Abstract
Purpose
Determine intra- and inter-examination repeatability of magnitude-based magnetic resonance imaging (MRI-M), complex-based magnetic resonance imaging (MRI-C), and magnetic resonance spectroscopy (MRS) at 3T for estimating hepatic proton density fat fraction (PDFF), and using MRS as a reference, confirm MRI-M and MRI-C accuracy.
Methods
Twenty-nine overweight and obese pediatric (n = 20) and adult (n = 9) subjects (23 male, 6 female) underwent three same-day 3T MR examinations. In each examination MRI-M, MRI-C, and single-voxel MRS were acquired three times. For each MRI acquisition, hepatic PDFF was estimated at the MRS voxel location. Intra- and inter-examination repeatability were assessed by computing standard deviations (SDs) and intra-class correlation coefficients (ICCs). Aggregate SD was computed for each method as the square root of the average of first repeat variances. MRI-M and MRI-C PDFF estimation accuracy was assessed using linear regression with MRS as a reference.
Results
For MRI-M, MRI-C, and MRS acquisitions, respectively, mean intra-examination SDs were 0.25%, 0.42%, and 0.49%; mean intra-examination ICCs were 0.999, 0.997, and 0.995; mean inter-examination SDs were 0.42%, 0.45%, and 0.46%; and inter-examination ICCs were 0.995, 0.992, and 0.990. Aggregate SD for each method was <0.9%. Using MRS as a reference, regression slope, intercept, average bias, and R2, respectively, for MRI-M were 0.99%, 1.73%, 1.61%, and 0.986, and for MRI-C were 0.96%, 0.43%, 0.40%, and 0.991.
Conclusion
MRI-M, MRI-C, and MRS showed high intra- and inter-examination hepatic PDFF estimation repeatability in overweight and obese subjects. Longitudinal hepatic PDFF change >1.8% (twice the maximum aggregate SD) may represent real change rather than measurement imprecision. Further research is needed to assess whether examinations performed on different days or with different MR technologists affect repeatability of MRS voxel placement and MRS-based PDFF measurements.
Keywords: Proton density fat fraction, PDFF, Hepatic steatosis, Obesity, Intra-examination repeatability, Inter-examination repeatability, Magnitude-based MRI, Complex-based MRI, Magnetic resonance spectroscopy, Quantitative imaging biomarkers
Proton density fat fraction (PDFF) is being developed as a non-invasive magnetic resonance (MR)-based quantitative imaging biomarker of hepatic steatosis [1–4]. Hepatic PDFF may be estimated by magnetic resonance spectroscopy (MRS), magnitude-based MRI (MRI-M) or complex-based MRI (MRI-C). These techniques have the same underlying MR physics, and exploit differences in resonant frequencies between water and fat proton signals to quantitatively measure the proton density fat-fraction [1, 2].
MRS allows the water and fat peaks to be directly identified to calculate liver fat fraction. Advanced MRS techniques provide a T1-independent, T2-corrected estimate of PDFF by acquiring long TR spectra at multiple TEs in a single breath-hold [1]. MRS is considered the non-invasive reference standard for hepatic PDFF estimation [5, 6], However, it requires expertise, can only be obtained at a limited number of locations in the liver, and lacks anatomic information [7–9].
MRI-M and MRI-C are advanced chemical shiftbased techniques that separate the MR signal into its water and fat components by acquiring images at multiple TEs. To minimize confounders, they estimate PDFF by acquiring images with low flip angle to minimize T1 weighting and multiple echoes to permit measurement and correction of T2* decay [1, 2, 10]. The images are then reconstructed using a multi-peak fat spectral model to correct for the multi-frequency interference effects of fat protons [1, 2, 11]. MRI-C allows measurement of signal fat-fraction between 0% and 100%, while MRI-M can measure between 0 and ~50% [1]. Both methods estimate hepatic PDFF accurately in children and adults, using MRS as a reference [7–9, 12–21].
In addition to demonstrating accuracy, imaging biomarker validation requires precision and repeatability [22]. Two kinds of repeatability for MR-based biomarkers have been described by Negrete et al. [21]: (a) Intra-examination repeatability, which refers to closeness of repeated measurements within a single MR examination (without subject or coil repositioning) under identical conditions and (b) Inter-examination repeatability, which refers to closeness of repeated measurements made in different MR examinations (with both subject and coil repositioning) over a time frame in which physiologic conditions are assumed constant.
