Abstract
μ-Opioid receptors (μOR) are G protein coupled receptors (GPCRs) that are activated by a structurally diverse spectrum of natural and synthetic agonists including endogenous endorphin peptides, morphine and methadone. The recent structures of the μOR in inactive1 and agonist-induced active states (companion article) provide snapshots of the receptor at the beginning and end of a signaling event, but little is known about the dynamic sequence of events that span these two states. Here we report the use of solution-state NMR to examine the process of μOR activation. We obtained spectra of the μOR in the absence of ligand, and in the presence of the high-affinity agonist BU72 alone, or with BU72 and a G protein mimetic nanobody. Our results show that conformational changes in transmembrane segments (TM) 5 and 6, which are required for the full engagement of a G protein, are almost completely dependent on the presence of both the agonist and the G protein mimetic nanobody revealing a weak allosteric coupling between the agonist binding pocket and the G protein coupling interface (TM5 and TM6) similar to what has been observed for the β2-adrenergic receptor2. Unexpectedly, in the presence of agonist alone, we observe larger spectral changes involving intracellular loop 1 (ICL1) and helix 8 (H8), when compared to changes in TM5 and TM6. These results suggest that one or both of these domains may play a role in the initial interaction with the G protein, and that TM5 and TM6 are only engaged later in the process of complex formation. The initial interactions between the G protein and ICL1 and/or H8 may play a role in G protein coupling specificity as has been suggested for other family A GPCRs.
Type mu-Opioid receptors (μOR) are membrane proteins belonging to the G protein-coupled receptor (GPCR) superfamily that are responsible for the analgesic and addictive properties of morphine3, a natural alkaloid contained in opium4. In fact, due to the clinical importance of opioids in pain management and addiction, five decades of medicinal chemistry generated a very chemically diverse opioidergic ligand system (reviewed in5) that can modulate the μOR intracellular signaling, namely the inhibitory heterotrimeric Gi protein and arrestins pathways6. The pharmacology of these different compounds are very well characterized and they present a wide spectrum of activity with respect to efficacy towards G protein and/or arrestin from very weak partial agonism to full agonism6,7. Evidence suggests that activation of the arrestin pathway may be responsible for some of the adverse clinical effects of the opioids drugs currently on the market6.
Intrinsic efficacy is a parameter used to define the effect of a ligand on GPCR activation of specific signaling pathways8. To understand intrinsic efficacy it is necessary to understand the process of GPCR activation. So far this process has only been described for three GPCRs using biophysical and crystallographic analyses of both inactive and active states: rhodopsin9, the β2-adrenergic receptor (β2-AR)10,11 and muscarinic M2 receptor (M2R)12. The most complete characterization of the GPCR activation process by diffusible ligands came from NMR studies on the β2-AR13–17 showing that the binding of agonist alone is associated with conformational heterogeneity in the receptor and is not sufficient to stabilize the active β2-AR conformation. Whether this observation is generalizable to other GPCR has not been determined.
Crystal structures are now available for the μOR in both inactive and fully agonist-induced states, representing the beginning and end of a signaling event (see companion article). Here, in an effort to better understand the process of μOR activation upon binding of opioid agonists, we took advantage of the versatility of NMR spectroscopy (as recently described for the β2-AR18) to analyze the activation signal propagation occurring in different receptor domains (See Extended data Fig.1 and methods for further technical details).
Dimethylamines give peaks with different intensities in the 1H-13C Heteronuclear Multiple Quantum Coherence (HMQC) spectrum of unliganded 13C-dimethylated μOR (13Cm2-μOR) preparations (Fig. 1a) that we unambiguously assigned to ten lysines and to the shortened N-terminus (starting at G52) using a systematic mutagenesis and proteolysis experiments (Extended data Fig. 2). It is interesting to note the coexistence of broad and irregular peaks (i.e. K98ICL1, K100ICL1 and K3448.51) with sharp and intense signals (K209ECL2, K2696.24) in the apo-state. Because a peak linewidth directly reports on the homogeneity of the local fluctuating magnetic field which depends on the dynamics of the labeled domain, this observation qualitatively indicates that the different domains under investigation i.e TM6 and ICL1-H8 present different dynamic properties (in the µs to ms range). Indeed, broad peaks suggest that the ε-N[13CH3]2 moieties detect two or more conformations (environments) that exchange on an intermediate time scale (low ms), while sharp peaks may represent a single conformation or two or more conformations that exchange on either a faster or slower time scale. The absence of a signal for a specific lysine may represent a lack of side chain dynamics or a dynamic timescale in the "slow intermediate" regime19. We have focused our analysis on a specific set of NMR correlation peaks that were separated in two groups of sensors: the extracellular (G52N-ter and K209ECL2) and intracellular (K98 ICL1, K100 ICL1, K2605.66, K2696.24, K2716.26 and K3448.51) lysines (Fig. 1b, Fig. 1c and Fig. 1d). The signals for other probes are very weak (K174ICL2, K2335.40, K3036.58) or even not observable (K1413.26 and K1854.43) and spectral modifications are difficult to interpret (Extended data Fig. 2).
