Abstract
In this study, we trace developmental stages using epigenome changes in human embryonic stem cells (hESCs) treated with drugs modulating either self-renewal or differentiation. Based on microscopy, qPCR and flow cytometry, we classified the treatment outcome as inducing pluripotency (hESC, flurbiprofen and gatifloxacin), mesendoderm (sinomenine), differentiation (cyamarin, digoxin, digitoxin, selegeline and theanine) and lineage-commitment (RA). When we analyzed histone PTMs that imprinted these gene and protein expressions, the above classification was reassorted. Hyperacetylation at H3K4, 9, 14, 18, 56 and 122 as well as H4K5, 8, 12 and 16 emerged as the pluripotency signature of hESCs. Methylations especially of H3 at K9, K20, K27 and K36 characterized differentiation initiation as seen in no-drug control and fluribiprofen. Sinomenine-treated cells clustered close to “differentiation initiators”, consistent with flow cytometry where it induced mesendoderm, along with cyamarin and possibly selegnine. Neurectoderm, induced by RA and theanine manifested methylations on H3 shifts to H3.3. By both flow cytometry and histone PTM clustering, it appears that cells treated with gatifloxacin, flurbiprofen, digitoxin and digoxin were not yet lineage-committed or mixed cell types. Taken together, our moderate-throughput histone PTM profiling approach highlighted subtle epigenetic signatures that permitted us to predict divergent lineage progression even in differentiating cells with similar phenotype and gene expression.
Keywords: histones, mass spectrometry, embryonic stem cells, differentiation, self-renewal
1 Introduction
The ability of human embryonic stem cells (hESCs) to self-renew as well as to differentiate into derivatives of the three embryonic germ layers (endoderm, ectoderm and mesoderm) both in vitro and in vivo, a property called pluripotency has offered valuable avenues to study embryonic development. The loss of pluripotency is the necessary first step to both spontaneous and lineage-directed differentiation, achieved by both monolayer growth and embryoid body formation. Growth factors and small molecule modulators have been shown in vitro to mimic the spatial and temporal expression patterns of germ layer markers, facilitating cellular differentiation [1]. In a high-throughput analysis for regulators of hESC self-renewal, Desbordes et al (2009) found several known therapeutic agents to promote pluripotency (flurbiprofen, gatifoxacin, sinomenine and theanine) and differentiation [digitoxin, digoxin, cymarin, selegiline and retinoic acid (RA)][2]. Compounds such as IDE1, IDE2 and indolactam V have been reported to steer endoderm cells to pancreatic lineage [3], stauprimide to synergize with activin A to promote endoderm differentiation of hESCs [4] and TGF-β receptor inhibitor, SB431542 to act with Noggin to induce neural differentiation in hESCs [5]. These studies demonstrated that small molecules can be used to regulate hESC self-renewal and differentiation.
Recently, epigenetic factors have emerged as important determinants of biological processes. Small molecule epigenetic modulators such as valproic acid (histone deacetylase inhibitor), BIX-01294 (H3K9 histone methyltransferase G9a inhibitor) and RG108 (DNA methyltransferase inhibitor) as well as chromatin-modifying enzymes have been found to facilitate reprogramming of somatic cells to pluripotent cells inducing ectopic Oct4 and KLF expression [6, 7, 8]. While these and numerous other reports confirm that remodeling of the epigenetic landscape is necessary to cell-fate programming [9], its role in development and cell fate determination is poorly understood.
Most differentiation protocols for pluripotent cells result in heterogeneous cell populations, yielding small percentage of the desired phenotype. A case in point for small molecule-directed differentiation is protein kinase C activator, indo lactam V (ILV) guiding definitive endoderm from hESCs into pancreatic progenitors [10]. While the available protocols focus on stepwise timely addition of cytokines and small molecules in response to expression of specific genes, the efficiency of the methodologies are far from optimal and rarely result in functional phenotypes. Low yield of pure cell populations have so far allowed only amplification-based methods for characterization of the epigenetic landscape of the chromatin. Using chromatin immunoprecipitation-sequencing (ChIP-seq), relative abundance of histone post-translational modifications (PTMs) such as H3K4me3, H3K27me3, and H3K36me3 were observed to change in five key cardiovascular developmental stages: undifferentiated hESCs, mesodermal progenitors, specified tripotential progenitors, committed cardiovascular cells, and definitive cardiovascular cells [11]. This confirmed that distinct histone PTM patterns are associated with cell fate decisions.
Mass spectrometry (MS) has been applied to study proteomics and epigenetics in human induced pluripotent cells (iPSCs) and hESCs [12, 13]. Moreover, MS has continuously evolved towards higher throughput and flexibility, allowing not only identification and quantification of single histone PTMs, but also their combinatorial patterns and even characterization of the intact proteins [reviewed in 14, 15]. Due to the high mass accuracy and sensitivity, MS has become the technique of choice, outperforming antibody-based strategies to study known and novel global histone PTMs even in low stoichiometry. In this study, we used MS to examine the global histone PTM landscape of hESCs exiting self-renewal and initiating differentiation. We screened nine drugs reported to modulate these events, validating their differentiation state by monitoring expression of gene and protein markers. Our study showed that enriched acetylation at H3K4, 9, 14, 18, 56 and 122 as well as H4K5, 8, 12 and 16 marked the pluripotent hESCs, while loss/decrease of acetylation marked differentiation. Besides this, both exit from pluripotency and lineage commitment (mesendoderm and neurectoderm) induced in this model system also seem to have a unique epigenome. Thus, the imprinted PTM states proved to be a valuable adjunct to gene/protein expression to characterize developmental stages.
