more than Sca1high HSCs. Guided by these observations, prospective isolation in combination with competitive transplantation allowed us to confirm the distinct functional activities of these subsets (Grinenko et al., 2014, Shin et al., 2014). Taken together, these data therefore highlight proliferation history as a marker to further refine populations previously considered homogenous.
In conclusion, our work adds aspects of HSC proliferation to other recent studies that have approached steady-state HSC function (Busch et al., 2015, Sun et al., 2014). Whereas undoubtedly essential in the transplantation setting, by which HSCs have traditionally been functionally evaluated, HSCs appear to only be a minor contributor to steady-state hematopoiesis, and we here reveal that this is directly reflected by their proliferative history. As a consequence, other more rapidly proliferating progenitors must have a higher degree of self-perpetuation than previously appreciated from transplantation studies, a fact underscored by the extensive cell production necessary to maintain appropriate homeostasis in the hematopoietic system.
Experimental Procedures
Mice
To track proliferation of HSPCs in vivo, Col1a1-tetO-H2B-mCherry mice (Egli et al., 2007; Jax mice stock number 014602) were used. Except for experiments with Col1a1-tetO-H2B-mCherry-Fgd5-ZsGreen mice and initial labeling experiments in which comparison of heterozygous and homozygous animals were made, homozygous animals were used throughout the study. Mice were maintained at the animal facilities at BMC at Lund University, and all animal experiments were performed with consent from a local ethical committee.
H2B-mCherry Labeling
H2B-mCherry labeling in Col1a1-tetO-H2B-mCherry mice was induced by administration of high-concentration DOX (2 g/kg; Ssniff Spezialdiäten) containing food pellets, except in initial labeling experiments when labeling with a lower concentration of DOX (200 mg/kg) was evaluated. For labeling, mice were fed DOX-containing food pellets for 1 week. Mice with the same genotype and gender as experimental animals, but fed with normal food, were used as negative controls.
qRT-PCR
For evaluation of H2B-mCherry expression in vivo, RNA was extracted from HSPCs isolated from non-induced Col1a1-tetO-H2B-mCherry mice or from mice kept for 7 days on Dox-containing (2 g/kg) food pellets followed by no and 7 days of chase. For qRT-PCR, total RNA was isolated with an RNeasy-micro mRNA purification kit (QIAGEN) and used for reverse-transcription with random hexamers using SuperScript III, according to the manufacturer’s instructions (Invitrogen). Real-time PCR reactions were performed with 1,000 cell equivalents of RNA using a MyiQ single color detection system (Bio-Rad) using a SYBR Green mix (Invitrogen).
Immunophenotyping and FACS
For HSPC analysis, single-cell suspensions of BM or spleen cells were lineage depleted using a cocktail of biotinylated antibodies CD11b (M1/70), Gr-1 (RB6-8C5), Ter119 (Ter119), CD8a (53-6.7), CD4 (GK1.5), B220 (RA3-6B2), with CD11b excluded from the lineage cocktail in mobilization experiments, and streptavidin-conjugated magnetic beads (according to manufacturer’s instructions, Miltenyi Biotec). Lineage-depleted cells were stained with fluorescent-conjugated streptavidin and antibodies. BM cells from mobilized mice were analyzed without lineage depletion. Erythrocytes were lysed with ammonium chloride before antibody staining. To determine the absolute number of the indicated cell fractions, overall organ cellularity was assessed using a Sysmex KX-21 N machine (Sysmex) and correlated with the frequencies of specific subsets obtained from flow cytometric analyses. HSPC phenotypes were analyzed as described (Ugale et al., 2014). BM and spleen cells were sorted and/or analyzed on a FACS Aria III cell sorter (Becton Dickinson). For blood analysis, peripheral blood was collected from the tail vein and red blood cells were sedimented with 2% Dextran T-500. Remaining red blood cells were lysed with ammonium chloride before staining with antibodies. Blood was analyzed on a FACS Aria III cell sorter or an LSRII (Becton Dickinson). Fluorescently labeled antibodies were directed against CD19 (1D3), Gr-1 (RB6-8C5), Sca1 (D7), c-Kit (2B8), CD45.1 (A20), CD45.2 (104), CD41 (MWReg30), CD48 (HM48-1), CD150 (TC15-12F12.2), and CD105 (MJ7/18). The analyzed subsets were distinguished by the following cell surface marker expression: HSC, Lin−cKit+Sca1+CD48−CD150+; MPP, Lin−cKit+Sca1+CD48−CD150−; GMLP, Lin−cKit+Sca1+CD48+CD150−; MyP, Lin−cKit+Sca1−CD150−; and MkP, Lin−cKit+Sca1−CD150+CD41+.