Figure 1.
HMQC spectrum of unliganded 13C-dimethylated μOR and positions of lysine residues in the inactive (cyan blue) and active (orange) μ-opioid receptor. (a) Position and identity of eleven correlated 1H-13C peaks corresponding to the μOR dimethyllysines and amino-terminus. (b) Superposition of inactive (cyan blue, 4DKL) and active (orange, #PDB) conformation of μOR highlighting the lysines (sticks) and the ε-NH2 (spheres) positioned in the extracellular face (yellow box) and in the intracellular face (blue box) of the μOR. Also shown are the BU72 ligand (green spheres) and the G protein mimetic nanobody (Nb) position (salmon surface). Extracellular (c) and intracellular (d) view of the μOR showing the reorganization of the lysine side chains upon receptor activation from inactive (cyan spheres) to fully active (orange spheres). The shortened amino-terminal domain (N-terminus) starting at G52 was not modeled in the inactive state structure but makes contact with the 7TM bundle to cap the binding pocket in the agonist-induced state.
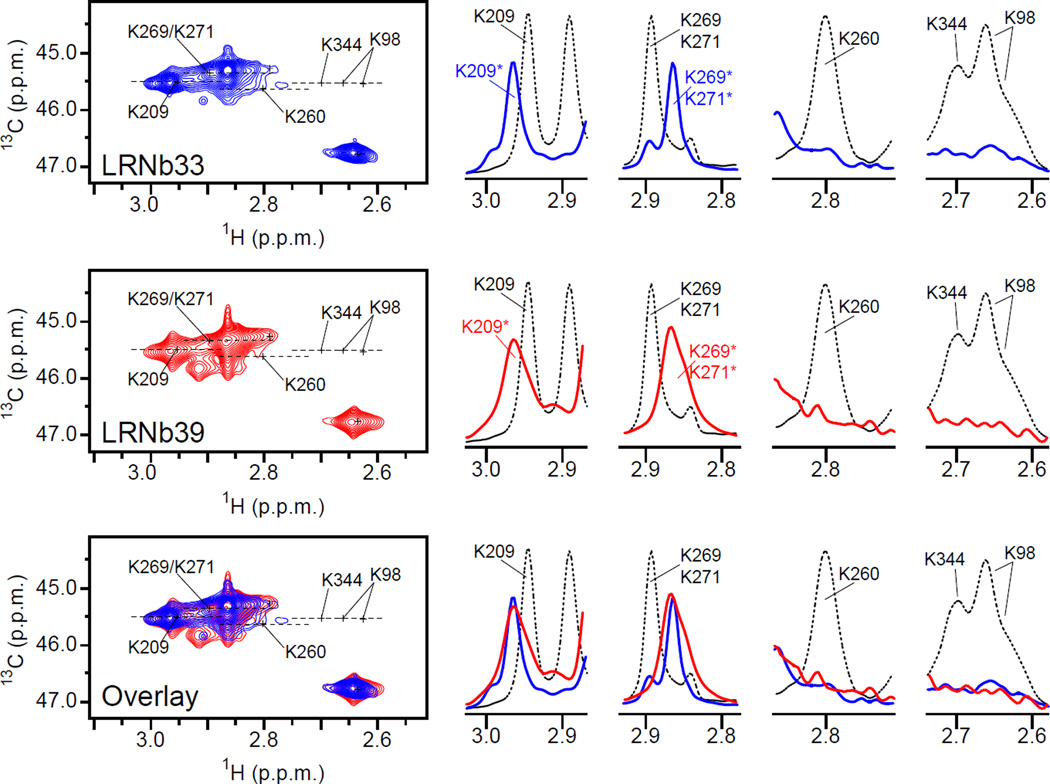
We first analyzed the effect of BU72, a high-efficacy μOR agonist20, on the NMR signals using a saturating amount of ligand and/or a saturating amount of a G protein mimetic nanobody (Nb33) (Fig. 2). We have selected BU72 because of its extremely high efficacy/potency and its slower dissociation rate relative to other agonists. Moreover this ligand was successfully used in the crystallization studies (see companion article). Nb33 is closely related to Nb39, the nanobody used to stabilize and capture the μOR agonist-induced state in crystallization studies (see companion article). Nb33 and Nb39 differ by only two amino acids in a region that does not directly interact with the receptor, and they have similar G protein-like effects on the affinity of agonist for the μOR. They also have similar effect in NMR experiments (Extended data Fig. 3). We used Nb33 throughout this study because it can be purified in much larger quantities than Nb39 (see methods).