2 Materials and methods
2.1 Human embryonic stem cells tissue culture and quantitative PCR
Human embryonic stem cell (hESC) strain WA09-Oct4-pGZ (also called H9-Oct4-GFP) was purchased from WiCell (Wisconsin). The cells expressed green fluorescent protein (GFP) upon activation of Oct4 promoter by mTeSR™1 Complete medium (Stem Cell Technologies), when grown at 37°C on matrigel (BD Biosciences). Once cells reached 80% confluency, they were detached using Accumax (Millipore) and washed with mTESR. The cells were replated 1:4 in mTESR1 complete medium; on day 2, the medium was changed to differentiation medium containing DMEM/F12 medium supplemented with 20% (vol/vol) KnockOut serum replacer, 0.1 mM nonessential amino acids, 1 mM L-glutamine (all from Invitrogen), 0.1 mM β-mercaptoethanol (Sigma), and treated with drugs at concentration range indicated in Table 1. Cells were examined for Oct4-GFP using fluorescent microscopy (Nikon Eclipse TE2000-U) daily and conditions that had no GFP were stained with Hoechst 33342 (Life Technologies) on day 6. Cell viability was assessed using propidium iodide exclusion in flow cytometry (BD LSRII, BD Biosciences) to determine a non-toxic concentration for further studies (Table 1). Briefly, cells were washed twice in PBS and incubated for 20 minutes at 37°C in cell dissociation buffer (Invitrogen). Cells were then dissociated by gentle pipetting, collected and resuspended in PBS for analysis.
Table 1.
Candidate compounds influencing human embryonic stem cell self-renewal and differentiation, based on gene and protein analysis.
| Therapeutic agents | Reported effect | Observed effect | Conc tested* | Non-toxic conc used |
|---|---|---|---|---|
| Flurbiprofen | Self-renewal | Self-renewal | 10uM-1mM | 50uM |
| Gatifoxacin | Self-renewal | Self-renewal | 10uM-1mM | 5uM |
| Sinomenine | Self-renewal | Differentiation | 10uM-1mM | 50uM |
| Theanine | Self-renewal | Differentiation | 10uM-1mM | 10uM |
| Digitoxin | Differentiation | Differentiation | 100pM-10nM | 5nM |
| Digoxin | Differentiation | Differentiation | 100pM-10nM | 1nM |
| Cymarin | Differentiation | Differentiation | 10pM-1nM | 1nM |
| Selegiline | Differentiation | Differentiation | 10uM-1mM | 100uM |
| Retinoic Acid** | Differentiation | Differentiation | 100nM-10uM | 1uM |
10uM concentrations used by Desbordes et al., 2008;
Positive control for differentiation
At day 6, about 106 cells each were washed in PBS for mRNA expression studies and flow cytometry, while approximately 107 washed cells were snap-frozen in liquid nitrogen for histone extraction. For qPCR, total mRNA was extracted from harvested cells using an RNeasy Mini kit and treated with RNase-free DNase (Qiagen, Valencia, CA). Reverse transcription was performed with the first-strand cDNA synthesis kit (Invitrogen). Quantitative PCR was performed in triplicate on a Roche Lightcycler 480 using LightCycler 480 SYBR Green I master mix (Roche, Indianapolis, IN) and 500 nM primer concentrations. All gene expression levels were normalized to the housekeeping gene GAPDH.
2.2 Flow cytometry
Cells were washed in PBS, dissociated into single cells using dispase (Invitrogen), filtered through 40μm nylon cell strainers and labeled for different lineages using anti-Brachyury (Abcam; ab20680), anti-FoxA2 (Abcam; ab5074), anti-human nestin conjugated to Alexa 488 (ebioscience; 53-9843-80) and anti-human NCAM-PE (ebioscience; 12-0567-41). Unconjugated primary antibodies were visualized using anti-rabbit Alexa 488 (Invitrogen; A21206), anti-rabbit A647-PE (Invitrogen; A20991) and anti-goat Alexa 647 (Invitrogen; A21447). Briefly, cells were fixed in 4% paraformaldehyde, permeabilized in 0.3% Triton X-100 in PBS and blocked in 3% horse serum. A minimum of 25,000 cells were stained per condition in primary antibodies for 30 min on ice, washed, followed by secondary staining for 30 min in dark on ice. Flow cytometric analysis was performed on BD LSRII multilaser analyzer with FACSDiVa software (BD Biosciences).
2.3 Histone extraction and digestion
Histones were acid-extracted and processed with two cycles of chemical derivatization, each before and after trypsin digestion, followed by desalting as previously described [16]. Briefly, histones were acid-extracted from nuclei with 0.2 M H2SO4 for 2 hours and precipitated with 33% trichloroacetic acid (TCA) overnight. Protein concentration was calculated using the Bradford assay. Purified histones were then dissolved in 30 μl of 50 mM NH4HCO3, pH 8.0. Derivatization reagent was prepared by mixing propionic anhydride with 2-propanol in the ratio 1:3 (v/v) and added to the histone sample in the ratio of 1:2 (v/v) for 15 min at 37°C. This reaction was performed twice to obtain complete labeling. Histones were then digested with trypsin (enzyme:sample ug ratio of 1:20, 6 hours at 37°C) in 50 mM NH4HCO3. After digestion, the derivatization reaction was performed again twice to cap peptide N-termini. Samples were then desalted by using C18 stage-tips [17].
2.4 Nano-liquid chromatography and tandem mass spectrometry for histone PTM analysis
Samples were analyzed through 75 μm ID × 17 cm Reprosil-Pur C18-AQ (3 μm; Dr. Maisch GmbH, Germany) in a nano-column using an EASY-nLC nanoHPLC (Thermo Scientific, Odense, Denmark). The HPLC gradient was 0–35% solvent B (A = 0.1% formic acid; B = 95% MeCN, 0.1% formic acid) over 30 min and from 34% to 100% solvent B in 20 minutes at a flow-rate of 250 nL/min. LC was coupled with a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Full scan MS spectrum (m/z 290–1650) was performed in the Orbitrap with a resolution of 30,000 (at 400 m/z) with an AGC target of 1×10e6. The MS/MS events included both data-dependent acquisition and targeted peptides. The targeted peptides were the ones with isobaric mass to enable MS/MS-based quantification. Fragmentation was performed by using higher-energy collisional dissociation (HCD) with normalized collision energy of 35, an AGC target of 5×10e4 and a maximum injection time of 120 ms. MS/MS data were collected in centroid mode in the orbitrap mass analyzer (resolution 7,500 at 400 m/z).
