Abstract
The PIWI-interacting RNA (piRNA) pathway is a conserved defense mechanism that protects the genetic information of animal germ cells from the deleterious effects of molecular parasites, such as transposons. Discovered nearly a decade ago, this small RNA silencing system comprises PIWI-clade Argonaute proteins and their associated RNA-binding partners, the piRNAs. In this review, we highlight recent work that has advanced our understanding of how piRNAs preserve genome integrity across generations. We discuss the mechanism of piRNA biogenesis, give an overview of common themes as well as differences in piRNA-mediated silencing between species, and end by highlighting known and emerging functions of piRNAs.
Keywords: PIWI proteins, piRNA biogenesis, ping-pong amplification loop, transposon control, transcriptional gene silencing (TGS), post-transcriptional gene silencing (PTGS)
Trends
Transcription and stabilization of piRNA precursors in Drosophila germ cells require the Rhino-Deadlock-Cutoff complex.
Primary piRNA biogenesis takes place at Yb bodies and depends on the endonuclease Zucchini and several additional mitochondria-anchored factors.
A tightly controlled, concerted interplay of Tudor proteins is necessary within the ping-pong cycle to ensure appropriate targeting of piRNA amplification.
Secondary piRNA processing through the ping-pong cycle and primary piRNA biogenesis are interconnected. Transposon slicing results in the generation of phased piRNA populations that associate with Piwi and mediate transcriptional gene silencing.
Transcriptional gene silencing via Piwi depends on Asterix and Panoramix, linking the piRNA pathway to the general silencing machinery.
The piRNA Pathway: Concepts and Discovery
Since the discovery of RNA interference (RNAi) during the late 1990s, the diversity of regulatory small RNAs has been continuously growing. Based on their processing mechanisms and partner Argonaute proteins, these small RNAs are categorized into three major classes: miRNAs, small interfering RNAs (siRNAs), and piRNAs. The biogenesis of miRNAs and siRNAs typically depends on RNase III type enzymes that convert their double-stranded RNA precursors into functional small RNAs. By contrast, piRNAs derive from single-stranded RNAs and, consequently, require alternative processing engines 1, 2.
Historically, piRNAs were first found through work carried out in Drosophila. In 2001, it was revealed that testes-expressed small RNAs derived from the Su(Ste) locus target and silence Stellate transcripts, thereby enabling proper spermiogenesis [3]. Cataloguing of small RNAs from different tissues and developmental stages identified 23- to 30-nucleotide (nt) RNAs that predominantly match transposable elements and repeats [4]. Remarkably, these RNAs were only found in male and female reproductive tissues. Later studies in flies, fish, and mammals showed a conserved association of these small RNAs with PIWI-clade Argonaute proteins (throughout this Review, we use ‘PIWI’ to refer to the protein family, while ‘Piwi’ is used for the fly protein that founded this protein family) and led to their classification as piRNAs 1, 5, 6, 7, 8, 9. Remarkably, long before piRNAs and their roles in protecting germ cell genomes were discovered, the flamenco locus, which is now known as one of the major Drosophila piRNA clusters, was identified genetically as a regulator of gypsy family transposons 10, 11.
Extensive small RNA profiling along with genetics and biochemical approaches have provided a framework of the piRNA pathway, with many aspects conserved throughout animal evolution. Generally speaking, two distinct versions of the pathway exist: one genetically encoded that produces primary silencing triggers, capable of detecting and keeping resident mobile elements in check or required for switching off genic transcripts after meiosis 6, 12, 13, and an adaptive one that is used to specifically amplify piRNAs and repress active transposons. The latter is now known as the ping-pong cycle and we refer to piRNAs produced via this route as secondary piRNAs 6, 8, 14. Amplification of silencing triggers via the ping-pong pathway is analogous to the activity of RNA-dependent RNA polymerases (RdRP) in worms and plants, enzymes that are not present in the genomes of flies and mammals. The major sources of primary piRNAs in flies are transcripts derived from piRNA clusters, genomic loci that resemble transposon graveyards [6]. Recent work uncovered an interesting interconnection between ping-pong amplification and the production of phased, primary piRNAs loaded predominantly into Piwi 15, 16, 17, 18, 19. This phased piRNA processing mechanism generates small RNA molecules that spread into downstream transcript regions, thereby allowing the targeting of diverse sequences that lie in proximity to the threat originally detected.
Conceptually, primary piRNA biogenesis can be broken down into several successive steps: piRNA cluster transcripts must be generated, followed by licensing to produce piRNA molecules, and export to the processing sites located in the cytoplasm. There, cluster transcripts are cleaved into piRNA intermediates, which are then loaded into PIWI proteins and subsequently trimmed and methylated to yield mature piRNA-PIWI complexes. Alternatively, Zucchini (Zuc)-dependent 3′ end formation of PIWI-associated piRNA intermediates can result in the production of trimming-independent, phased piRNAs 15, 16, 17, 18, 19.
Mature piRNA-PIWI complexes have different fates depending on the protein involved. Drosophila Piwi will enter the nucleus, whereas Aubergine (Aub), which is also loaded with primary piRNAs, localizes to the perinuclear cytoplasm [6]. Similarly, murine MILI and MIWI are cytoplasmic proteins, whereas MIWI2 localizes to nuclei 14, 20. The localization of these Argonaute proteins greatly influences the mode of action by which they carry out silencing. Target repression by piRNAs also comes in two forms: transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS). Typically, piRNA-mediated PTGS is sequence specific and depends on catalytically active enzymes 21, 22, 23, 24. The ping-pong cycle, which is discussed later in this review, not only utilizes PTGS to destroy transposon mRNAs, but also harnesses this mechanism to amplify silencing triggers targeting active elements 6, 8. In addition, the concomitant production of phased piRNAs allows sequence diversification via spreading and feeds back to TGS-competent PIWI proteins 15, 16, 17, 18, 19, 25. By contrast, TGS does not rely on target RNA catalysis 21, 26. Instead, piRNA-directed TGS leads to target shutdown through chromatin modifications that include repressive histone marks and DNA methylation. We discuss both strategies in detail in this review.