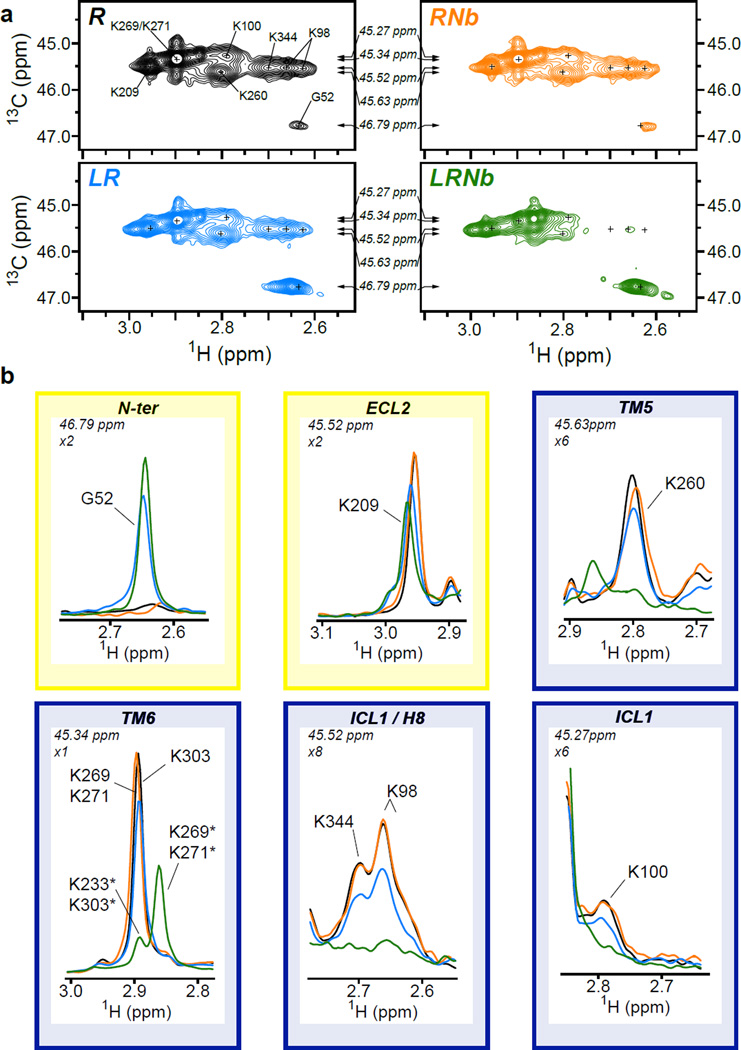
Figure 2.
Activation of μOR by BU72. (a) HMQC spectra of unliganded 13C-dimethylated μOR (30 µM, R, black), 13C-dimethylated μOR treated with a saturating concentration of BU72 (150 µM, LR, blue), 13C-dimethylated μOR treated with BU72 and a saturating concentration of Nb33 (60 µM, LRNb, green) and of unliganded 13C-dimethylated μOR treated with a saturating concentration of Nb33 (60 µM, RNb, orange). The small crosses indicate the position of peaks visible in the apo-state that are reduced in intensity or no longer visible in the other spectra. Arrows indicate the position of the 1D slice represented in (b). (b) 1D slice of HMQC spectra in the 1H dimension highlighting the effect of BU72 alone (blue) and BU72 + Nb33 (green) on the N-terminus (G52) and on indicated set of dimethyllysines (ECL2, TM5, TM6, ICL1 and H8 lysines). The yellow and blue boxes indicate the positions in the extracellular and intracellular domains respectively. The zoom level for each peak and the 13C chemical shifts are indicated in the top left of each panel. Asterisks indicate the chemical shifts of methyl probes in the presence of both BU72 and Nb33.
In the extracellular surface, the peak corresponding to K209ECL2 was slightly modified upon treatment with BU72 (Fig. 2a, LR panel) and, as better observed in the 1H dimension, was decreased in intensity and shifted downfield (Fig. 2b, ECL2 panel). As discussed above, this spectral modification indicates a change in the conformational state of the μOR most likely due to a modification of the K209ECL2 microenvironment, as the beta-hairpin containing this residue undergoes a change upon receptor activation (Fig. 1c). When the sample was treated with a saturating concentration of the Nb33 only, no significant changes were observed (Fig. 2a, RNb panel), suggesting an absence of environment changes around K209ECL2 in the absence of agonist. When both BU72 and Nb33 were added to the 13Cm2-μOR, the K209ECL2 peak intensity decreased further as compared to the sample treated with BU72 alone (Fig. 2a LRNb panel and Fig. 2b ECL2 panel). We also observed changes in the N-terminus upon binding of BU72 alone; however, in this case the peak intensity increases. BU72 and Nb33 co-treatment slightly increased the effect observed for BU72 alone (Fig. 2b, N-ter panel). It is of interest that the active-state crystal structure of the μOR reveals that the N-terminus folds into the binding pocket and interacts with BU72 (companion article).