2.5 Data analysis of histone PTM signatures
The relative abundance of histone peptides was calculated by using in-house software EpiProfile [18], with 10 ppm tolerance for extracting the ion chromatogram. Only histone H3 and H4 peptides were considered in the analysis. A list of commonly searched peptides for canonical histone H3 PTM analysis was generated using Matlab. Extracted ion chromatogram was performed to determine the abundance of the various peptides. The relative abundance of a modified form was calculated by dividing the abundance of a specific modified form by all forms of the same given peptide (which was considered as 100%). This type of normalization corrects for minimal errors in sample injection, since histone PTMs are estimated as a relative number. In case of isobaric peptides (e.g. KacSTGGKAPR and KSTGGKacAPR) we performed targeted MS/MS in order to extract the relative ratio of the two species using unique fragment ions. Only charge state 1+ was considered for product ions.
3 Results
The objective of this study is to describe epigenetic characteristics of hESCs in two conditions: (i) maintenance of self-renewal and (ii) alternative differentiation states that potentially mirror exit from self-renewal. Oct4 was used as the major deterministic marker to investigate the influence of selected small molecules on self-renewal. We cultured Oct4-GFP-reporter hESC line, H9-Oct4-pGZ as monolayer in self-renewing condition and the next day (day 1 of differentiation), switched to non-directed differentiation medium with drugs, monitoring the expression of the Oct4 for 6 days (Fig. 1A). Morphologically, the compact hESCs seen in tight colonies and uniformly expressing GFP started enlarging by day 2, and colonies spread out into more confluent monolayer by day 3. This morphology has been associated with the epigenomic “openness” of the hESC, reflected in small nucleus in which nuclear factors are minimally compartmentalized. This is in contrast to differentiation, where nuclei enlarge, become more variable in shape and get compartmentalized, consistent with changes in chromatin structure and nuclear organization [19] and cease to express GFP, corresponding to downregulation of pluripotency.
Figure 1.
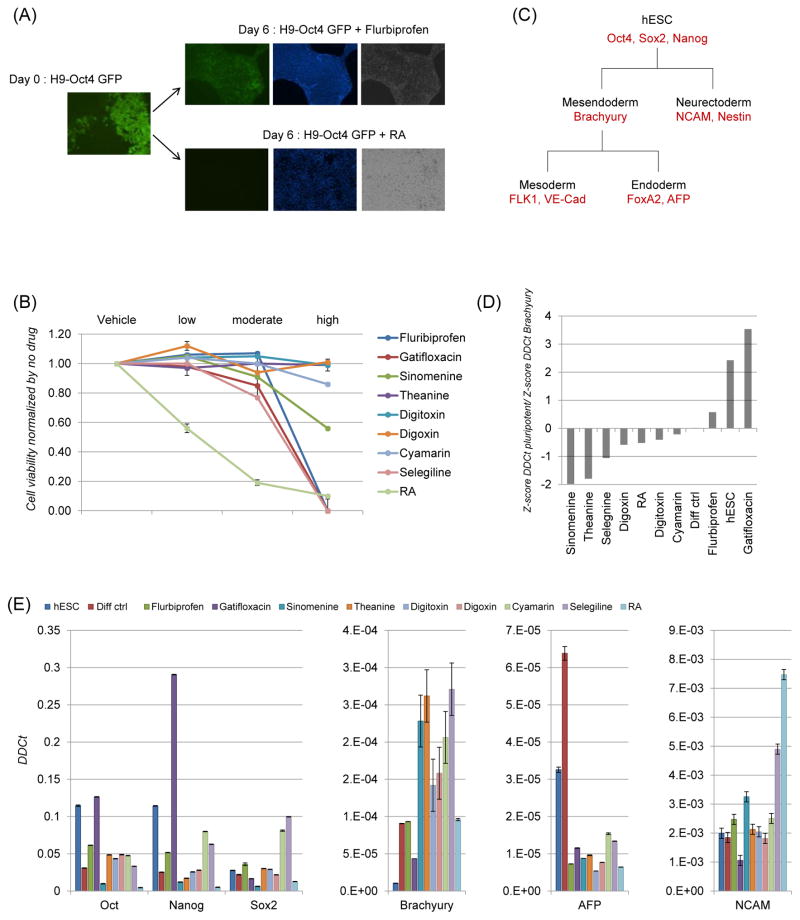
Cell viability, Oct4-GFP expression and gene markers for pluripotency. (A) Fluorescent microscopy shows a colony of self-renewing H9-Oct4-GFP (green) uniformly. On differentiating for 6 days in the presence of drugs (two informative examples, flurbiprofen and retinoic acid, RA shown) in medium lacking pluripotent signals, cells were fairly Oct-4-positive in flurbiprofen while Oct4-GFP expression was extinguished in RA, showing clearly that flurbiprofen supported self-renewal. For comparison, nuclei stained by Hoechst 33342 (blue) and brightfield images are shown. (B) Human ESC cell line, H9 grown to self-renew in monolayer were switched to differentiation medium and treated with various concentrations of drugs (high concentration log-titrated to moderate and low doses, Table 1) for 6 days with media + drug change every other day. On day 6, flow-cytometric analysis of cell viability was done for propidium iodide exclusion (% propidium-iodide-positive, i.e., dead). Values for each condition and concentration were normalized by no drug treatment. Cells grown in medium + DMSO served as vehicle control. Non-toxic concentrations listed in Table 1 were used for further studies. (C) Flow chart showing possible commitment and lineage progression of H9 cells grown in differentiation medium. Gene and protein markers used for quantitative PCR and flow cytometry are indicated (red). (D) Expression of the genes denoted were analyzed by qPCR and expression was normalized by z-score across the different drug treatments, and then plotted as the ratio of combined (Oct4+Nanog+Sox2) pluripotent/Brachyury. This plot classifies ES, flurbiprofen and gatifloxacin-treated conditions together and apart from the rest of the conditions. (E) Expression of the genes denoted was analyzed by qPCR. Normalized expression is shown as the mean±SD from three experiments. hESC is control for pluripotency, Diff control is drug is drug-free differentiation control, RA is the positive differentiation control.