Here, we draw on recent findings to summarize our current understanding of piRNA biogenesis, silencing, and function, and elaborate on open questions for future research. Due to space limitations, we mainly focus on insect and mammalian pathways.
piRNA Clusters and the Definition of Precursors
Deep-sequencing and computational analyses of fly and mammalian piRNA populations identified a pronounced clustering of multiple piRNAs at distinct genomic loci, resulting in sites with high piRNA coverage, thus the term ‘piRNA cluster’ was born 5, 6, 7, 9. Cytologically, mouse clusters reside in euchromatic domains, whereas clusters in flies are embedded in heterochromatin and predominantly found at pericentromeric and subtelomeric regions 5, 6, 7. Interestingly, piRNA clusters rarely contain full-length copies of transposons. Instead, they harbor remnants and nested fragments that are unable to transpose, thereby providing a genetic memory of past transposon challenges. Clusters come in several versions (Figure 1). They can produce piRNAs from both genomic strands by convergent transcription, as seen for most Drosophila germline clusters (‘dual-strand’ clusters). Alternatively, some loci, including the flamenco cluster in Drosophila follicle cells and murine pachytene piRNA clusters 5, 6, 7, 9, give rise to piRNAs from only one genomic strand and, hence, are termed ‘uni-strand’ clusters. Most meiotic piRNA clusters are considered uni-strand clusters, although they are transcribed from promoters that fire bidirectionally, because the produced piRNA are restricted to individual, nonoverlapping genomic strands.
Figure 1.
PIWI-interacting RNA (piRNA) Clusters and Transcription of piRNA Sources. The body of uni-strand piRNA clusters (left, top) carries repressive histone 3 lysine 9 trimethylation (H3K9me3) marks. Yet, clusters resemble coding genes in that their promoters contain histone 3 lysine 4 dimethylation (H3K4me2) marks. piRNA clusters also produce transcripts via RNA Polymerase II (Pol II) that have 5′ caps, undergo (alternative) splicing and are terminated by canonical poly(A) signals. Meiotic piRNA clusters show similar characteristics, yet often produce piRNAs from two nonoverlapping genomic strands (left, bottom). These clusters are transcribed from bidirectional promoters that are activated by the A-MYB transcription factor. By contrast, dual-strand clusters in Drosophila germ cells (right, top) carry H3K9me3 modifications, but show no H3K4me2 signatures typical for active promoters. Instead, these piRNA sources utilize Pol II-dependent noncanonical read-through transcription from neighboring coding genes. It is hypothesized that 3′ end processing of the nascent RNAs results in the release of uncapped piRNA precursors. Cutoff (Cuff), a component of the Rhino-Deadlock-Cutoff (Rhi-Del-Cuff) complex that is anchored on piRNA cluster bodies via the H3K9me3-binding capacity of Rhi, associates with these uncapped transcripts and protects them from degradation, splicing, and transcription termination. Finally, recruitment of UAP56 allows the delivery to processing sites located across the nuclear envelope (right, bottom). Abbreviations: CBC, cap-binding complex; Pld, phospholipase D; TSS, transcription start site; UAP56, U2AF65–associated protein.
We are only beginning to understand the regulation and transcription of piRNA clusters. Although located in heterochromatic regions, which are generally thought to be transcriptionally silent [27], these clusters are readily expressed. Moreover, production of piRNAs from both uni- and dual-strand clusters in flies is impaired in Eggless/dSETDB1 mutants [28]. In normal cells, Eggless and its cofactor Windei initiate heterochromatin formation through deposition of histone 3 lysine 9 trimethylation (H3K9me3) marks, resulting in heterochromatic protein 1 [HP1, also known as Su(var)205] recruitment 29, 30. Thus, cluster expression either directly or indirectly requires methylated H3K9 residues. This hypothesis is further supported by a recent study that linked the Drosophila RNaseP protein p30 (Rpp30), an enzyme important for tRNA processing, to piRNA cluster maintenance. Rpp30 mutant ovaries show reduced H3K9me3 levels at dual-strand clusters, accompanied by lower amounts of precursor RNAs and a collapse of mature piRNAs [31]. However, how tRNA clusters and tRNA processing affect the heterochromatin status of nearby piRNA source loci remains to be understood.
Cluster transcripts are found predominantly in gonadal tissue, suggesting tissue-specific control. Indeed, production of pachytene (but not prepachytene) piRNA cluster RNAs in mice is dependent on the conserved A-MYB transcription factor [32]. Early work from Drosophila demonstrated that the uni-strand cluster flamenco is transcribed from a single promoter that produces RNAs approximately 200 kb in size [6]. More recently, it was shown that fly and murine uni-strand clusters are conventional RNA Polymerase II (Pol II) transcripts with histone 3 lysine 4 dimethylation (H3K4me2) marks at their transcription start sites, 5′ caps, and canonical polyadenylation and/or termination sites, and potentially undergo splicing 32, 33, 34.
New findings shed light on the more complex transcription of Drosophila dual-strand clusters. These loci lack H3K4me2 signatures typical for active promoters and their transcripts neither carry 5′ caps nor are spliced 34, 35. Instead, germline-specific clusters are thought to utilize noncanonical read-through transcription from neighboring genes. In this model, an interdependent protein complex comprising Rhino, Deadlock, and Cutoff (also known as the RDC complex) ensures the protection of nascent RNAs from degradation. Rhino, an HP1 homolog, specifically binds to repressive H3K9me3 heterochromatin marks present on clusters and, via the linker protein Deadlock, recruits Cutoff, a protein with similarity to the Rai1 transcription termination factor 34, 35, 36, 37, 38, 39. Cutoff is predicted to be catalytically inactive. Thus, rather than causing termination, Cutoff is believed to cotranscriptionally bind uncapped 5′ ends, stabilizing cluster transcripts and flagging them for piRNA processing 34, 35. Thus, dual-strand clusters appear to depend on transcriptional signals that direct expression of nearby loci rather than carrying their own.