The potentiating effects of Nb33 on BU72 are much more pronounced in the intracellular part of 13Cm2-μOR, as BU72 or Nb33 induced only small spectral changes by themselves (Fig. 2a LR and RNb panels), while formation of the ternary complex led to more dramatic spectral modifications (Fig. 2a, LRNb panel). The proton peak centered around 2.9 ppm, which corresponds to the lysines K2696.24, K2716.26 and K3036.58 positioned in TM6, almost disappeared (the leftover peak being K3036.58, see assignments in Extended data Fig 2) with a concomitant appearance of an intense peak at 2.85 ppm due to K2696.24 and K2716.26 alone (Fig. 2b, TM6 panel). This spectral shift, which is not observed in spectra from receptor treated with BU72 alone, is consistent with the 10Å outward movement of TM6 upon receptor activation that repositions the K2696.24 and K2716.26 side chains (Fig. 1d). The K2605.66 peak intensity is also markedly decreased by exposure to both BU72 and Nb33, and thus represents a sensor for the receptor conformational change in TM5 (Fig. 2b, TM5 panel). These results show that binding of the high-affinity agonist BU72 alone is not sufficient to fully stabilize the active conformations of TM5 and TM6 and support the notion of a weak connection between the ligand binding and the G protein coupling domains of the μOR. This weak coupling was previously observed for the β2AR, and was thoroughly investigated more recently using NMR and DEER spectroscopy2. Of interest, the results obtained with the β2AR and the μOR stand in contrast to rhodopsin for which a stronger link between photoisomerization of retinal and structural changes at the cytoplasmic end of TM6 was observed21.
In addition to the TM5 and TM6 probes, we also characterized the conformational transitions in the ICL1 (K98ICL1 and K100 ICL1) and H8 domains (K3448.51) of μOR that also undergo conformational changes upon μOR activation (Fig. 1d). Comparison of the R panel with the LRNb panel in Fig. 2a revealed a near complete loss of intensity of K98ICL1, K100ICL1 and K3448.51 peaks upon μOR treatment with BU72 and Nb33 (Fig. 2a and Fig. 2b, ICL1/H8 panels, orange trace). While a complete loss of intensity for these peaks was only observed with both BU72 and Nb33, we did observe a reduction in intensity with BU72 alone (Fig. 2b, ICL1/H8 panels, red trace). The loss of peak intensity could originate from a change in the local dynamic of ICL1 and H8 domains in the µs to ms timescale that broadens the correlation peaks beyond the detection limit, a well-characterized phenomenon in NMR spectroscopy22, and suggests that receptor activation is modifying significantly the structural dynamic of these domains. It should be noted that none of the lysines discussed here directly interact with the related Nb39 in the crystal structure. Changes in the NMR signals intensity therefore reflect the μOR activation process. Moreover, the effects of agonist and Nb33 can be reversed by treatment with the antagonist naloxone (Extended data Fig. 4).
Taken together our data demonstrate that binding of the G protein surrogate is required for the agonist to fully stabilize the active conformation in both extracellular and intracellular domains, suggesting the existence of a two-way allosteric coupling between the extracellular μOR ligand binding domain and the G protein coupling interface. These data are in agreement with what was observed with the isotopically labeled β2-AR13 and suggest that this behavior could be a common feature of the GPCR activation process.