3.1 Drugs affected cell viability, morphology and Oct4-GFP expression
Initially, a dose-response experiment was performed treating hESCs to increasing concentrations of selected drugs to determine the minimal dose needed for cell viability (Table 1; Fig. 1B). The nontoxic doses used in the current study were different from the 10μM used in the previous study utilizing non-GFP H9 cells, except for theanine where the same concentration was used [2]. Some drugs had to be titrated to nanomolar concentrations (digitoxin, digoxin and cymarin) or lower micromolar concentrations (gatifloxacin and RA). However, drugs such as flurbiprofen, sinomenine and selegiline were well-tolerated at higher than the reported concentrations. Both morphology of colonies and GFP fluorescence of Oct4-promoter was monitored during drug treatment to determine loss of self-renewal and pluripotency in the hESCs. Different drug treatments resulted in various percentages of GFP expression; while it was somewhat sustained in flurbiprofen in differentiation condition, the rest of the drugs markedly extinguished Oct4-GFP expression, indicating that flurbiprofen fostered self-renewal even in a differentiation environment (Fig. 1A). This effect of flurbiprofen is strengthened by the fact that in a matched experiment, all other drugs except RA failed to induce differentiation of hESCs in self-renewal medium (data not shown). This suggests that withdrawal of self-renewal factors in mTESR and not the presence of drugs are the strongest signal for loss of pluripotency. Therefore, drug treatment may only enhance the commitment of the differentiating cells to particular lineages. In this light, flurbiprofen effectively substituting the self-renewal cues in differentiation medium seems significant.
3.2 Gene expression profiles for determination of pluripotency and differentiation
The pluripotency genes Oct4, Nanog and Sox2 are characteristic biomarkers of hESCs cultured in self-renewal medium. While exogenous Oct4 and Sox2 induce pluripotency in differentiated cells (as in induced pluripotent cells, iPSCs), it is well recognized that the ratio of Sox2 and Oct4 is critically controlled and even small increases in levels triggered differentiation [20]. Therefore, in order to accurately assess pluripotency, we also included Nanog as a third marker. Theoretically, most of the cells exiting self-renewal stage may express early lineage genes such as Brachyury and neural cell adhesion molecule, NCAM (Fig. 1C). Brachyury is expressed in the nascent mesendoderm and is required for primitive streak development [21] while NCAM is a pan-neuronal marker [22]. As differentiation progresses, Brachyury+ cells can diverge into mesoderm or endoderm and can be tracked by gene markers FLK1/VE-Cad for mesoderm progenitors [23] and FoxA2 for definitive endoderm/AFP for viscera endoderm [24]. First, we confirmed the correlation of Oct4-GFP expression to pluripotency by determining gene expression profile of hESCs before and after drug treatment using quantitative polymerase chain reaction (qPCR). Since the levels of expression of pluripotency genes and Brachyury were of different magnitude, we normalized expression using z-score across the different drugs treatment, and then plotted the ratio of obtained score for pluripotent markers to Brachyury (Fig. 1D). This comparison unequivocally established that hESCs, flurbiprofen and gatifloxin had a transcriptional profile typical of self-renewing cells, consistent with GFP expression and morphology. Therefore, multiple gene markers and analytical strategies proved valuable in understanding confounding transcript profiles. Compared to spontaneously differentiating cells, hESCs treated with cyamarin still had some residual pluripotency gene transcripts (Fig. 1E). However, the cells had completely lost Oct4-GFP by microscopy (data not shown) and expressed Brachyury transcripts indicating that they were poised to differentiate into mesendoderm. Similarly, cells cultured in selegiline had lingering Nanog and Sox2 mRNA, while completely devoid of Oct4 transcript; these cells expressed both Brachyury and NCAM indicating mixed mesendodermal and neurectodermal populations respectively (Fig. 1E). In short, maintenance of pluripotency markers, along with absence of Brachyury or lineage markers, indicated pluripotency, while the reverse meant differentiation. Interestingly, both sinomenine and theanine extinguished Oct4-GFP, while turning on Brachyury (Fig. 1D and 1E), contrary to self-renewing potential reported earlier [2] (Table 1). All other drugs were found to have effects consistent with previous work (Table 1).