One of the most intriguing questions in the field is how transcripts, specifically those from uni-strand clusters, are licensed for piRNA biogenesis. What distinguishes these RNAs from canonical gene-coding transcripts? While surrounding chromatin appears to at least indirectly influence this process for dual-strand clusters via RDC complex recruitment [34], uni-strand clusters do not seem to rely on specialized chromatin structure. This was exemplified by piRNA source loci that were artificially taken out of their endogenous genomic context, yet readily produced piRNAs [40]. Of note, numerous gene-coding transcripts are known to give rise to piRNAs, and this has been observed in several species 41, 42. Recently, sequence stretches present at the 5′ end of the flamenco cluster transcript or located within the 3′ untranslated region (UTR) of tj were shown to be sufficient to recruit Yb and the piRNA processing machinery to heterologous reporters 19, 43. However, the precise nature of the signals (such as RNA sequence motifs, secondary structure, or RNA modification) that direct these genic and uni-strand cluster transcripts to the piRNA machinery remain to be determined by future research.
Biogenesis of Primary piRNAs
Transcripts assigned for the production of piRNAs require transport across the nuclear envelope to the processing sites that reside in the cytoplasm. In germ cells, piRNA processing is thought to happen at perinuclear, multiprotein structures called ‘nuage’, and delivery of cluster transcripts to processing sites requires the DEAD box helicase U2AF65-associcated protein (UAP56, also known as Hel25E) 35, 44. By contrast, in follicle cells, piRNA production occurs at so-called ‘Yb bodies’ 45, 46. Screenings for genes essential for transposon silencing identified several conserved export factors and nuclear pore components 36, 47, 48, although the precise molecular function of these proteins in piRNA biogenesis remains an open question.
Following export to the cytoplasmic piRNA production centers, cluster transcripts are processed into piRNA intermediates (Figure 2). A recent study found that the RNA-helicase MOV10L1 (the mouse homolog of Drosophila Armitage, Armi) associates with piRNA precursors [49]. Furthermore, the ATP-dependent unwinding activity of MOV10L1 was shown to be required for piRNA production, suggesting a function in remodeling secondary structures within piRNA precursors [49]. Based on structural work, combined with biochemistry and analyses of small RNA populations from mutants, Zucchini/MITOPLD emerged as the candidate nuclease producing the 5′ ends of piRNAs in flies and mice 50, 51. In addition, faithful primary biogenesis in these organisms depends on the function of the conserved factors Minotaur (Mino)/GPAT2 and Gasz 36, 47, 52, 53, 54. Interestingly, all of these proteins, except Armi/MOV10L1, show an evolutionary conserved localization to the outer mitochondrial membrane, hinting at a key function of mitochondria in primary piRNA processing 36, 47, 53, 54, 55.
Figure 2.
De Novo Biogenesis of PIWI-interacting RNAs (piRNAs) in Somatic Cells. Following export from the nucleus, transcripts carrying signals of unknown nature (red question mark) are recognized as piRNA precursors and transported to the processing sites Yb bodies. There, Zucchini (Zuc) and its co-factors produce piRNA intermediates with a 5′ uracil. This processing step requires Vreteno (Vret), Minotaur (Mino), and Gasz, although the precise molecular mechanisms are not fully understood. Armi seems to be involved in resolving secondary structures of these piRNA intermediates before they are loaded into Piwi and undergo further processing. The 3′ end formation is carried out either by another cleavage event by Zuc, which results in the formation of phased Piwi-associated piRNAs, or, alternatively, piRNA intermediates are resected to their mature size by a putative exonuclease named ‘trimmer’ and its co-factor Papi. Following methylation by Hen1, mature piRNA-Piwi complexes enter the nucleus to exert silencing. Most factors required for de novo piRNA biogenesis are anchored in the outer mitochondrial membrane, suggesting a central role of mitochondria in this process.
Our current model for piRNA biogenesis suggests that the characteristic size of mature piRNAs is a consequence of loading of piRNA intermediates into PIWI-clade proteins 6, 56, followed by trimming by a yet to be identified exonuclease or through alternative 3′ end formation by Zuc 15, 16, 17, 18, 57. The Tudor-domain protein Yb, which is eponymous for Yb bodies, is required for piRNA biogenesis in follicle cells 45, 46. A recent report found that Yb directly binds piRNA intermediates via its N-terminal domain, which shows homology to DEAD-box RNA helicases [58]. While germ cells lack Yb, its function is likely carried out by two homologs called Brother of Yb (BoYb) and Sister of Yb (SoYb) [59]. Another Tudor-domain protein, Vreteno, was shown to be essential for piRNA production in both tissues 59, 60, although its precise mechanistic role is not fully resolved. Loading of piRNA precursors into PIWI proteins requires Heat shock protein 90 (Hsp90) and the co-chaperone Shutdown 61, 62, both widely conserved factors. However, the loading process is not understood at the molecular level.
Sequence analysis of insect primary and mouse pachytene piRNAs revealed a remarkable bias towards a 5′ terminal uridine (U), also known as 1U bias 6, 23, 63. Consistent with sequencing data, in vitro loading experiments using Bombyx Siwi also found a preferential binding of precursor RNAs that begin with U [57]. Recent work demonstrated that this bias is due to the structure of the PIWI-clade Argonaute MID domain, which harbors the 5′ terminal nucleotides [64]. Once 1U-containing piRNA intermediates are loaded into PIWI proteins, they are resected to mature size, with distinct PIWI proteins showing characteristic differences in their length. Although experimental evidence is scant, PIWI protein-specific RNA-binding pockets in combination with processing that occurs after loading could provide an elegant explanation of signature footprints 57, 65. How are piRNA 3′ ends produced? Biochemical studies suggested resection by an Mg2+-dependent 3′ to 5′ exonuclease, trimmer, yet this activity remains to be definitively related to the responsible protein [57]. Of note, Nibbler (Nbr), which was previously reported to trim certain miRNAs 66, 67, has emerged as a candidate enzyme that potentially participates in exonucleolytic maturation of piRNAs [68]. Expression of catalytically inactive Nbr in Drosophila ovaries results in a subset of longer sized piRNAs [68]. However, further biochemical studies are necessary to characterize trimming of piRNAs at the molecular level. In mice and flies, the mitochondria-localized Tudor-domain protein TDRKH (in Drosophila named Papi) has been implicated in 3′ end formation, potentially by acting as adapter between trimmer and PIWI-RNA complexes 17, 18, 69. Alternatively, 3′ end formation can be catalyzed by Zuc-mediated endonucleolytic cleavage, potentially followed by additional trimming 15, 16, 17, 18. Interestingly, while Zuc shows no sequence specificity in vitro 50, 51, it appears to cleave preferentially in U-rich RNA stretches in vivo, which is likely aided by unknown cofactors. Cleavage immediately upstream of a U nucleotide would further reinforce the 1U bias of phased primary piRNAs produced downstream. In a final step, Hen1 methylates PIWI-associated mature piRNAs at their 3′ termini 70, 71. This RNA modification is thought to be protective and is found commonly among small RNAs that guide Argonaute proteins to target sequences via near-perfect sequence complementarity, resulting in cleavage of target transcripts. While exonucleolytic trimming and Hen1-catalyzed 2′-O-methylation are tightly coupled in a cell-free system [57], it remains to be investigated whether this is also the case in vivo.