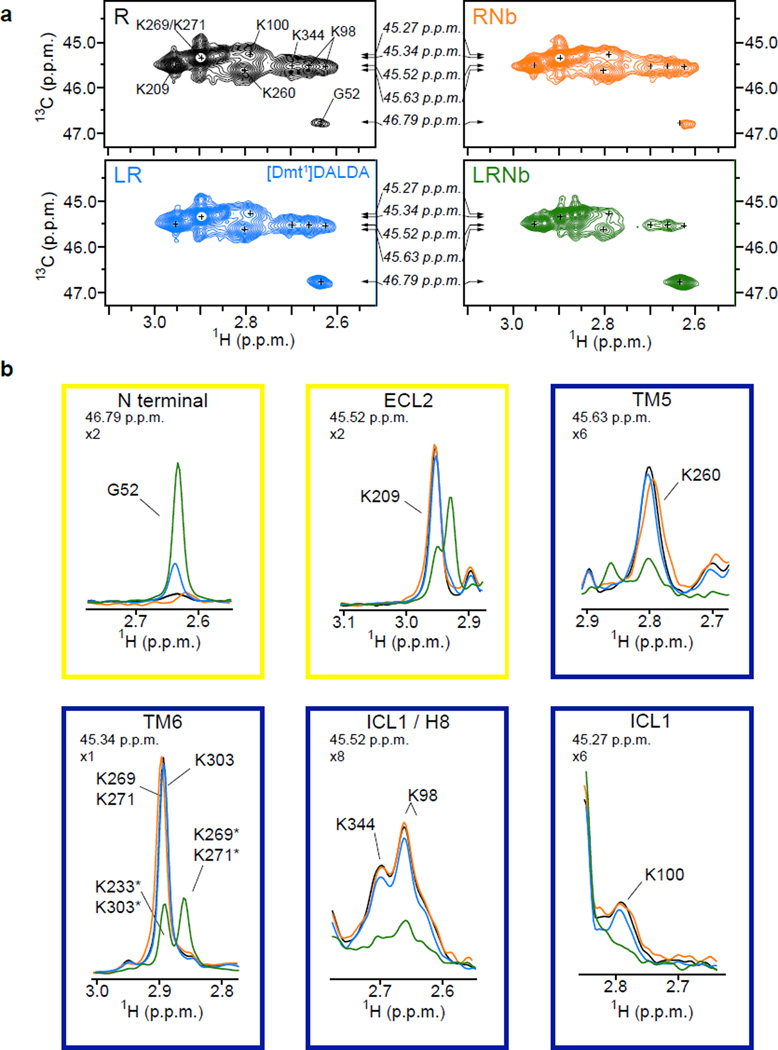
We next quantitatively analyzed the effect of BU72 alone (LR) and together with Nb33 (LRNb) on the volumes of the different intracellular lysine peaks (from TM5, TM6, ICL1 and H8 domains). Of particular interest, we found a notable difference in the effect of BU72 alone on TM5 and TM6 compared to its effect on ICL1 and H8 domains (Fig. 3a). While BU72 alone had a slight effect on TM5 and TM6 signal intensity, it produced larger spectral changes for the ICL1 and H8 sensors (Fig. 3a), suggesting that the allosteric coupling from the agonist binding pocket to ICL1/H8 is stronger than the coupling to TM5 and TM6. This effect is not specific to BU72 as we observed the same phenomenon with two other agonists [Dmt1]-DALDA (2',6'-dimethyltyrosine-D-Arg-Phe-Lys-NH2) and DAMGO (Tyr-D-Ala-Gly-NMe-Phe-Gly-ol) (Fig. 3a, Extended data Fig. 5 and Extended data Fig. 6). Notably, the formation of the ternary complex leads to larger volume changes for the different domains under investigation (Fig. 3b) and, together with Fig. 3a, demonstrates that the fully agonist-induced state can only be achieved in the presence of an agonist and a G protein mimetic nanobody.
Figure 3.
Quantitative analysis of spectral changes in intracellular domains of μOR and a structural interpretation. Percent of maximal effect in the volume of indicated peaks in sample treated with agonist alone (BU72, [Dmt1]-DALDA and DAMGO) (a) or agonists together with Nb33 (b) highlighting the stronger effect of agonist in absence of Nb33 on the ICL1 and H8 domains as compared to TM5 and TM6 domains. Error bars represent the uncertainty in volume determination due to random noise (see methods). (c) Model proposing the propagation of conformational changes from the agonist binding pocket to the ICL1 and H8 domains initiating interactions with the G protein Gi (represented by the α5 C-terminal domain of Gi, yellow circle) (middle panel) before the fully agonist-induced state of the μOR is reached (bottom panel), with the direct interaction of the Giα5 with TM5 and TM6. Numbered circles represent the identity and the position of the TM domains view from the intracellular side of the membrane.
While we observe that agonists alone have larger effects on ICL1 and H8 reporters than on TM5 and TM6 reporters, our steady-state experiments do not provide information about the temporal sequence of these conformational changes; however, we speculate that a G protein may first engage ICL1 and/or Helix 8 before engaging TM5 and TM6. In fact, the H8 domain is critical for G protein coupling as observed for rhodopsin23 and for other GPCRs (see24). More interestingly, the H8 of the muscarinic M3R was shown to cross-link with the Gαq protein in an agonist-dependent manner25 suggesting a close proximity between the receptor H8 domain and the G protein. Our results support a model where G protein could initially interact with the H8 and/or ICL1 domains of the receptor before docking in the 7TM core and stabilizing the TM5 and TM6 domains in an open conformation (Fig. 3c).