3.3 Flow cytometry confirms emergence of lineages
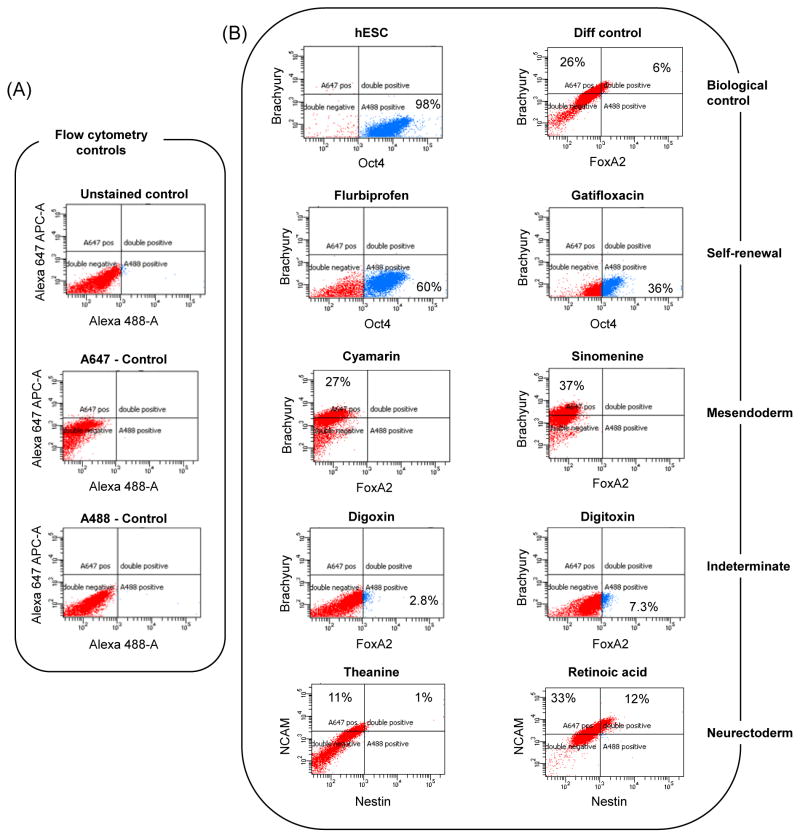
Next, we looked for protein expression of differentiation markers such as Brachyury (mesoendoderm), FoxA2 (endoderm), NCAM and nestin (neurectoderm) using two-color flow cytometric analysis, gating with flow cytometric controls: unstained cells (control) and fluorochrome-matched isotype controls (A647- and A488 controls) (Fig. 2A). Human ESCs with 98% cells expressing Oct4-GFP and none expressing Brachyury served as positive control for pluripotency; hESC grown drug-free in differentiation medium was the negative control for drug effects. On day 6, cells grown with flurbiprofen and gatifloxacin had 60% and 36% cells expressing Oct4, respectively aligning closer to hESCs. Cyamarin and sinomenine had considerable Brachyury expression (27% and 37%) consistent with gene transcript levels indicating that they were differentiating although not into definitive endodermal progenitors (FoxA2-negative). Digoxin and digitoxin did not express either Brachyury or FoxA2. Cells treated with cyamarin, sinomenine, digoxin and digitoxin appear to be in various developmental stages of a particular lineage or most likely had a mixed population of progenitors from various lineages. On the other hand, RA induced nestin and NCAM robustly, while theanine also expressed, although to a lesser degree. Overall, taking into account gene transcript and flowcytometric protein analyses, we could cluster the drug treatments as follows: hESCs (pluripotency control), flurbiprofen and gatifloxacin (self-renewal), cyamarin, selegnenine and sinomenine (mesoendoderm or indeterminate), drug-free control, digoxin and digitoxin (indeterminate) and theanine and RA (neurectoderm).
Figure 2.
Flow cytometric analysis for pluripotent and lineage markers expressed on day 6 of differentiation in the presence of drugs. Alexa 647-APC and Alexa 488 antibodies were used to double-stain cells to look for pluripotency (Oct4+/Brachury-), mesendoderm (Oct4-/Bra+, Bra+/FoxA2-), endoderm (FoxA2+) and neurectoderm (NCAM+, NCAM+/nestin+). (A) Antibody controls using RA-treated cells. (B) Lineage-specific marker analysis.
3.4 Quantification of histone PTMs before and after drug treatment
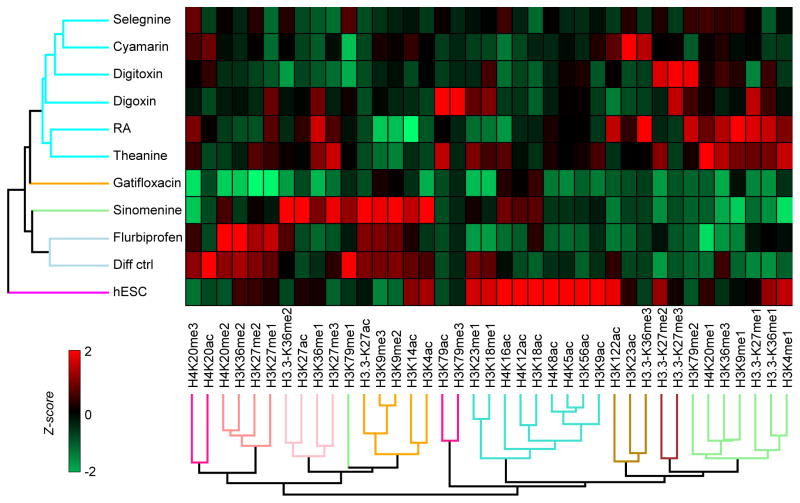
Following gene and protein profiling for differentiation, we sought to define the histone H3 and H4 PTMs in drug-induced pluripotency, mesendoderm and neurectoderm gene/protein clusters, with hESC as positive control and conditions producing mixed lineages (indeterminate) as negative control. Histone PTMs were clustered by performing Z-score transformation for individual marks between treatments (Fig. 3). We arbitrarily grouped the different drug treatments into five clusters based on the Euclidean distance of the generated tree. As expected, hESCs had very unique PTM patterns, including the highest relative abundance of several acetyl marks. The permissive mark H3K9ac was most abundant in hESCs and the repressive mark H3K9me1 was most abundant in RA-treated cells. Histone H3K9ac, along with H3K4me3, has been reported to activate Oct4 in hESCs while methylations on H3K9 and H3K27 suppressed Oct4 in definitive endoderm cells and hepatocytes [25]. H3K18ac, a mark with a robust peak at the transcription start site (TSS) of active and poised genes in ChIP-seq studies [26], was also most abundant in hESCs. Similarly, acetylation of H3K56, H4K5, H4K8, H4K12 and H4K16 were mostly abundant in hESCs, reflecting the generally permissive nature of the pluripotent chromatin [27]. Among the hyperacetyl marks in hESC, H3K56ac has been shown to correlate positively with the binding of Nanog, Sox2, and Oct4 transcription factors at their target gene promoters [27]. On the other hand, the global acetylation patterns in gatifloxacin and flurbiprofen do not resemble hESCs, in spite of Oct4 and Brachyury expression at gene transcript and protein levels. However, both these conditions are defined as single element in unique clusters, with flurbiprofen having the least deviation from hESCs as implied by the length of the branches in the hierarchical tree. This clearly reflects the epigenetic changes being set up for the next developmental stage which still does not show up in the transcriptome and proteome.