While intra-examination repeatability has been reported for MRI-M and MRS at both 1.5 and 3T [8, 9], and inter-examination repeatability has been demonstrated for MRI-M (3T) [21], MRI-C (1.5T) [23], and MRS (1.5T) [23], they have not been directly compared in the same cohort. Hence, the purpose of this study was to compare intra- and inter-examination repeatability of MRI-M, MRI-C, and MRS at a single site, in a single subject cohort. Our secondary aim was to confirm accuracy of MRI-M and MRI-C, using MRS as a reference.
Methods
This prospectively designed, cross-sectional, single-site observational clinical study was approved by an Institutional Review Board and was compliant with the Health Insurance Portability and Accountability Act (HIPAA).
Pediatric and adult subjects were recruited from August to October 2009 by physician referral from hepatology and obesity clinics, and self-referral in response to informational flyers posted at our institution. Eligibility criteria were body mass index (BMI) greater than 28 kg/m2, age ≥8 years, and willingness to participate. Subjects with contraindication to MR, known claustrophobia, or pregnancy were excluded. Adult subjects gave informed consent; pediatric subjects gave assent with parental informed consent. Age, sex, and BMI were recorded.
Twenty-nine subjects meeting eligibility criteria were enrolled (mean age 24.2 years, range 12–59 years; mean BMI, 37 kg/m2; BMI range, 28.1–51.1 kg/m2), with twenty children (5 female, 15 male; mean age 15.9 years) and nine adults (1 female, 8 male; mean age 36.0 years). Mean hepatic PDFF measured by MRS was 13.8% (range 0.4–34.3%) (Table 1).
Table 1.
Hepatic PDFF measured by reference MRS
| MRS PDFF (%) | |
|---|---|
| All subjects | |
| Mean ± SD | 13.8 ± 9.0 |
| Range | 0.4–34.3 |
| Adults (n = 9) | |
| Mean ± SD | 10.8 ± 10.7 |
| Range | 0.4–33.8 |
| Children (n = 20) | |
| Mean ± SD | 15.2 ± 8.0 |
| Range | 1.5–34.3 |
Nine MRS PDFF values were averaged for each subject to yield a single per-subject value. This table summarizes the range, mean, and standard deviation (SD) of these per-subject values across the entire study cohort, and separately for adults and children
MR examinations
Subjects were scanned supine with an eight-channel torso phased-array receive surface coil at 3T (GE Signa EXCITE HDxt, GE Medical Systems, Milwaukee, WI) by one of two MR technologists. For any given subject, the technologist was the same. A dielectric pad was placed between the surface coil and abdominal wall.
Subjects underwent three consecutive, same-day MR examinations over a 75- to 90-min period (Fig. 1). Each examination was ~20 min long and included three MRI-M, three MRI-C, and three MRS acquisitions. Between examinations, subjects were removed for 5–10 min, and then repositioned on the scanner table; the phased-array surface coil was reattached, the dielectric pad repositioned, and the next examination performed [15]. A 3 × 3 × 3 acquisition design was used because three measurements are the minimum required for a nontrivial estimation of variance. Using this design, we had an adequate number of measurements to assess both inter- and intra-examination variability. Techniques are summarized below; additional details are provided in the Supplement.
Fig. 1.
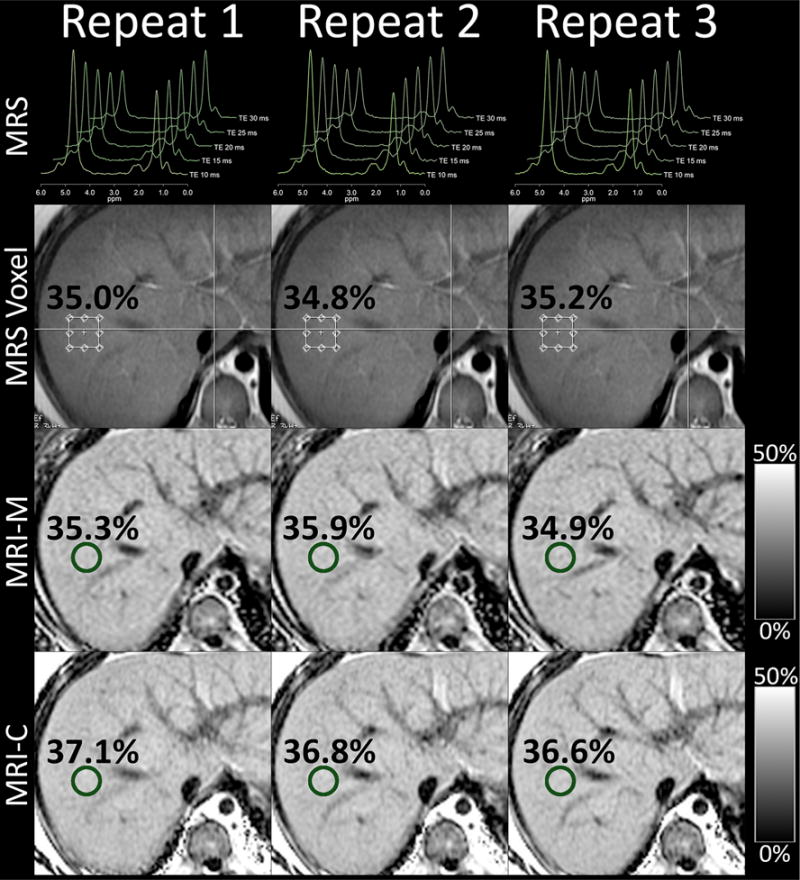