Here we describe the measurement of conformational changes associated with μOR activation in both the extracellular and intracellular domains. Our data offer insights into agonist regulation of μOR subdomain conformations revealing a possible allosteric pathway of activation from the agonist binding pocket to the ICL1 and H8 domains to initiate the ternary complex formation and associated changes in TM5 and TM6. Importantly our data complement the high-resolution structural data of the μOR agonist-induced state bound to BU72 described in the companion article and offer unprecedented insights into the structural mechanisms of opioid receptor activation. A better understanding of the structural basis for opioid receptor activation may lead to new therapeutic approaches with fewer side effects.
METHODS
Protein expression and purification
Nanobody (Nb) sequences were subcloned into a pMalp2x vector containing an N-terminal, 3C protease-cleavable maltose binding protein (MBP) tag and a C-terminal 8×His tag. Plasmids were transformed into BL21(DE3) cells and protein expression induced in LB by addition of IPTG to 1 mM at an OD600 of 0.8. After 24 h of incubation at 20 °C, cells were collected and sonicated in the presence of lysozyme and benzonase. MBP–nanobody fusions were purified by Ni-NTA chromatography and MBP was removed using 3C protease. Cleaved MBP was separated from the nanobody by additional amylose purification and size exclusion chromatography in a buffer containing 20 mM HEPES pH 7.5 and 0.1 M NaCl. Of note, because we needed large amount of Nb for the NMR experiments, we decided to use Nb33 throughout this study as it was produced and purified in much larger quantities than Nb39 (almost ten times as much for the same volume of BL21(DE3) cell culture).
We generated a μ-OR mouse construct with features designed to enhance stability for NMR spectroscopy. A tobacco etch virus (TEV) protease recognition site was introduced after residue 51, and a human rhinovirus 3C protease site after residue 358. A FLAG epitope tag was added to the amino-terminus and an octa-histidine tag was appended to the carboxy terminus. Receptor expression was largely improved by using a M72T single point mutation as assessed by naloxone binding. The final construct (μOR–2x) is shown in Extended data Fig. 1.
The μOR–2x construct was expressed in Sf9 cells using the Bestbac (Expression systems) baculovirus system in the presence of 3 µM naloxone. For all other mutants (assignments) we used the bac-to-bac system (Life technologies). Sf9 cell cultures (Life technologies) were grown to a density of 4×106 cells per ml, infected with baculovirus containing the μOR-2x genes, shaken at 27°C for 48 hr, and cell pellets were harvested and stored at −80 °C. The receptor was purified as previously described1 and eluted from the anti-FLAG M1 affinity resin with a buffer containing 0.01% MNG, 0.001% CHS, 20 mM HEPES pH 7.5, 0.1 M NaCl, 1 μM naloxone, 0.2 mg/ml FLAG peptide and 2 mM EDTA.
To remove flexible amino and carboxy termini, TEV and 3C protease were added at a 1:5 and 1:10 Protease:μOR-2x ratio by weight. The sample was incubated at 4°C overnight in the presence of 100 µM of TCEP. We then used a negative Ni-NTA chromatography step to remove TEV and 3C proteases.
μOR Reductive methylation
Receptor preparation from the Ni-NTA flow through were incubated at 4°C overnight with 10 mM 13C-formaldehyde and 10 mM NaBH4CN. Excess of reagent was eliminated by dialysis and 13Cm2-μOR was further purified by SEC chromatography in a buffer containing 0.01% MNG, 0.001% CHS, 20 mM HEPES pH 7.5, 0.1 M NaCl.
The monodisperse peak was then concentrated to 20 to 40 µM final, and dialyzed in 98.85% D2O buffer with 0.01% MNG, 0.001% CHS, 20 mM HEPES-d18 pH 7.4 (uncorrected) and 40 mM NaCl.
NMR spectroscopy
Final samples (≈270 µL at 20–40 µM) were loaded into Shigemi microtubes susceptibility matched to D2O. All data for ligands and mutant studies were acquired on 700 MHz and 500 MHz Bruker Avance III spectrometers, respectively (Bruker, Rheinstetten, Germany), both equipped with 5 mm cryogenic H/C/N/D probes with Z-axis gradient. 1H-13C correlation spectra were recorded using heteronuclear multiple-quantum coherence (HMQC) experiments in echo anti-echo mode. 13C and 1H chemical shifts and peak linewidths in the HMQC spectra reveal the chemical and magnetic environments of the 13C-methyl probes as well as their dynamic properties. Spectral widths in ω1 and ω2 were 8417.5 Hz (6009.6 Hz) and 3519.6 Hz (2515.2 Hz) at 700 MHz (500 MHz), respectively centered at 40 ppm in the 13C dimension. 13C decoupling was performed with a GARP4 sequence. Typically, 134 (81) complex points with 32–48 scans per FID were recorded, to ensure a 27 Hz resolution per point at 700 MHz (500 MHz) before zero filling. The relaxation delay was set to 1.5 s. 32 steady-state scans preceded data acquisition. Total collection time varied between 3 and 4 Hrs, depending on the sample concentration. Spectra were processed using NMRPipe/NMRDraw software26 and visualized using CCPNMR27.