Transposon Slicing, Ping-Pong Amplification, and the Production of Phased piRNAs
First discovered in Drosophila, the ping-pong cycle is the best understood piRNA-dependent PTGS mechanism. When small RNA-binding partners of the three PIWI-clade Argonautes were profiled, prominent orientation biases were observed 6, 8. Furthermore, Piwi- and Aub-bound sequences showed a pronounced tendency to start with 1U, whereas Argonaute3 (Ago3)-associated piRNAs featured an adenine at position 10 (10A bias) and no 5′ bias. Computational analysis revealed significant 10-nt overlaps between Aub- and Ago3-associated sequences. This seminal work led to the proposal of the ping-pong model and, later on, similar amplification loops were identified in silkworm, fish, mouse, and many other organisms.
Ping-pong amplification is unique because it couples piRNA biogenesis to target silencing. In fly germ cells, Aub, associated with a cluster-derived primary piRNA, detects and, through its slicer activity, cuts active transposon transcripts (Figure 3). These cleavage events produce the 5′ ends of new piRNAs that are in sense orientation to transposons. Following loading into Ago3 and maturation through either trimming by an unknown nuclease or Zuc-dependent 3′ end formation, Ago3-piRNA complexes in turn recognize and cleave cluster transcripts to generate more antisense piRNAs with sequences identical (or near-identical) to the original primary small RNA triggers. Maturation of Aub-associated intermediates can occur via trimming or by Zuc-mediated cleavage, with the latter also resulting in production of phased piRNAs from the downstream transcript area 17, 18. These phased piRNAs predominantly associate with Piwi and allow target adaptation via sequence diversification 15, 16. Studies of catalytically inactive and mutant PIWI proteins in various species have confirmed the general framework of ping-pong amplification 21, 22, 24. In addition to the 1U bias, which is at least in part a consequence of the MID domain structure of Piwi, Aub, Siwi, and MIWI (discussed earlier [64]), recent work suggests that the 10A bias seen in ping-pong partner PIWI proteins is also dictated by intrinsic properties of Aub, Siwi and MILI, respectively [24]. Computational analysis indicated the preferential binding of Aub and Siwi to mRNA targets with an adenosine (A) at the position opposite to the first base of their piRNA partner (described as t1A). Upon target slicing and subsequent piRNA maturation, t1A then becomes 10A in a ping-pong-derived piRNA [24]. Recent structural data are in strong support of this model, because human Argonaute2 was found to contain a binding pocket that selects for t1A-bearing targets [72]. This t1A-binding pocket is structurally conserved in the PIWI clade proteins Aub, Siwi, and MILI, explaining the signature 10A bias in ping-pong pairs [72].
Figure 3.
The Ping-Pong Amplification Loop and Phased PIWI-interacting RNA (piRNA) Production. Argonaute3 (Ago3) associated with a sense piRNA recognizes and cleaves piRNA cluster transcripts to produce piRNA intermediates with a 5′ uracil (U), which are loaded into Aubergine (Aub), likely assisted by Spindle-E (Spn-E) (A). Maturation of piRNAs can follow two nonexclusive roads: cleavage by Zuc results in 3′ end formation of Aub-bound piRNAs, as well as production of phased primary piRNAs that associate with Piwi. Alternatively, 3′ end formation can occur via Papi-dependent trimming (B). The combination of both mechanisms is likely. Following Hen1-mediated methylation (C), mature Aub complexes receive symmetric dimethyl-arginine (sDMA) modifications at their amino-termini (D). sDMA-Aub is recruited by Krimper (Krimp), which also interacts with unloaded Ago3, thus bringing both factors in close proximity (E). Subsequent to piRNA-Aub-dependent detection and slicing of transposon RNAs, the 3′ cleavage product is loaded into Ago3 aided by Vasa (F). Maturation of Ago3-loaded intermediates via Zuc or trimming (G), followed by methylation by Hen1 (H) results in mature Ago3 complexes that in turn cleave cluster transcripts to start yet another cycle (I). In Drosophila ovaries, the ping-pong amplification loop is the predominant determinant that specifies piRNA precursors for Piwi (J). Capsuleen (Csul) and its cofactor Valois (Vls) add symmetric dimethyl-arginines (sDMA) modifications to Aub and Ago3. Qin inhibits homotypic Aub:Aub ping-pong by preventing Aub-sliced transcripts from becoming substrates for Aub (inset in center).