Cy/G-CSF Mobilization
Col1a1-tetO-H2B-mCherry mice were H2B-mCherry labeled and chased for 4 weeks before mobilization. Mice were injected intraperitoneally with 200 mg/kg Cy (Baxter Medical AB) and then for 5 consecutive days subcutaneously with 250 μg/kg of human G-CSF (Amgen). Control mice were sham injected with PBS. Mice were analyzed 1 day after the last injection.
In Vivo Antibody Depletion
B cell depletion was conducted according to a previously described protocol (Keren et al., 2011) by intraperitoneally injecting 150 μg/mouse rat anti-mouse CD19 and CD22 (clones 1D3 and CY34.1; Bio X Cell) and rat anti-mouse B220 (clone: RA3-6B2; eBioscience). Forty-eight hours later, 150 μg/mouse of anti-rat kappa light chain (clone: MAR18.5; Bio X Cell) was injected intraperitoneally. Neutrophil depletion was achieved by injecting 500 μg of anti-mouse Ly-6G (clone: RB6-8C5; Bio X Cell) intraperitoneally into mice on days 0 and 5. Analysis was performed on the indicated days after the last injection. In all antibody depletion experiments, DOX pellets were removed when the first injections were made.
Transplantation Experiments
3 × 105 or 1 × 107 unfractionated BM cells from uninduced Col1a1-tetO-H2B-mCherry mice were transplanted into lethally irradiated (800–950 rad) C57Bl/6 CD45.1 mice. For secondary transplantations, 3 × 106 unfractionated BM cells from primary recipients were transplanted into lethally irradiated C57Bl/6 CD45.1 recipients or F1 C57Bl/6 CD45.1 × C57Bl/6 CD45.2 mice, together with 3 × 106 previously transplanted C57bl/6 CD45.1 competitor cells (Rundberg Nilsson et al., 2015). When total white blood cell values had reached normal levels in primary or secondary transplanted recipient mice (16 or 18 weeks after transplantation), H2B-mCherry was induced and followed by 4 weeks chase before assessing HSPC proliferation.
Multiplex qRT-PCR Analysis Using the Fluidigm Biomark HD Platform
Individual HSCs isolated from 2 or 5 weeks chased Col1a1-tetO-H2B-mCherry mice were index sorted, using a Lin−Sca1+c-kit+CD48−CD150+ phenotype, into individual wells of a 96-well PCR plate containing 5 μl lysis buffer (10 mM TrisHCl [pH 8.0], 0,1 mM EDTA, 0.1 units/μl SUPERase-In [Life Technologies], and 0.5% NP40 [Sigma]). Plates with sorted cells were snap frozen on dry ice and stored at −80°C until further processing. For cDNA synthesis, qScript cDNA supermix (Quanta Bioscience) was used. Specific target amplification (STA) of genes of interest were performed in multiplex using 96 Delta Gene assays and TATAA preAmp GrandMaster Mix (Tataa Biocenter) using cycling conditions of 95°C for 10 min followed by 22× (96°C for 15 s; 60°C for 6 min). After STA, samples were treated with Exonuclease I (New England Biolabs) at 37°C for 30 min to remove excess primers. Thereafter, inactivation of the enzyme was performed at 80°C for 15 min. The cDNA was diluted five times with TE low buffer (10 mM TrisHCl [pH 8.0] and 0.1 mM EDTA) prior to qPCR on the BioMark HD (Fluidigm). All primers were used at 500 nM in the final reaction, and EvaGreen Supermix with Low ROX (Bio-Rad) was used for detection of the qPCR products. The qPCR was performed using the 96.96 Dynamic Array Integrated Fluidic Circuits (IFC) (Fluidigm) chip in combination with the BioMark HD with the following thermal cycler program: thermal mix 70°C for 4 min followed by 60°C for 30 s, hot start 95°C for 60 s, and PCR cycle 30× (96°C 5 s; 60°C for 20 s), followed by melting curve analysis.