All ligands were dissolved in deuterated DMSO to 10 mM and directly added to the sample in the Shigemi tube at a final concentration of 150 µM. Nb 33 and Nb 39 were concentrated to 0.6 mM and dialyzed in 100 % D2O buffer with 0.01% MNG, 0.001% CHS, 20 mM HEPES-d18 pH 7.4 (uncorrected) and 40 mM NaCl. The Nbs were added directly in the Shigemi tubes at a final concentration of 60 µM before data acquistion.
Peak volume analysis
All NMR spectra were processed using the suite of programs provided in the NMRPipe/NMRDraw software distribution26. The spectra were normalized using DSS (2,2-dimethyl-2-silapentane-5-sulfonic acid) as an internal reference. For peak fitting analysis, spectra were processed with a squared sine-bell window function in each dimension, and zero-filled to 4096 × 1024 data points in time domain data t2 and t1, respectively. Spectra were fit with the program nlinLS, provided as a part of nmrDraw package. Gaussian models were used for the fitting in each dimension, starting from values obtained from the peak-peaking routine in nmrDraw. The quality of the fits was examined visually by estimating the residual difference between the experimental data and the results of the model calculations. Peak volumes in apo-state, ligands alone (BU72, [Dmt1]-DALDA, and DAMGO) and ternary complexes spectra were extracted from the peak fitting. Errors in the peak volume were calculated based on the effect of random noise for the peak height estimated by nlinLS.
To directly compare the ligand effects on the apo-state relative to the maximal effect obtained with BU72-Nb33 treatment (Fully agonist-induced state), we normalized the peak volumes with the volume difference between the apo-state and ternary complex condition (BU72-Nb33) as the 100% (Vapo−VBU72-Nb33). Ligand effects are represented as percent of the maximal effect: 100 × (Vapo−VL or L-Nb33 / (Vapo−V BU72-Nb33). For the TM6 data, because of the overlapping peaks in the area of interest, we considered the peak volume of K2696.24 and K2716.26 in the ternary complex with BU72 as the 100% (indicated as K269* and K271* in Fig. 2) and in this case the percent of maximal effect is 100 × (VL or L-Nb33 /V BU72-Nb33). The uncertainty was determined by (ΔVL or L-Nb332 + ΔVapo2 + ΔVBU72-Nb332)1/2.
Extended Data
Extended data Figure 1.
Overall strategy for the preparation of 13C-dimethylated μOR and quality control. The wild-type μOR contains a total of twelve lysine residues strategically positioned to sense conformational dynamics in both extracellular and G protein coupling domains upon receptor activation. To monitor the μOR activation process in solution in an amphiphile membrane-like environment, we exploit the sensitivity of two 13C-methyl groups covalently bound to the ε-NH2 of lysine side chains (ε-N[13CH3]2-lysines) and to the α-NH2 of the receptor N-terminus as NMR probes, an approach recently validated in the GPCR field (ref 17 main text). We slightly modify the wild-type sequence to facilitate receptor purification and to remove flexible N and C-termini for improvement of sample stability in NMR measurements. (a) Snake plot presenting the μOR-2x construct and highlighting the twelve endogenous lysines (red circles), the protease cleavable motifs (black circles) and the six-histidine and Flag tags (grey circle). Extracellular and intracellular surfaces are colored in yellow and blue respectively. (b) The different biochemical steps were analyzed by SDS-PAGE (left panel) and a typical final size exclusion chromatography highlighting the monodispersity of 13C-dimethylated μOR is shown (right panel). (c) Shown is the reductive methylation of the N-terminus and of lysine side chains after reaction with 13C-formaldehyde (13C in green) in the presence of sodium cyanoborohydride (NaBH3CN). Amino acids are shown in ball-and-stick representation and for clarity side-chains of unmodified amino acids are replaced by large spheres. This reaction is known to minimally affect protein structure and function and, as previously observed with β2-AR (see reference 16 of the main text), it did not affect the functionality of μOR as 13C-dimethylated μOR binds both antagonist (Naloxone) and agonist (BU72) with a similar affinity than unlabeled μOR (d). In addition, we observed similar agonist-induced interaction with the Nb39 between μOR or 13C-dimethylated μOR using pull-down experiments (e) demonstrating that methylation does not prevent the agonist-dependent interaction with the Nb39. Saturation binding experiments on soluble μOR or 13C-dimethylated μOR (both at 100 nM) were done in the presence of increasing amount of radiolabeled naloxone (up to 1 µM) and non-specific binding was determined in the presence of 100 µM of naloxone. Competition assays on soluble μOR or 13C-dimethylated μOR were done with 100 nM of radiolabeled naloxone and increasing amount of BU72. Free and receptor bound radioligands were separated using gel-filtration columns. Total binding was plotted as a function of [3H]-naloxone or BU72 concentration and data were analyzed using Prism with saturation or competitive binding analyses.