How are Aub- and Ago3-cleaved transcripts specifically funneled into designated protein complexes, as evident from the pronounced strand biases? In addition, why are these RNAs not degraded rapidly? It was hypothesized that additional proteins integral to nuage, the location where secondary piRNA production occurs, help in this process. Indeed, numerous factors, including the DEAD-box RNA helicase Vasa, and a battery of Tudor-domain proteins, including Spindle-E (Spn-E), Krimper, Tejas, and Tapas, are required for proper ping-pong amplification to occur, because lesions in these genes resulted in a collapse of secondary piRNA production 73, 74, 75. Mechanistically, Tudor domains bind symmetric dimethyl-arginines (sDMAs), modifications added onto PIWI proteins by the methyltransferase Capsuleen (Csul, also known as dPRMT5) and its cofactor Valois (Vls) 76, 77. These sDMA modifications are conserved from flies to mammals and are thought to serve as scaffolding agents. Indeed, recent work in cultured silkworm cells uncovered a hierarchical network of Tudor proteins termed the ‘amplifier complex’ and shed light on the molecular roles of some of these factors 78, 79. Vasa interacts with Siwi, the Bombyx Aub homolog, and clamps onto its target RNAs, preventing degradation. Upon target cleavage, Vasa specifically hands over the 3′ cleavage product (carrying the 5′ end of the future secondary piRNA) to Bombyx Ago3. This step, which is crucial for the generation of secondary piRNAs, was shown to depend on the ATPase activity of Vasa 78, 79. Interestingly, Siwi was found to form another complex that is different from the amplifier complex. Here, Siwi interacts with Spn-E and Qin, yet another Tudor protein. This complex is exclusively required for loading of Siwi with piRNAs resembling the primary silencing trigger [78]. Furthermore, two recent reports suggest that, in fly ovaries, Krimper interacts with Aub and Ago3 to ensure proper loading of Ago3 80, 81. Thus, the concerted interplay between several protein complexes ensures handover of cleavage products and continuous ping-pong looping. Of note, mutations in Drosophila Qin also lead to erroneous piRNA amplification. Instead of heterotypic Aub:Ago3 ping-pong, these mutants show faulty, homotypic Aub:Aub interactions [82]. Recent work suggests that Qin prevents Aub-sliced transcripts from becoming substrates for Piwi or Aub, thereby enforcing Aub:Ago3 ping-pong [15]. Similar measures seem to be in place in murine germ cells, where RNF17, a putative functional analog to Qin, generally suppresses ping-pong amplification in meiotic stages [83].
Piwi-Dependent Transcriptional Silencing
Small RNA-mediated TGS has been studied extensively in yeast and plants. The existence of nuclear PIWI proteins and the coincidental loss of repressive histone marks and/or DNA methylation in certain piRNA pathway mutants spurred speculations about PIWI-dependent transcriptional silencing in animals. However, direct evidence for transcriptional silencing of transposons via the piRNA pathway emerged only recently. Combined analyses of RNA Pol II occupancy at mobile elements, nascent transcription, steady-state mRNA levels, and H3K9me3 marks in fly gonadal cells demonstrated that Piwi mainly acts at the transcriptional level 26, 84, 85, 86. Depletion of Piwi resulted in an increase of Pol II occupancy at promoters, elevated nascent and steady-state RNA levels, and a decrease in H3K9me3 over transposon bodies. In accord with these findings, transposable elements remained suppressed in flies carrying only catalytically inactive Piwi [26]. Together, these studies suggest that mature Piwi complexes enter the nucleus and scan for, likely nascent, transposon transcripts [87] (Figure 4). Following detection, transcriptional repression is enforced through formation of heterochromatin, although a detailed mechanistic understanding of this process is lacking.
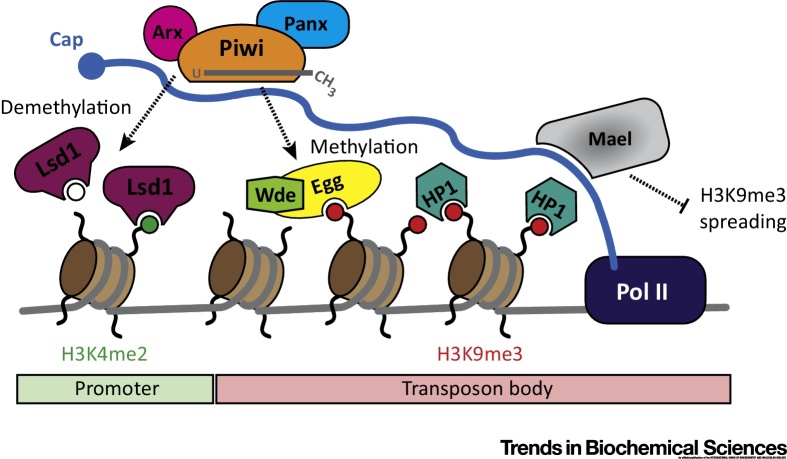
Figure 4.
PIWI-interacting RNA (piRNA)-Mediated Transcriptional Silencing. In the Drosophila ovary, piRNA-Piwi/Asterix (Arx) complexes scan for, and detect, nascent transposon transcription. Upon target engagement, Piwi likely undergoes conformational changes that lead to the recruitment of Panoramix (Panx). This piRNA-protein (comprising Piwi, Arx, and Panx,) complex induces co-transcriptional repression through recruitment of general silencing machinery components. As a consequence, transposon bodies receive repressive histone 3 lysine 9 trimethylation (H3K9me3) marks, a modification produced by Eggless (Egg) and its cofactor Windei (Wde). Subsequent recruitment of HP1 to H3K9me3 leads to heterochromatin formation. In addition, Lysine-specific demethylase 1 (Lsd1) likely removes active histone 3 lysine 4 dimethylation (H3K4me2) marks from transposon promoter regions, leading to efficient suppression of transposons at the transcriptional level. Maelstrom (Mael), a putative single-stranded RNA-binding protein, is required for transcriptional silencing and blocks H3K9me3 spread.
In mammals, transposon silencing via TGS is not limited to histone modifications. Instead, de novo DNA methylation functions as a heritable silencing mark 14, 88. In embryonic testes, MIWI2 is loaded through interaction with cleavage-competent MILI [21], and subsequently enters the nucleus to promote the establishment of CpG DNA methylation on transposons 14, 88. Recent work uncovered a complex formed by the RNA-binding protein EXD1 and the Tudor protein TDRD12. This complex is important for loading of MIWI2 with piRNAs in a process that shares similarities with the production of phased piRNAs in flies [25]. The ping-pong cycle persists in Miwi2 mutants, while MILI impacts DNA methylation by an unknown MIWI2-independent mechanism [89]. These results point to a more complex interplay between MILI and MIWI2 and suggests additional functional diversification. Several factors of the ‘general silencing machinery’, including DNMT3L and LSD1, are engaged in the DNA methylation-establishing process [14], yet the precise cascade of events that lead to transcriptional silencing in mammals is not entirely understood.