Intra-examination repeatability in a 14-year-old boy. Shown are three repeated magnetic resonance spectroscopy (MRS), magnitude-based magnetic resonance imaging (MRI-M), and complex-based magnetic resonance imaging (MRI-C) acquisitions within a single examination. The anatomic T2W single-shot fast spin echo image used to guide MRS voxel placement is shown. MRS voxel and MRI region of interest locations are overlain as well as the corresponding PDFF values. Notice close agreement between all methods and acquisitions (collages for Figs. 1 and 2 were created using Osirix 5.8 and Photoshop CS6).
MRI-M
MRI-M was performed as described previously [9, 15]. Briefly, a two-dimensional multi-echo spoiled gradient-recalled echo (SPGR) sequence with low flip angle (10°) was acquired with >125-ms repetition time (TR) in a single 18- to 30-s breath-hold. Six echoes were obtained per TR at serial, nominally in- and out-of-phase echo times (TEs) of 1.15, 2.3, 3.45, 4.6, 5.75, and 6.9 ms. Source images were processed offline using a custom plug-in open-source Osirix algorithm (Osirix Foundation, Geneva, Switzerland) to generate PDFF maps [20]. Correction was made for confounding effects due to exponential T2* decay. A spectral model was applied to address the multi-frequency interference effects of protons in fat [9].
MRI-C
MRI-C was performed as described previously [10]. Briefly, an investigational version of a water-fat separation method [Iterative Decomposition of water and fat with Echo Asymmetry and Least-squares estimation (IDEAL)] was implemented as a three-dimensional multi-echo SPGR sequence with low flip angle (3°) at 7-ms TR in a single 21-s breath-hold with two-dimensional Autocalibrating Reconstruction for Cartesian imaging, and with an effective net acceleration factor of 2.2. Six echoes were obtained per TR at TEs of 1.0, 1.8, 2.6, 3.4, 4.2, and 5.1 ms. Images were processed online with an investigational IDEAL algorithm that generated water and fat images from complex source data using a regiongrowing approach to avoid water-fat swapping [14]. Correction was made for confounding effects due to exponential T2* decay [10, 11, 23], noise bias [24], and eddy currents [25]. The same spectral model was applied as for MRI-M [19, 21, 26]. PDFF maps were generated from corrected water and fat images as [F/(F + W)].
MRS
Avoiding major vessels, bile ducts and liver margins, a single 20 × 20 × 20 mm voxel was placed in the right hepatic lobe (Couinaud segments V–VIII) for the first MRS acquisition of the first MR examination, and duplicated on the first MRS acquisition of the second and third examinations using internal landmarks for guidance. The MRS voxel was shimmed automatically, and its location was overlaid on the corresponding axial localization image, and saved for imaging-spectroscopy co-localization. Shimming was redone at the beginning of each examination.
MRS was performed as described previously [9, 14]. Briefly, a stimulated-echo acquisition mode (STEAM) sequence was acquired with long TR (3500 ms) and 5-ms mixing time. Five STEAM spectra were acquired at TEs of 10, 15, 20, 25, and 30 ms in a single 21-s breath-hold. Signals recorded at eight array elements were combined by singular value decomposition, and saved for offline analysis [9].