Figure 3.
Characterization of histone PTM signatures. Hierarchical clustering and heat map visualization of histone PTMs normalized by using z-score between conditions.
Both differentiation control and flurbiprofen that represent exit from pluripotency had higher expression of the repressive mark H3K9me2/me3, as well as H3K27me1/me2, H3K36me2 and H4K20me2. Cyamarin, sinomenine, digoxin and digitoxin clustered together primarily based on absence of any notable histone PTM pattern, consistent with indeterminate protein expression. We also observed many invariant PTMs that possibly are “redundant histone codes”, that provide both stability and plasticity to the epigenome [reviewed in 28]; it should also be recognized that not all local rearrangements on the chromatin are reflected at global levels at a particular time point.
Another interesting finding is that H3.3 characterized by the unique peptide 27–40 had more variability of PTMs compared to H3 counterpart. Since H3.3 normally replaces H3 at transcriptionally active regions [29], we propose that the H3/3.3 peptide 27–40 may feature prominently in divergent repertoire of genes transcribed in hESCs and differentiated cells.
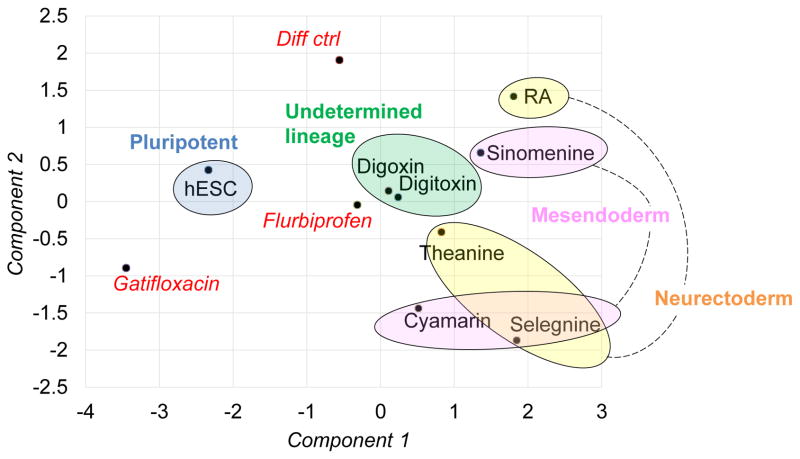
In short, to provide a simpler overview of histone PTM/phenotype correlation, we plotted our dataset using Principal Component Analysis (PCA) (Fig. 4). Results highlighted that the histone PTM profile mirrored the partial germ layer grouping by qPCR (Fig. 1D and 1E) and flow cytometry (Fig. 2). Human ESCs, mesendoderm and neurectoderm were each defined by unique epigenomes that imprinted pluripotency and lineage-restricted gene/protein changes. Spontaneously differentiating cells (no drug/differentiation control) and the rest of the treatments yielded partially differentiating cells that had subtle histone changes intermediate between pluripotent and lineage-committed cells. As histone codes precede gene and protein expression in the temporal sequence of programming, epigenome will look more differentiated than pheno- and genotype at some early timepoint. In general, since most differentiation protocols result in heterogeneous cell populations that get committed asynchronously and progress into different/same lineages at different rates, histone PTM analysis of sorted cells or at single cell level will resolve indeterminate cell types when the technology is available. Also, the seemingly indeterminate histone PTMs even in well-characterized cell populations may be part of combinatorial codes and may cross-talk for regulation of differentiation.
Figure 4.
Principal component analysis of histone PTM relative abundances with highlights of functional analysis from flow cytometry and qPCR (in ellipses). The blue ellipse highlights the pluripotent stage. Cells treated with gatifloxacin and flurbiprofen and no drug (Diff ctrl) have characteristics of cells exiting pluripotency (in red italic font). Digoxin and digitoxin have differentiated features that do not fall within lineages analyzed by limited gene and protein markers used in the study (green ellipse; undetermined lineage); selegnine shares some common traits with both mesendoderm and neurectoderm. Cyamarin and sinomenine represent typical mesoendoderm cells (pink ellipse), while theanine and RA represent neuroectoderm cells (yellow ellipse).