Random forest clustering was performed using R functions located at https://labs.genetics.ucla.edu/horvath/RFclustering/RFclustering.htm, which at its core uses the “randomForest” package, which is hosted at the official R repository at https://cran.r-project.org/web/packages/randomForest/index.html. Using these functions, as documented without modification, will perform random forest clustering as described in the manuscript. The parameters we used in the random forest dist function were mtry1 = 3 no.tree = 433 and no.rep = 224. Before random forest clustering (Shi and Horvath, 2006), data from failed assays and non-expressed genes (expressed in <20% of the cells; 14 genes) were excluded. Data from no template controls and from failed cells were also removed. Random forest clustering is a process by which decision trees are generated and used to define the distance between variables (in this case cells). Random forest clustering is an adaptation of its use as a classification tool that has been used extensively to analyze biological data due to its accuracy and speed. Random forest clustering offers several advantages over other clustering methods such as hierarchical and k-means for the following reasons: first, random forest clustering is rank based, which means transformations made to the data have no effect on the outcome, as long as the ranks are preserved. Second, random forest clustering does not rely on user-specified thresholds. There is no need to supply linkage methods, the number of partitions expected, and cutoffs to define similarity. This means random forest clustering is truly unsupervised and removes user bias. Third, random forests are more resistant to outliers and weight each variable individually, leading to increased sensitivity. Random forest clustering was performed multiple times on the dataset with very similar results. The heatmap in Figure 6B was prepared using Single Cell expression Visualizer (Lang et al., 2015; http://stemsysbio.bmc.lu.se/SCexV). Significance values were calculated by using Tukey’s multiple comparisons test in Graphpad Prism or with Student’s two-tailed t test (Figure 2B) in Microsoft Excel (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Statistical Analysis
Data were analyzed using Microsoft Excel (Microsoft) and Graphpad Prism (GraphPad Software). All FACS analyses were performed using Flowjo software (TreeStar).
Author Contributions
Conceptualization, P.S. and D.B.; Methodology, P.S., S.L., P.M., D.J.R., S.S., and D.B.; Software, S.L. and S.S.; Validation, P.S. and D.B.; Formal Analysis, P.S. and D.B.; Investigation, P.S.; Resources, P.S., S.L., P.M., D.J.R., S.S., and D.B.; Data Curation, S.L. and S.S.; Writing – Original Draft, P.S. and D.B.; Writing – Review & Editing, P.S., S.L., P.M., D.J.R., S.S., and D.B.; Visualization, P.S. and D.B.; Supervision, D.B.; Project Administration, P.S.; Funding Acquisition, D.B.
Acknowledgments
We thank Gerd Sten and Eva Erlandsson for expert technical support. This work was supported by grants from the Swedish Cancer Society, the Swedish Medical Research Council, and ERC Consolidator grant 615068 to D.B.
Published: March 17, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes one figure and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.02.073.
Accession Numbers
The accession number for the Fluidigm data reported in this paper is GEO: GSE77477.
Supplemental Information
Fluidigm data from all HSCs presented in figure 6. 2 plates with HSCs sorted after 2 weeks chase and 2 plates with HSCs sorted after 5 weeks chase were analyzed.