MR analysis
MRI-M and MRI-C
A single MRI analyst (trained research fellow, 6 months experience) manually placed a 20-mm diameter circular region of interest (ROI) on an out-of-phase image (fifth echo, TE = 5.75 ms) for MRI-M, and on a water image for MRI-C, co-localizing each ROI to the center of the MRS voxel based on anatomic landmarks. For each MRI method, the ROI was propagated to the corresponding PDFF map for the same slice, the slice below and the slice above. Thus, for each MRI method, PDFF values for three ROIs were calculated, centered on the MRS voxel, and the mean of those three values was considered the PDFF estimate for each acquisition [14]. ROIs were placed on anatomic images rather than PDFF maps to avoid potential feedback bias.
MRS
MR spectral analyses were performed as per Hamilton et al. [27, 28] using the Advanced Method for Accurate, Robust and Efficient Spectral fitting of MRS data (AMARES) included in Java-based magnetic resonance user interface software (http://sermn02.uab.es/mrui) [29, 30].
As described previously, T2-corrected areas of the water (4–6 ppm) and the fat (0–3 ppm) were estimated as there is insufficient spectral resolution in vivo to accurately characterize the individual fat peaks, or to distinguish water from two nearby fat peaks. The contribution to the water peak from neighboring fat peaks was corrected using a previously derived fat spectrum post-T2 correction, which reassigned these fat peaks from water to fat signal.
The MRI analyst and the MR spectroscopist were blinded to each other’s results. There were three MRI-M, three MRI-C, and three MRS acquisitions in each of the three MR examinations, and so a total of nine MRI-M, nine MRI-C PDFF, and nine MRS PDFF values were recorded per subject.
Statistical analyses
Statistical analyses were performed by a team of a staff and a faculty biostatistician, using R version 3.0.2 (R Foundation for Statistical Computing, Vienna, Austria, 2013).
Intra-examination repeatability
For each method (MRI-M, MRI-C, and MRS), the SDs of the PDFF values for the three acquisitions within each examination were calculated and their mean (intra-examination SD) was computed for each subject; the mean intra-examination SD across all subjects was then calculated. Intra-class correlation (ICC) for the three acquisitions within each MR examination, and their mean (intra-examination ICC) were computed for each method.
Inter-examination repeatability
For each method, the mean PDFF value for the three acquisitions within each MR examination, and their SD (inter-examination SD) was computed for each subject; the mean inter-examination SD across subjects was then calculated. The inter-examination ICC for each method was computed using the first acquisition of each MR examination.
Aggregate SD
For each subject and each method, the variance of the three first repeats was computed. For each method, aggregate SD, an estimate of overall average inter-exam variability of the sample, was defined as the square root of the average of the first repeat variances.
Comparison across methods
Inter- and intra-examination SDs were compared pairwise across methods using paired t-tests. ICCs of each method were compared pairwise using a bootstrap-based test. Bonferroni adjustment for multiple comparisons was applied to each set of three comparisons.
Accuracy
To assess MRI-M and MRI-C accuracy relative to MRS, MRS was modeled as a function of MRI-M or MRI-C in separate linear regressions. Four accuracy metrics were obtained for the first acquisition of the first MR examination: intercept and slope of the regression line, average bias (square root of the mean squared difference between the regression line and the identity line), and regression coefficient of determination (R2). Bootstrap-based confidence intervals were computed around each accuracy parameter. Bootstrap-based tests with piece-wise Bonferroni adjustment were used to compare MRI-M and MRI-C accuracy parameters.
Results
Intra-examination repeatability
Mean intra-examination SDs were 0.25, 0.42, and 0.49%, for MRI-M, MRI-C, and MRS, respectively (Table 2). Intra-examination SD of MRI-M was lower than that of MRI-C (p = 0.0014) and MRS (p < 0.0001), but intra-examination SDs of MRI-C and MRS were not significantly different (p = 0.192). A representative example is shown in Fig. 1 in a 14-year-old boy, demonstrating the high intra-examination repeatability using the different methods.
Table 2.
Intra- and inter-examination repeatability for hepatic PDFF estimation of each method
| Method | Mean intra-examination SD | Mean inter-examination SD | Mean intra-examination ICC | Inter-examination ICC |
|---|---|---|---|---|
| MRI-M | 0.25 [0.21,0.30] | 0.42 [0.28,0.55] | 0.999 [0.997,0.999] | 0.995 [0.989,0.999] |
| MRI-C | 0.42 [0.33,0.50] | 0.45 [0.32,0.59] | 0.997 [0.987,0.999] | 0.991 [0.959,0.997] |
| MRS | 0.49 [0.42,0.57] | 0.46 [0.35,0.56] | 0.995 [0.989,0.998] | 0.990 [0.976,0.995] |
Values in brackets are 95% confidence intervals, parametric for SDs, bootstrap for ICCs
MRS = magnetic resonance spectroscopy; MRI-M = magnitude-based magnetic resonance imaging; MRI-C = complex-based magnetic resonance imaging; SD = standard deviation; ICC = intraclass correlation coefficient
Mean intra-examination ICCs for MRI-M, MRI-C, and MRS were 0.999, 0.997, and 0.995, respectively. Intra-examination ICC of MRI-M was higher than that of MRI-C or MRS (p < 0.0001 in both cases), but intra-examination ICCs of MRI-C and MRS were not different (p = 0.45).