Extensions of the aforementioned work have uncovered additional factors required for piRNA pathway-mediated TGS. In Drosophila, establishment of H3K9me3 at transposon loci requires the zinc finger protein Asterix (Arx)/Gtsf1, which directly interacts with Piwi 48, 90, 91. The piRNA pathway component, Maelstrom (Mael), was shown to be required for transcriptional silencing, because RNA Pol II occupancy and nascent transcripts increased upon its loss [26]. Interestingly, Mael is not exclusively nuclear but rather shuttles between the nucleus and cytoplasm or nuage [92], suggesting a more complex role or even multiple functions. Indeed, structural reconnaissance of the MAEL domain revealed a putative nuclease fold, leading to the proposal that Mael acts as a single-stranded RNA-binding protein [93], or as an endonucleolytic RNase [94], depending on the studied species. However, the catalytic activity of Drosophila Mael was dispensable for piRNA-mediated silencing [94]. Thus, in addition to engaging in TGS, fly Mael might also transport piRNA precursors to the biogenesis machinery. This was suggested for murine MAEL, which was shown to associate with precursors of pachytene piRNAs, as well as with MIWI and TDRD6 [95]. Recent work identified Panoramix (Panx, also called Silencio) as another crucial player in TGS 36, 47, 96, 97. Panx mutants show strong derepression of transposons at both the nascent and steady-state level. Surprisingly, tethering of Panx to artificial reporters led to heterochromatin formation at the targeted locus, including reduced nascent and steady-state transcripts as well as de novo deposition of H3K9me3 marks 96, 97. Remarkably, Panx triggers silencing regardless of whether it is tethered to DNA [96] or RNA 96, 97. Panx interacts with Piwi and induces heterochromatin formation through the H3K9 methyltransferase, Eggless, and its co-factor, Windei. Yet, how the Piwi/Panx complex recruits the general silencing machinery remains enigmatic.
Methylation of H3K9 usually leads to recruitment of HP1, resulting in heterochromatin establishment and shutdown of transcription. HP1 knockdown in germ cells results in transposon derepression similar to Piwi [98]. In Drosophila, Piwi and HP1 were reported to interact directly [99]; however, residues necessary for this complex are not conserved in mammals, leaving the role of this specific interaction in silencing to be investigated. Unlike Arx or Panx mutants, cells depleted of Mael show no reduction of H3K9me3 levels but rather spreading of this mark into downstream regions. These results suggest that H3K9 methylation per se is not sufficient as a final silencing mark and that Mael acts as effector protein downstream of Piwi. Furthermore, Piwi prevents transcription at some transposon loci through a mechanism independent of H3K9me3 [84], potentially through instructing the removal of activating H3K4me2 marks, as seen in mouse two- to eight-cell embryos [100]. In accord with this hypothesis, depletion of the Lysine-specific demethylase 1 (Lsd1) in Drosophila ovaries results in upregulation of a subset of transposons [36]. Furthermore, Lsd1-depleted cells resisted silencing upon tethering of Panx [97], suggesting a central role of this enzyme in cotranscriptional transposon control. Overall, uncovering the complex interplay between all of the factors involved in piRNA directed silencing holds the exciting potential to reveal the molecular mechanism of TGS in detail.
Functions of piRNAs: Transposon Control and Beyond?
Given that germ cells carry the genetic information for future generations, animals have evolved sophisticated mechanisms to protect those lineages from harm. The piRNA pathway arms animals with a powerful system to prevent transposon propagation. While piRNA clusters provide a record of past exposure to mobile elements (‘genetic memory’), the ping-pong feed-forward amplification mechanism is thought to be important to generate sufficient amounts of silencing triggers against highly active elements and provide a mechanism for sequence diversification via downstream phased piRNA production.
Why do animals engage in both PTGS and TGS? One can argue that either alone is not enough to face the many challenges that transposons present. Indeed, the interplay between destruction of eminent threats by PTGS and stable repression at the chromatin level through TGS in theory sets multiple barriers to transposon mobilization. However, while this is true for germ cells, the somatic compartment of Drosophila gonads relies only on a TGS pathway. These so-called ‘follicle cells’ are thought to require Piwi to prevent gypsy family transposons from propagation via virus-like particles capable of infecting neighboring germ cells [101]. Additionally, piRNA-PIWI protein complexes can be inherited from the mother and induce PTGS and TGS in offspring, thereby ensuring silencing of transposable elements in subsequent generations.
The importance of maternally deposited piRNAs for efficient transposon silencing was exemplified by studies of the phenomenon ‘hybrid dysgenesis’. Hybrid dysgenesis describes the differential fertility of offspring from crosses between specific Drosophila strains, wherein fertility depends upon the strain of the mother. A sterile progeny phenotype has been associated with a single paternally inherited transposon. Analysis of maternally deposited small RNAs revealed an essential and fundamental role of transgenerationally inherited piRNAs in effective transposon silencing 102, 103. These studies showed that hybrid dysgenesis arises from a lack of inherited piRNAs able to target the paternally transmitted transposon. Furthermore, maternally inherited piRNAs also seem to help specify loci as sources of piRNA production. In fact, certain clusters showed a property known as ‘paramutation’, wherein a heritable epigenetic state can be transmitted from one allele to another within the same animal [104]. Here, piRNA-generating clusters convert other homologous sequences located elsewhere in the genome into loci that produce substantial amounts of piRNAs. Thus, transgenerationally inherited piRNAs themselves are involved in the specification of piRNA clusters in the next generation. This process likely requires Piwi, which is also maternally deposited, to instruct the methylation of H3K9 residues, which in turn triggers the recruitment of the RDC complex 34, 38. Recent work has shown that silencing by piRNAs produced from the paramutagenic cluster requires Rhino and Cutoff (cluster transcription), Zuc (piRNA processing), as well as the ping-pong factor Aub [105].
Drosophila females deposit both Piwi and Aub, but not substantial quantities of Ago3 [102]. The details of whether Aub-mediated PTGS or Piwi, which acts by TGS, or a combination of both mechanisms are the main forces to ensure silencing over newly acquired elements remains to be established, although interlinked mechanisms involving ping-pong-primed production of piRNAs seem likely. By analogy to erasure and remethylation of DNA during mammalian embryonic development [106], maternally deposited piRNAs might serve this function in flies because they set suitable chromatin environments at piRNA cluster loci that allow for piRNA production while ensuring stable repression of euchromatic transposon insertions throughout the genome. However, it is currently not clear whether repressive chromatin marks at euchromatic transposons are propagated through the germ lineage or re-established in each generation.