References
- Bryder D., Rossi D.J., Weissman I.L. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am. J. Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., Rodewald H.R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Cheshier S.H., Morrison S.J., Liao X., Weissman I.L. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Rosains J., Birkhoff G., Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- Fleming W.H., Alpern E.J., Uchida N., Ikuta K., Spangrude G.J., Weissman I.L. Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. J. Cell Biol. 1993;122:897–902. doi: 10.1083/jcb.122.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R., Mandal P.K., Ebina W., Ben-Zvi A., Nombela-Arrieta C., Silberstein L.E., Rossi D.J. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 2014;211:1315–1331. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimm H., Oh I.H., Eaves C.J. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0) Blood. 2000;96:4185–4193. [PubMed] [Google Scholar]
- Gratwohl A., Baldomero H., Aljurf M., Pasquini M.C., Bouzas L.F., Yoshimi A., Szer J., Lipton J., Schwendener A., Gratwohl M., Worldwide Network of Blood and Marrow Transplantation Hematopoietic stem cell transplantation: a global perspective. JAMA. 2010;303:1617–1624. doi: 10.1001/jama.2010.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinenko T., Arndt K., Portz M., Mende N., Günther M., Cosgun K.N., Alexopoulou D., Lakshmanaperumal N., Henry I., Dahl A., Waskow C. Clonal expansion capacity defines two consecutive developmental stages of long-term hematopoietic stem cells. J. Exp. Med. 2014;211:209–215. doi: 10.1084/jem.20131115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibian H.K., Peters S.O., Hsieh C.C., Wuu J., Vergilis K., Grimaldi C.I., Reilly J., Carlson J.E., Frimberger A.E., Stewart F.M., Quesenberry P.J. The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. J. Exp. Med. 1998;188:393–398. doi: 10.1084/jem.188.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan C.T., Astle C.M., Zawadzki J., Mackarehtschian K., Lemischka I.R., Harrison D.E. Long-term repopulating abilities of enriched fetal liver stem cells measured by competitive repopulation. Exp. Hematol. 1995;23:1011–1015. [PubMed] [Google Scholar]
- Keren Z., Naor S., Nussbaum S., Golan K., Itkin T., Sasaki Y., Schmidt-Supprian M., Lapidot T., Melamed D. B-cell depletion reactivates B lymphopoiesis in the BM and rejuvenates the B lineage in aging. Blood. 2011;117:3104–3112. doi: 10.1182/blood-2010-09-307983. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., He S., Ashkenazi R., Gentry S.N., Teta M., Kushner J.A., Jackson T.L., Morrison S.J. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Morrison S.J. CD150- cells are transiently reconstituting multipotent progenitors with little or no stem cell activity. Blood. 2008;111:4413–4414. doi: 10.1182/blood-2007-12-129601. author reply 4414–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., He S., Yilmaz O.H., Kiel M.J., Morrison S.J. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Cook P.R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Ugale A., Erlandsson E., Karlsson G., Bryder D., Soneji S. SCExV: a webtool for the analysis and visualisation of single cell qRT-PCR data. BMC Bioinformatics. 2015;16:320. doi: 10.1186/s12859-015-0757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone G., Leotta S., Indelicato F., Mercurio S., Moschetti G., Di Raimondo F., Tornello A., Consoli U., Guido G., Giustolisi R. G-CSF alone vs cyclophosphamide plus G-CSF in PBPC mobilization of patients with lymphoma: results depend on degree of previous pretreatment. Bone Marrow Transplant. 2003;31:747–754. doi: 10.1038/sj.bmt.1703912. [DOI] [PubMed] [Google Scholar]