Inter-examination repeatability
Mean inter-examination SDs were 0.42%, 0.45%, and 0.46% for MRI-M, MRI-C, and MRS, respectively (Table 2). There were no significant differences between them. MRI-M vs. MRI C: p = 0.697, MRI-M vs. MRS: p = 0.654, MRI-C vs. MRS: p = 0.992. A representative example is shown in Fig. 2 in the same 14-year-old boy, demonstrating the high inter-examination repeatability using the different methods.
Fig. 2.
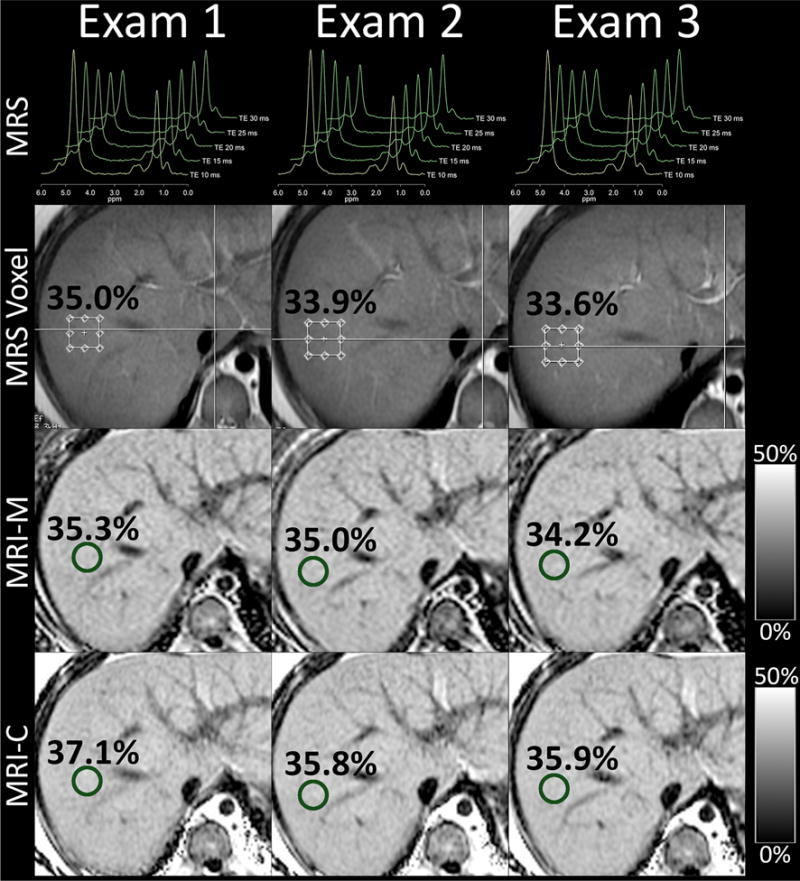
Inter-examination repeatability in a 14-year-old boy. Shown are first acquisitions of magnetic resonance spectroscopy (MRS), magnitude-based magnetic resonance imaging (MRI-M), and complex-based magnetic resonance imaging (MRI-C) for each of the three examinations. The anatomic T2W single-shot fast spin echo image used to guide MRS voxel placement is shown. MRS voxel and MRI region of interest locations are overlain as well as the corresponding PDFF values. Notice close agreement between all methods and examinations (collages for Figs. 1 and 2 created using Osirix 5.8 and Photoshop CS6).
Inter-examination ICCs for MRI-M, MRI-C, and MRS were 0.995, 0.992, and 0.990, respectively. Inter-examination ICC of MRI-M was higher than that of MRS (p < 0.001). Inter-examination ICC of MRI-C was not different from MRI-M or MRS (p = 0.34 and 0.9, respectively).
Aggregate SD
The aggregate SDs for MRI-M, MRI-C, and MRS were 0.64, 0.89, and 0.88%, respectively.