In addition to the conserved ping-pong amplification system, an increasing number of noncanonical PTGS mechanisms for piRNAs, besides transposon silencing, have been reported in flies, mammals, and other species. Numerous pseudo-genes are known targets of piRNA-mediated silencing, with a prominent example being the interaction between Stellate and Su(Ste), which led to the discovery of piRNAs [3]. Work in Drosophila also implicated piRNAs in clearance of maternally deposited nanos mRNA in embryos [107]. While sequences within the 3′ UTR of the nanos transcript with similarity to transposons are targeted by Aub-bound piRNAs, the mRNA is not cleaved conventionally, but instead decayed via deadenylation by the CCR4-Not complex, similar to the mode of action of miRNAs. Recent Aub iCLIP (individual-nucleotide resolution ultraviolet crosslinking and immunoprecipitation) data suggest a broader role for the piRNA-mediated decay of transcripts during the maternal-to-zygotic transition [108], although the precise underlying mechanism (deadenylation-dependent decay or direct Aub-cleavage) remains to be investigated. Interestingly, a similar role for MIWI-associated pachytene piRNAs in late stages of spermiogenesis was reported [109]. At the elongating spermatid stage, MIWI stimulated broad mRNA deadenylation and decay via selective interaction with CAF1, a component of the CCR4-Not complex. This function is essential for sperm maturation. In addition, direct cleavage of mRNAs by pachytene piRNAs associated with MIWI was demonstrated recently 12, 13. Finally, silkworm has repurposed the piRNA system to play a crucial role in sex determination [110]. A single piRNA derived from the female-determining W chromosome controls a chromosome Z-linked target transcript, which is responsible for masculinization in male embryos.
Concluding Remarks
Although much progress has been made since the original discovery of piRNAs nearly a decade ago, many aspects of piRNA biology remain poorly understood. This may be attributable to two main barriers. First, the piRNA biogenesis and silencing machineries are more complex than those of other small RNA pathways (i.e., miRNA and siRNA). Second, the restriction of the piRNA system to gonadal tissues has long hampered a systematic analysis ex vivo. However, the introduction and detailed characterization of cell lines containing primary or secondary piRNA pathways will greatly enhance studies of piRNA biogenesis and silencing 42, 63, 111, 112. Indeed, the first in vitro systems that recapitulate aspects of the pathway have now been established 57, 78. Furthermore, recently published large-scale screens likely reveal the complete catalog of factors involved in piRNA-mediated genome defense 36, 47, 48, now allowing a systematic mechanistic annotation of each pathway protein. Emerging technical advances in proteomics and microscopy in combination with CRISPR/Cas9-mediated genome editing will aid in dissecting the molecular steps that generate piRNAs and provide detailed insight into transcriptional and post-transcriptional silencing by PIWI-piRNA complexes (see Outstanding Questions for important questions for future research).
Outstanding Questions.
Definition of the piRNA repertoire: how do piRNA clusters evolve and what is the full collection of factors required for cluster maintenance and transcription? How precisely are cluster transcripts protected from degradation and dispatched to processing centers, while other RNAs escape piRNA production? We are missing a clear understanding of what happens between nuclear transcript generation and piRNA processing in the cytoplasm.
piRNA biogenesis: how does piRNA biogenesis and loading of piRNA precursors into PIWI proteins work at the molecular level? What enzymes are involved in 3′ end formation of piRNAs? Do mitochondria contribute to piRNA biogenesis beyond acting as scaffolds for the processing machinery? Although we have produced a rough framework, the exact interplay and hierarchy of all involved factors remains to be established.
The ping-pong cycle: how does nuage ensure effective ping-pong amplification? What controls the interconnection to the nuclear PIWI pathways? Through genetic analysis, we have probably uncovered most of the players involved, and we are beginning to recapitulate steps of this pathway in vitro. The big challenges will be to piece together the molecular processes into a cascade of events that resembles the endogenous pathway.
Mechanisms of piRNA-directed TGS: how does transcriptional gene silencing function at the molecular level? How precisely does Panx connect the specific piRNA pathway and the general silencing machinery? It will be important to identify all factors involved and to establish a hierarchy of events that leads to transcriptional shutdown of transposons.
Emerging functions of piRNAs: what roles do piRNAs have beyond transposon control? Some hints suggest widespread roles outside of the germline. A thorough examination of piRNA functions at multiple developmental time points and in various tissues will be required to explore the full breath of this pathway.
Note added in proof
While this article was in production, Wang et al. reported that the putative trimmer, Nibbler (Nbr), and Piwi interact at the protein level. In addition, the authors identified antagonistic functions for Nbr and the methyltransferase Hen1 in piRNA 3′ end formation, further highlighting that a concerted interplay of factors is required for proper piRNA biogenesis [113].
Acknowledgments
We thank members of the Hannon group for helpful discussion and comments on the manuscript. Work in the Hannon laboratory is supported by CRUK. We apologize to all colleagues whose work could not be referenced owing to space limitations.
Contributor Information
Benjamin Czech, Email: benjamin.czech@cruk.cam.ac.uk.
Gregory J. Hannon, Email: greg.hannon@cruk.cam.ac.uk.