- Moignard V., Macaulay I.C., Swiers G., Buettner F., Schütte J., Calero-Nieto F.J., Kinston S., Joshi A., Hannah R., Theis F.J. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Weissman I.L. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Wright D.E., Weissman I.L. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc. Natl. Acad. Sci. USA. 1997;94:1908–1913. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundberg Nilsson A., Pronk C.J., Bryder D. Probing hematopoietic stem cell function using serial transplantation: seeding characteristics and the impact of stem cell purification. Exp. Hematol. 2015;43:812–817. doi: 10.1016/j.exphem.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Nygren J.M., Bryder D. A novel assay to trace proliferation history in vivo reveals that enhanced divisional kinetics accompany loss of hematopoietic stem cell self-renewal. PLoS ONE. 2008;3:e3710. doi: 10.1371/journal.pone.0003710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren J.M., Bryder D., Jacobsen S.E. Prolonged cell cycle transit is a defining and developmentally conserved hemopoietic stem cell property. J. Immunol. 2006;177:201–208. doi: 10.4049/jimmunol.177.1.201. [DOI] [PubMed] [Google Scholar]
- Oguro H., Ding L., Morrison S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orschell-Traycoff C.M., Hiatt K., Dagher R.N., Rice S., Yoder M.C., Srour E.F. Homing and engraftment potential of Sca-1(+)lin(-) cells fractionated on the basis of adhesion molecule expression and position in cell cycle. Blood. 2000;96:1380–1387. [PubMed] [Google Scholar]
- Passegué E., Wagers A.J., Giuriato S., Anderson W.C., Weissman I.L. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Qiu J., Papatsenko D., Niu X., Schaniel C., Moore K. Divisional history and hematopoietic stem cell function during homeostasis. Stem Cell Reports. 2014;2:473–490. doi: 10.1016/j.stemcr.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel V.I., Miller C.L., Eaves C.J., Lansdorp P.M. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood. 1996;87:3500–3507. [PubMed] [Google Scholar]
- Rebel V.I., Miller C.L., Thornbury G.R., Dragowska W.H., Eaves C.J., Lansdorp P.M. A comparison of long-term repopulating hematopoietic stem cells in fetal liver and adult bone marrow from the mouse. Exp. Hematol. 1996;24:638–648. [PubMed] [Google Scholar]
- Seita J., Sahoo D., Rossi D.J., Bhattacharya D., Serwold T., Inlay M.A., Ehrlich L.I., Fathman J.W., Dill D.L., Weissman I.L. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS ONE. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Horvath S. Unsupervised learning with random forest predictors. J. Comput. Graph. Stat. 2006;15:118–138. [Google Scholar]
- Shin J.Y., Hu W., Naramura M., Park C.Y. High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J. Exp. Med. 2014;211:217–231. doi: 10.1084/jem.20131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo K., Ema H., Morita Y., Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., Camargo F.D. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H., Regoes R.R., Boddupalli C.S., Bonhoeffer S., Manz M.G. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 2011;208:273–284. doi: 10.1084/jem.20101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R.I., Coffman R.L., Leder P. An eosinophil-dependent mechanism for the antitumor effect of interleukin-4. Science. 1992;257:548–551. doi: 10.1126/science.1636093. [DOI] [PubMed] [Google Scholar]
- Ugale A., Norddahl G.L., Wahlestedt M., Säwén P., Jaako P., Pronk C.J., Soneji S., Cammenga J., Bryder D. Hematopoietic stem cells are intrinsically protected against MLL-ENL-mediated transformation. Cell Rep. 2014;9:1246–1255. doi: 10.1016/j.celrep.2014.10.036. [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Kent D.G., Buettner F., Shehata M., Macaulay I.C., Calero-Nieto F.J., Sánchez Castillo M., Oedekoven C.A., Diamanti E., Schulte R. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O.H., Kiel M.J., Morrison S.J. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood. 2006;107:924–930. doi: 10.1182/blood-2005-05-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluidigm data from all HSCs presented in figure 6. 2 plates with HSCs sorted after 2 weeks chase and 2 plates with HSCs sorted after 5 weeks chase were analyzed.