Accuracy
Regression slope, intercept, average bias, and R2 were, respectively, 0.99%, 1.73%, 1.61%, and 0.986 for MRI-M, and 0.96%, 0.43%, 0.40%, and 0.991 for MRI-C (Fig. 3). Intercept and average bias of MRI-C were lower than for MRI-M (p = 0.004 and p < 0.001, respectively; Bonferroni-adjusted p < 0.05), but slope and R2 were not significantly different (p = 0.014 and p = 0.96, respectively; Bonferroni-adjusted p > 0.05 in both cases).
Fig. 3.
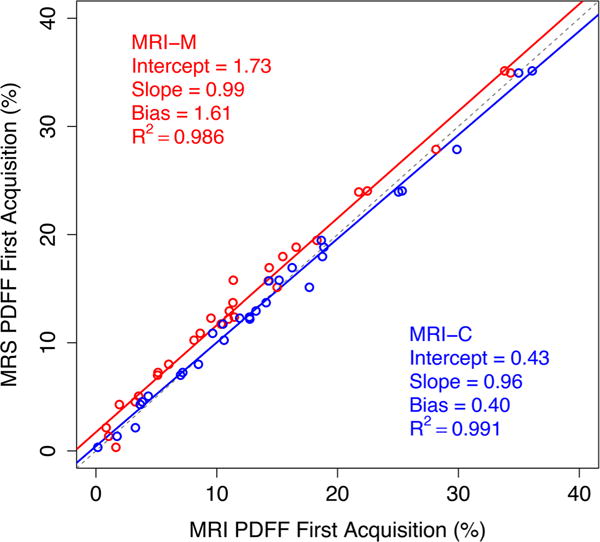
Linear regression of proton density fat fraction (PDFF) estimated by magnitude-based magnetic resonance imaging (MRI-M) and complex-based magnetic resonance imaging (MRI-C) against reference PDFF measured by magnetic resonance spectroscopy (MRS). Regression parameters are shown on the plot (Fig. 3 created using R 3.2.1 and re-sized in Photoshop CS6).
Discussion
As MR usage for detection and quantification of fatty liver disease has spread, need has grown for a reliable fat quantification method that is reproducible across platforms and robust to imaging parameters and techniques. Intra-examination repeatability has been reported for MRI-M and MRS at both 1.5 and 3T [8, 9, 12, 16], and inter-examination repeatability has been demonstrated for MRI-M (3T) [21], MRI-C (1.5T) [26], and MRS (1.5T) [26]. MR-based estimation of PDFF has shown to be equivalent with MRI-C and MRI-M across 1.5 and 3T [12], and across different platforms [15].
We performed a prospective, cross-sectional, single-site clinical study in overweight and obese children and adults to estimate intra- and inter-examination repeatability of MRI-M, MRI-C, and MRS to estimate hepatic PDFF. We kept as many variables constant as possible, using the same scanner, surface coil, dielectric pad, MR, MRI analyst, and spectroscopist across examinations. We also performed the examinations on the same day. Doing so allowed us to keep interpretation of variability focused strictly on inter- and intra-examination repeatability. We also assessed the accuracy of MRI-M and MRI-C to confirm prior reports.
We found both MRI methods to have high intra- and inter-examination repeatability, with standard deviations below 0.5% and ICCs above 0.990 for all analyses. Intra- and inter-examination repeatability were similar, suggesting that variability introduced by subject and coil positioning was negligible. Although intra-examination SDs were lower for MRI-M, this was unlikely to be clinically meaningful as SDs for both MRI methods were below 0.5%. Finally, we confirmed that MRI-M and MRI-C accurately estimated hepatic PDFF, using MRS as a reference.
The high intra- and inter-examination repeatability observed in this study is in accord with previously published studies that separately assessed but did not directly compare MRI-M and MRI-C. Those studies reported intra-examination SDs of <1% for MRI-M and MRI-C [8, 9], and inter-examination SDs of <1% for MRI-M [21] and MRI-C [26]. Additionally, using MRS as a reference, high PDFF estimation accuracy was confirmed for both methods, also as previously reported [7, 8].
The aggregate SD <0.9% for all three methods suggests that in longitudinal studies, a change in hepatic PDFF >1.8% (i.e., >twice the conservatively chosen maximum aggregate SD), represents true change rather than measurement imprecision. This is similar to the 1.6% value reported by Negrete et al. [21] for MRI-M. However, these values are estimates based on cross-sectional aggregate SDs, which assume that measurement techniques remain stable over time, and that there is no temporal variability in true hepatic PDFF. For these reasons, values should be applied with caution in designing longitudinal clinical trials. A better estimate of longitudinal variability, addressing the possibilities of both temporal drift in measurement technique and temporal variability in true hepatic PDFF, would require a longitudinal design with assessment of measurement repeatability on different days, diurnal variation in hepatic PDFF, and physiologic effects of feeding, hydration, exercise, and other potential factors.