References
- 1.Houwing S. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Vagin V.V. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 3.Aravin A.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 4.Aravin A.A. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 5.Aravin A. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 7.Girard A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 8.Gunawardane L.S. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 9.Lau N.C. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 10.Pelisson A. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prud’homme N. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goh W.S. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015;29:1032–1044. doi: 10.1101/gad.260455.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015;25:193–207. doi: 10.1038/cr.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravin A.A. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol. Cell. 2015;59:819–830. doi: 10.1016/j.molcel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senti K.A. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015;29:1747–1762. doi: 10.1101/gad.267252.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohn F. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han B.W. Noncoding RN.A. piRNA-guided transposon cleavage initiates Zucchini-dependent phased piRNA production. Science. 2015;348:817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homolka D. PIWI Slicing and RNA Elements in precursors instruct directional primary piRNA biogenesis. Cell Rep. 2015;12:418–428. doi: 10.1016/j.celrep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 20.Kuramochi-Miyagawa S. Two mouse piwi-related genes: miwi and mili. Mech. Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 21.De Fazio S. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 22.Li C. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter M. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 24.Wang W. The initial uridine of primary piRNAs does not create the tenth adenine that Is the hallmark of secondary piRNAs. Mol. Cell. 2014;56:708–716. doi: 10.1016/j.molcel.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol. Cell. 2015 doi: 10.1016/j.molcel.2015.11.009. Published online December 6, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sienski G. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermaak D., Malik H.S. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 2009;43:467–492. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 28.Rangan P. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch C.M. Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/Eggless in germ line development. PLoS Genet. 2009;5:e1000644. doi: 10.1371/journal.pgen.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon J. dSETDB1 and SU(VAR)3-9 sequentially function during germline–stem cell differentiation in Drosophila melanogaster. PLoS ONE. 2008;3:e2234. doi: 10.1371/journal.pone.0002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molla-Herman A. tRNA processing defects induce replication stress and Chk2–dependent disruption of piRNA transcription. EMBO J. 2015;34:3009–3027. doi: 10.15252/embj.201591006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X.Z. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell. 2013;50:67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goriaux C. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014;15:411–418. doi: 10.1002/embr.201337898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohn F. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czech B. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell. 2013;50:749–761. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klattenhoff C. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le, Thomas Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014;28:1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pane A. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30:4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muerdter F. Production of artificial piRNAs in flies and mice. RNA. 2012;18:42–52. doi: 10.1261/rna.029769.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robine N. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr. Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito K. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 43.Ishizu H. Somatic primary piRNA biogenesis driven by cis-acting RNA elements and trans-acting Yb. Cell Rep. 2015;12:429–440. doi: 10.1016/j.celrep.2015.06.035. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olivieri D. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito K. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Handler D. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell. 2013;50:762–777. doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muerdter F. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell. 2013;50:736–748. doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vourekas A. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29:617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ipsaro J.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishimasu H. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 52.Ma L. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiromoto Y. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA. 2013;19:803–810. doi: 10.1261/rna.038521.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagin V.V. Minotaur is critical for primary piRNA biogenesis. RNA. 2013;19:1064–1077. doi: 10.1261/rna.039669.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vourekas A. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawaoka S. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 58.Murota Y. Yb integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 2014;8:103–113. doi: 10.1016/j.celrep.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 59.Handler D. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zamparini A.L. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olivieri D. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell. 2012;47:954–969. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Preall J.B. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA. 2012;18:1446–1457. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawaoka S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15:1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cora E. The MID-PIWI module of Piwi proteins specifies nucleotide- and strand-biases of piRNAs. RNA. 2014;20:773–781. doi: 10.1261/rna.044701.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aravin A.A. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 66.Han B.W. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu N. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr. Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feltzin V.L. The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell. 2015;14:443–452. doi: 10.1111/acel.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saxe J.P. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horwich M.D. The Drosophila RNA methyltransferase DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 71.Saito K. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schirle N.T. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife. 2015;4:07646. doi: 10.7554/eLife.07646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim A.K., Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malone C.D. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patil V.S., Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 76.Kirino Y. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishida K.M. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishida K.M. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 2015;10:193–203. doi: 10.1016/j.celrep.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 79.Xiol J. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 80.Sato K. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol. Cell. 2015;59:553–563. doi: 10.1016/j.molcel.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 81.Webster A. Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by Krimper. Mol. Cell. 2015;59:564–575. doi: 10.1016/j.molcel.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Z. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wasik K.A. RNF17 blocks promiscuous activity of PIWI proteins in mouse testes. Genes Dev. 2015;29:1403–1415. doi: 10.1101/gad.265215.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klenov M.S. Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic Acids Res. 2014;42:6208–6218. doi: 10.1093/nar/gku268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le Thomas A. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rozhkov N.V. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Post C. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA. 2014;20:1977–1986. doi: 10.1261/rna.046300.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuramochi-Miyagawa S. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manakov S.A. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 2015;12:1234–1243. doi: 10.1016/j.celrep.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Donertas D. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013;27:1693–1705. doi: 10.1101/gad.221150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohtani H. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013;27:1656–1661. doi: 10.1101/gad.221515.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Findley S.D. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 93.Chen K.M. Metazoan Maelstrom is an RNA-binding protein that has evolved from an ancient nuclease active in protists. RNA. 2015;21:833–839. doi: 10.1261/rna.049437.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matsumoto N. Crystal structure and activity of the endoribonuclease domain of the piRNA pathway factor maelstrom. Cell Rep. 2015;11:366–375. doi: 10.1016/j.celrep.2015.03.030. [DOI] [PubMed] [Google Scholar]
- 95.Castaneda J. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 2014;33:1999–2019. doi: 10.15252/embj.201386855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sienski G. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015;29:2258–2271. doi: 10.1101/gad.271908.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu Y. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science. 2015;350:339–342. doi: 10.1126/science.aab0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang S.H., Elgin S.C. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. U.S.A. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brower-Toland B. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fadloun A. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- 101.Lecher P. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J. Gen. Virol. 1997;78:2379–2388. doi: 10.1099/0022-1317-78-9-2379. [DOI] [PubMed] [Google Scholar]
- 102.Brennecke J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khurana J.S. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Vanssay A. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 105.Hermant C. Paramutation in Drosophila requires both nuclear and cytoplasmic actors of the piRNA pathway and induces cis-spreading of piRNA production. Genetics. 2015;201:1381–1396. doi: 10.1534/genetics.115.180307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Molaro A. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 2014;28:1544–1549. doi: 10.1101/gad.244350.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rouget C. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barckmann B. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 2015;12:1205–1216. doi: 10.1016/j.celrep.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gou L.T. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24:680–700. doi: 10.1038/cr.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kiuchi T. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature. 2014;509:633–636. doi: 10.1038/nature13315. [DOI] [PubMed] [Google Scholar]
- 111.Lau N.C. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niki Y. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang H. Antagonistic roles between Nibbler and Hen1 modulate piRNA 3′ ends in Drosophila. Development. 2015 doi: 10.1242/dev.128116. Published online December 30. [DOI] [PMC free article] [PubMed] [Google Scholar]