4 Discussion
Human ES cells grow clonally and therefore are heterogeneous unless maintained in selective conditions as we did with zeocin-resistant H9-Oct4-pGZ cell line expressing GFP when Oct4 was active. In this homogenous pluripotent population with uniform epigenomes, we predicted an array of effects ranging from withdrawal of self-renewal signal, directed differentiation, early lineage progenies and sustenance of self-renewal using alternate cues, when drugs were used. As expected, we found drugs inducing a variety of early lineages, ranging from defined self-renewal to lineage-committed cells, with cells poised for lineages in between. Even though by transcriptome, hESCs, flurbiprofen- and gatifloxacin-treated cells appeared to be pluripotent, protein expression and epigenome analysis showed evidence for differentiation. Such distinction of pheno/genotype and histone modifications has been reported for Oct4-expressing ESC cultures [30, 31] since quantitative PCR and flowcytometry seems to be detecting the waning pluripotent gene and protein expression, while the epigenome is setting up for differentiation. With this multidimensional data, the similarities and differences were compressed in the PCA (Fig 4) and revealed lineage-poised, committed and indeterminate differentiation. Interestingly, Desbordes and coworkers (2008) reported that hESCs treated with self-renewal drugs (flurbiprofen, gatifloxacin, theanine and sinomenine) showed increased percentage of undifferentiated cells on repeated passaging suggesting positive selection of pluripotent cells of a particular pheno/genotype and their clonal expansion, suggesting that these drugs possibly are truly inducing self-renewal [2]. We did not maintain drug-treated cells in long-term culture and cannot comment on culture adaptation.
Generally, histone acetylation is found to be associated with gene activation, being found at the promoters of actively transcribed genes as well as throughout the active gene [6, 8]. In the current study, we found most H3 and H4 lysines acetylated in hESCs, consistent with earlier observations [32]; this correlates with increased chromatin plasticity and transcription [32]. HDAC inhibition by valproic acid and butyrate facilitated cellular iPSC reprogramming confirming histone acetylation may play an important role in pluripotency [9–12]. Melcer and coworkers showed that rather than directly modulating expression of pluripotency genes, histone acetylations indirectly acted to increase chromatin-bound protein accessibility and mobility [33]. However, other site-specific acetylations such as H3K23ac and H3K27ac, besides H4K20ac were increased in cyamarin, sinomenine and differentiation conditions respectively. Acetylation mark-specific permissive states can exist both during pluripotency and lineage commitment and rely largely on chaperones, transcription factors and DNA modifications [34]. HDACi especially suppressing HDAC3 activity promoted pluripotency gene transcription while those to HDAC1/5/8 promoted H3 acetylations favoring neurodevelopmental gene expression [35]. From the descriptive map we have provided herein of all histone PTMS at different cellular states, future studies can localize hyperacetylation marks to individual promoter regions and reveal the epigenetic mechanism involved.
Besides the hyperacetylation signature of pluripotent hESCs, the epigenome category that is interesting encompasses RA and theanine with high expression of methylations: H3K4me1, H3K9me1, H3K36me1, H3K36me3, H3K79me2, H3.3-K27me1, H3.3-K36me1 and H4K20me1. Both these drugs express gene and protein markers consistent with neural or mesectodermal derivatives. With theanine still expressing more Brachyury and less NCAM than RA, it appears that RA has definitely committed cells to neural lineage while theanine is either getting them there or driving them into mesectoderm; it is possible that H3K79me3 expressed more in theanine than RA is a marker of mesectoderm. The third intriguing epigenomic class comprising flurbiprofen and spontaneously differentiating cells had characteristic abundance of H3K27me1, H3K27me2, H3K36me2 and H4K20me2. Phenotypically, flurbiprofen still expresses Oct4-GFP, while the later has extinguished it, moving on to differentiation. Nevertheless, the increase in just methylation of H3 and H4 seem to indicate that these (i) facilitate loss of pluripotency (ii) activate differentiation genes (iii) repress non-lineage genes during differentiation. The fourth class of epigenome unique to sinomenine is characterized by H3K9me2, H3K9me3, H3K27me3, H3K79me1, H3.3-K36me2 as well as acetylations on H3K4, H3K14, H3K27 and H3.3-K27ac, aligning closer to drug-free differentiation and flurbiprofen.
Since the epigenomes were clustered not only based on expression of histone PTMs, but also the degrees to which other PTMs were downregulated or maintained, these PTMs become important correlates of functional signatures. For example, the co-occurrence of H3K14ac, H3K9me3 and H3K9me2 in RA-treated cells gains significance in light of the reported H3K9me2/H3K14ac crosstalk during G9a inhibition of MCF7 cells [36]. Based on the above results, it appears that the modification patterns of histone H3 are subject to dynamic changes and are fine-tuned in a cell type-specific manner during hESC differentiation. Shift in PTM-crosstalk and bivalent domains possibly facilitate the cell fate divergences of hESCs during differentiation. It is also possible that relative abundances of different histones, their variant forms as well PTMs on histones other than H3 and H4 may have important role in priming differentiation.
With mass spectrometry-enabled histone PTM analysis, we have extended the repertoire of tools available for monitoring stem cell differentiation. Even though the drug-induced differentiation is far from complete, we show that PTM analysis may be a useful complement to the gene/protein expression profiles for monitoring differentiation in vitro. Histone PTM analysis not only confirmed lineages classified by gene and protein expression, but also helped separate indeterminate differentiation conditions into cells not yet committed (gatifloxacin, flurbiprofen) as well as those that have mixed lineages (digitoxin and digoxin).
In conclusion, we highlight that in spite of a homeostasis in the global histone H3 and H4 abundance, pharmacological cues promoting varied cellular fates were characterized by histone signatures. Priming to different lineages were imprinted by the epigenetic changes that were evident in our analysis. The scope of this paper was to identify PTM profile that defined maintenance and exit from self-renewal, which unequivocally seems to be the acetylation patterns. Profiling of H3 and H4 PTMs on a genome-wide scale requires relatively homogeneous populations of cells which is a limitation for most differentiation protocols. Secondly, mass spectrometry requires large numbers of such populations purified by fluorescence-activated cell sorting with specific antibodies. In future, the combined power of efficient differentiation strategies and ability to perform mass spectrophotometry in small numbers of cells will greatly increase the potential of genome-wide epigenetic profiling approaches targeted at unmasking histone PTMs, cross-talk and integrated network of epigenetic modulators regulating development.
Supplementary Material
Statement of significance of the study.