A strength of this study was that it enrolled a cohort of overweight and obese pediatric and adult subjects with a clinically relevant PDFF range. Another strength was that our study was the first to directly compare repeatability and accuracy of MRI-M, MRI-C, and MRS in the same subject cohort and imaging setting.
Although we found inter-examination MRS repeatability to be similar to that for MRI-M and MRI-C, it is possible that our study design, with a short time interval between MR examinations and performed by the same technologist, led to overestimation of MRS repeatability. It is conceivable that MRS voxel placement would be less consistent on examinations performed on different days or by different technologists, which could impact repeatability.
Another limitation of our study was that we evaluated repeatability only at the MRS voxel location. However, Negrete et al. recently showed high inter-examination repeatability of MRI-M across all nine Couinaud segments [21]. While it is likely that MRI-C also shows high inter-examination repeatability across hepatic segments, this has not yet been empirically assessed. Also, precision has multiple components, many of which were not addressed, but which could be evaluated in future studies. These include intra-technologist and intra-reader repeatability, and inter-technologist and inter-reader reproducibility.
In conclusion, our findings show that MRI-M, MRI-C, and MRS have high intra- and inter-examination repeatability in estimating hepatic PDFF. Using MRS as a reference, MRI-C may be slightly more accurate than MRI-M, but the difference is small and unlikely to be clinically meaningful. Hence, we suggest both MRI methods as suitable for longitudinal monitoring in clinical trials. Based on inter-examination aggregate SDs, changes in PDFF >1.8% may represent true change rather than measurement imprecision, although this should be confirmed in a longitudinal study. Further research is needed to assess whether examinations performed on different days or with different MR technologists affect repeatability of MRS voxel placement and MRS-based PDFF measurements.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Institutes of Health; Contract grant numbers: NIDDK R01 DK075128, NIDDK R01 DK088925, NCMHD EXPORT P60 MD00220, NIH T32 EB005970, UL1TR000100.
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s00261-015-0542-5) contains supplementary material, which is available to authorized users.
References
- 1.Reeder SB, Cruite I, Hamilton G, Sirlin CB. Quantitative assessment of liver fat with magnetic resonance imaging and spectroscopy. J Magn Reson Imaging. 2011;34:729–749. doi: 10.1002/jmri.22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeder SB, Sirlin CB. Quantification of liver fat with magnetic resonance imaging. Magn Reson Imaging Clin N Am. 2010;18:337–357. ix. doi: 10.1016/j.mric.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Permutt Z, Le TA, Peterson MR, et al. Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2012;36:22–29. doi: 10.1111/j.1365-2036.2012.05121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le TA, Chen J, Changchein C, et al. Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: a randomized controlled trial. Hepatology. 2012;56:922–932. doi: 10.1002/hep.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomsen C, Becker U, Winkler K, et al. Quantification of liver fat using magnetic resonance spectroscopy. Magn Reson Imaging. 1994;12:487–495. doi: 10.1016/0730-725x(94)92543-7. [DOI] [PubMed] [Google Scholar]
- 6.Johnson NA, Walton DW, Sachinwalla T, et al. Noninvasive assessment of hepatic lipid composition: advancing understanding and management of fatty liver disorders. Hepatology. 2008;47:1513–1523. doi: 10.1002/hep.22220. [DOI] [PubMed] [Google Scholar]
- 7.Meisamy S, Hines CD, Hamilton G, et al. Quantification of hepatic steatosis with T1-independent, T2-corrected MR imaging with spectral modeling of fat: blinded comparison with MR spectroscopy. Radiology. 2011;258:767–775. doi: 10.1148/radiol.10100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoo T, Bydder M, Hamilton G, et al. Nonalcoholic fatty liver disease: diagnostic and fat-grading accuracy of low-flip-angle multiecho gradient-recalled-echo MR imaging at 1.5 T. Radiology. 2009;251:67–76. doi: 10.1148/radiol.2511080666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoo T, Shiehmorteza M, Hamilton G, et al. Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology. 2011;258:749–759. doi: 10.1148/radiol.10100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, McKenzie CA, Shimakawa A, et al. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging. 2007;26:1153–1161. doi: 10.1002/jmri.21090. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Shimakawa A, McKenzie CA, et al. Multiecho waterfat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med. 2008;60:1122–1134. doi: 10.1002/mrm.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artz NS, Haufe WM, Hooker CA, et al. Reproducibility of MR-based liver fat quantification across field strength: same-day comparison between 1.5T and 3T in obese subjects. J Magn Reson Imaging. 2015;42:811–817. doi: 10.1002/jmri.24842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guaraldi G, Besutti G, Stentarelli C, et al. Magnetic resonance for quantitative assessment of liver steatosis: a new potential tool to monitor antiretroviral-drug-related toxicities. Antivir Ther. 2012;17:965–971. doi: 10.3851/IMP2228. [DOI] [PubMed] [Google Scholar]
- 14.Johnson BL, Schroeder ME, Wolfson T, et al. Effect of flip angle on the accuracy and repeatability of hepatic proton density fat fraction estimation by complex data-based, T1-independent, T2*-corrected, spectrum-modeled MRI. J Magn Reson Imaging. 2014;39:440–447. doi: 10.1002/jmri.24153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang GH, Cruite I, Shiehmorteza M, et al. Reproducibility of MRI-determined proton density fat fraction across two different MR scanner platforms. J Magn Reson Imaging. 2011;34:928–934. doi: 10.1002/jmri.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn JP, Hernando D, Mensel B, et al. Quantitative chemical shift-encoded MRI is an accurate method to quantify hepatic steatosis. J Magn Reson Imaging. 2014;39:1494–1501. doi: 10.1002/jmri.24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ligabue G, Besutti G, Scaglioni R, Stentarelli C, Guaraldi G. MR quantitative biomarkers of non-alcoholic fatty liver disease: technical evolutions and future trends. Quant Imaging Med Surg. 2013;3:192–195. doi: 10.3978/j.issn.2223-4292.2013.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mashhood A, Railkar R, Yokoo T, et al. Reproducibility of hepatic fat fraction measurement by magnetic resonance imaging. J Magn Reson Imaging. 2013;37:1359–1370. doi: 10.1002/jmri.23928. [DOI] [PubMed] [Google Scholar]
- 19.Reeder SB, Robson PM, Yu H, et al. Quantification of hepatic steatosis with MRI: the effects of accurate fat spectral modeling. J Magn Reson Imaging. 2009;29:1332–1339. doi: 10.1002/jmri.21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang A, Tan J, Sun M, et al. Nonalcoholic fatty liver disease: MR imaging of liver proton density fat fraction to assess hepatic steatosis. Radiology. 2013;267:422–431. doi: 10.1148/radiol.12120896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negrete LM, Middleton MS, Clark L, et al. Inter-examination precision of magnitude-based MRI for estimation of segmental hepatic proton density fat fraction in obese subjects. J Magn Reson Imaging. 2014;39:1265–1271. doi: 10.1002/jmri.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.US Department of Health and Human Services. Analytical Procedures and Methods Validation for Drugs and Biologics: Guidance for Industry. Food and Drug Administration CDER & CBER; 2015. http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm386366.pdf. Accessed 23 Dec 2014. [Google Scholar]
- 23.Hines CD, Frydrychowicz A, Hamilton G, et al. T(1) independent, T(2) (*) corrected chemical shift based fat-water separation with multi-peak fat spectral modeling is an accurate and precise measure of hepatic steatosis. J Magn Reson Imaging. 2011;33:873–881. doi: 10.1002/jmri.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CY, McKenzie CA, Yu H, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med. 2007;58:354–364. doi: 10.1002/mrm.21301. [DOI] [PubMed] [Google Scholar]
- 25.Hernando D, Hines CD, Yu H, Reeder SB. Addressing phase errors in fat-water imaging using a mixed magnitude/complex fitting method. Magn Reson Med. 2012;67:638–644. doi: 10.1002/mrm.23044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noureddin M, Lam J, Peterson MR, et al. Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials. Hepatology. 2013;58:1930–1940. doi: 10.1002/hep.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton G, Middleton MS, Bydder M, et al. Effect of PRESS and STEAM sequences on magnetic resonance spectroscopic liver fat quantification. J Magn Reson Imaging. 2009;30:145–152. doi: 10.1002/nbm.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton G, Yokoo T, Bydder M, et al. In vivo characterization of the liver fat 1H MR spectrum. NMR Biomed. 2011;24:784–790. doi: 10.1002/jmri.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naressi A, Couturier C, Devos JM, et al. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001;31:269–286. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 30.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129:35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