(i) Pluripotency and self-renewal have been described thus far by transcriptome analysis. The current work is one of the few epigenetic studies that describes the histone H3 and H4 signatures of the uncommitted as well as differentiating human embryonic stem cells in a moderate-throughput manner. (ii) We used a novel strategy of priming multiple lineages using small molecules known to induce differentiation and found that their global histone profiles reflected the poised state of developmental genes. (iii) We found self-renewal being characterized by specific histone acetylation pattern consistent with chromatin openness. (iv) Importantly, we found cell stage-specific histone patterns that imprinted developmental gene and protein marker expression. (v) With this comprehensive map of over 40 histone PTMs for emerging lineage-committed cells, future studies looking at PTM crosstalk and bivalent chromatin domains will help understand the functional epigenetic modulation during development.
Acknowledgments
We gratefully acknowledge funding from NIH grant R01GM110174.
Abbreviations
- ac
acetylation
- AFP
Alpha feto-protein
- ChIP-seq
Chromatin immunoprecipitation-sequencing
- conc
concentration
- DMSO
Dimethyl sulfoxide
- FLK1
Fetal liver kinase1
- FoxA2
Forkhead box A2
- GFP
Green fluorescent protein
- HCD
Higher-energy collisional dissociation
- hESCs
human embryonic stem cells
- iPSCs
induced pluripotent cells
- KLFa
Kruppel-like factor a
- me1/me2/me3
mono/di/trimethylation
- MS
mass spectrometry
- NCAM
Neural cell adhesion molecule
- Oct4
Octamer-binding transcription factor 4
- PBS
Phosphate-buffered saline
- PTM
Post translational modification
- qPCR
Quantitative PCR
- RA
Retinoic acid
- Sox2
SRY-box 2
- TCA
trichloroacetic acid
- VE-Cad
vascular endothelial-cadherin
References
- 1.Zaret KS. Using small molecules to great effect in stem cell differentiation. Cell Stem Cell. 2009;4:373–374. doi: 10.1016/j.stem.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Desbordes SC, Placantonakis DG, Ciro A, Socci ND, et al. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiak M, Maehr R, Chen S, Chen AE, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H, Wurdak J, Wang CA, Lyssiotis EC, et al. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell. 2009;4:416–426. doi: 10.1016/j.stem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Desponts C, Do JT, Hahm HS, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Huangfu D, Maehr R, Guo W, Eijkelenboom A, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onder TT, Daley GQ. New lessons learned from disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev. 2012;22:500–508. doi: 10.1016/j.gde.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waddington CH. Gene regulation in higher cells. Science. 1969;166:639–640. doi: 10.1126/science.166.3905.639. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Borowiak M, Fox JL, Maehr R, et al. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 11.Paige SL, Thomas S, Stoick-Cooper CL, Wang H, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benevento M, Munoz J. Role of mass spectrometry-based proteomics in the study of cellular reprogramming and induced pluripotent stem cells. Expert Rev Proteomics. 2012;9:379–399. doi: 10.1586/epr.12.30. [DOI] [PubMed] [Google Scholar]
- 13.Phanstiel D, Brumbaugh J, Berggren WT, Conard K, et al. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young NL, Dimaggio PA, Garcia BA. The significance, development and progress of high-throughput combinatorial histone code analysis. Cellular and molecular life sciences : CMLS. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J Proteomics. 2012;75:3419–3433. doi: 10.1016/j.jprot.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res. 2006;5:988–994. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- 18.Yuan ZF, Lin S, Molden RC, Cao XJ, Bhanu NV, et al. EpiProfile quantifies histone peptides with modifications by extracting retention time and Intensity in high-resolution mass spectra. Mol Cell Proteomics. 2015;14:1696–1707. doi: 10.1074/mcp.M114.046011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler JT, Hall LL, Smith KP, Lawrence JB. Changing nuclear landscape and unique PML structures during early epigenetic transitions of human embryonic stem cells. J Cell Biochem. 2009;107:609–621. doi: 10.1002/jcb.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 22.Conley BJ, Trounson AO, Mollard R. Human embryonic stem cells form embryoid bodies containing visceral endoderm-like derivatives. Fetal Diagn Ther. 2008;19:218–223. doi: 10.1159/000076701. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa SI, Nishikawa S, Hirashima M, Matsuyoshi N, et al. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hematopoietic lineages. Development. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- 24.Wong JC, Gao SY, Lees JG, Best MB, et al. Definitive endoderm derived from human embryonic stem cells highly express the integrin receptors αV and β5. Cell Adhesion & Migration. 2010;4:39–45. doi: 10.4161/cam.4.1.10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H, Jang MJ, Kang MJ, Han YM. Epigenetic signatures and temporal expression of lineage-specific genes in hESCs during differentiation to hepatocytes in vitro. Hum Mol Genet. 2011;20:401–412. doi: 10.1093/hmg/ddq476. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Zang C, Rosenfeld JA, Schones DE, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan Y, Xue Y, Song C, Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maleszka R, Mason PH, Barron AB. Epigenomics and the concept of degeneracy in biological systems. Brief Funct Genomics. 2014;13:191–202. doi: 10.1093/bfgp/elt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong SH, Rampalli S, Lee JB, McNicol J, et al. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell. 2011;9:24–36. doi: 10.1016/j.stem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Meshorer E, Yellajoshula D, George E, Scambler PJ, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, et al. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nature Communications. 2012;3:910–933. doi: 10.1038/ncomms1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cedar H, Bergman Y. StemBook [Internet] Cambridge (MA): Harvard Stem Cell Institute; 2008–2009. Epigenetic silencing during early lineage commitment. [PubMed] [Google Scholar]
- 35.Qiao Y, Wang R, Yang X, Tang K, Jing N. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J Biol Chem. 2015;290:2508–2520. doi: 10.1074/jbc.M114.603761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, et al. Chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.