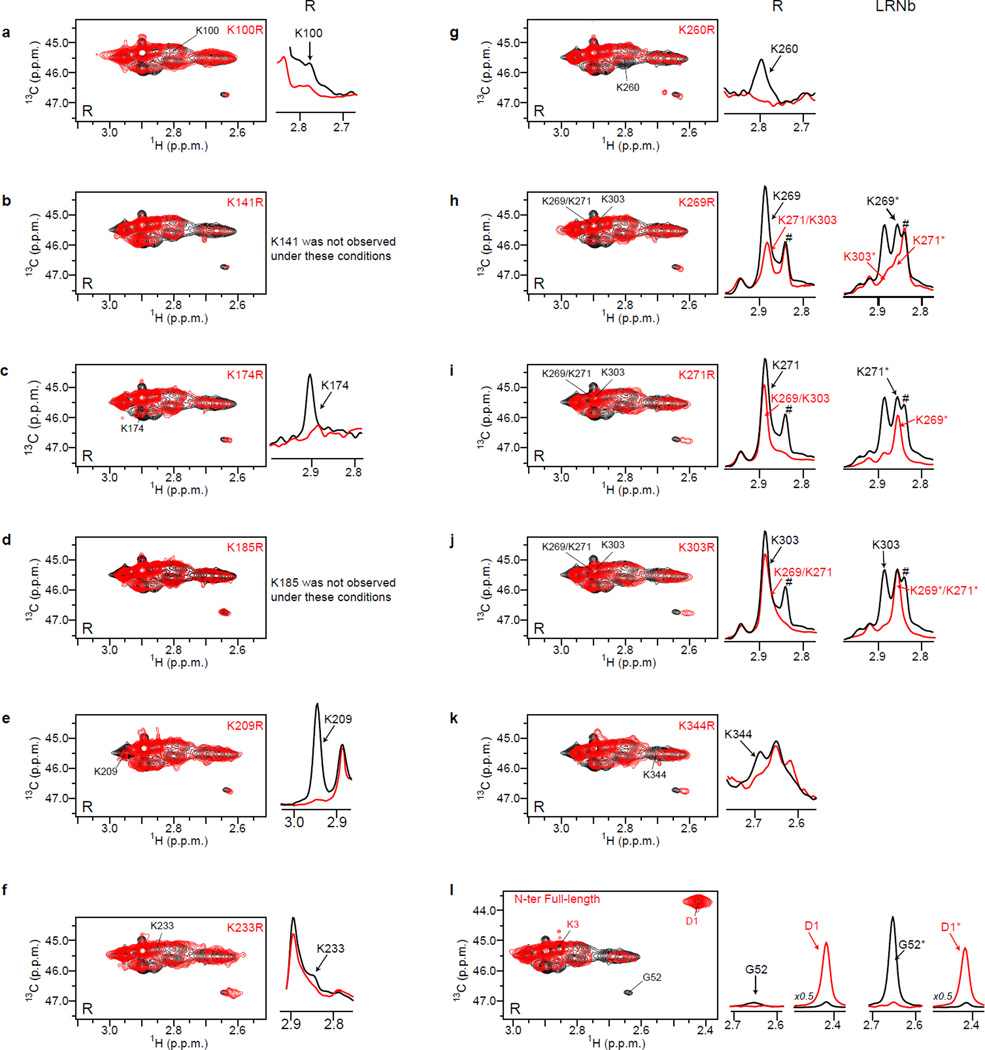
Extended data Figure 2.
Assignments of the N-terminus and dimethyllysine peaks. Each single mutants were expressed, purified and labeled as described for μOR-2x. For each mutant we analyzed the R and LRNb spectra. (a–k) Left panels represent the 2D spectra with the indicated lysine to arginine mutant in red. On the right panels, we highlight the peak disappearance for each mutant (red line) as compared to wt μOR preparation (black line) in the 1H dimension in the receptor alone condition (R). In some instances, peaks are overlapping in the area of interest and to assign these peaks we had to record the NMR spectra in the ternary complex situation (LRNb). This was the case for K269 (panel h), K271 (panel i), and K303 (panel j). In the panels h, i and j, the hashtag sign (#) indicates the position of the lysine peak from the FLAG tag that we measured in some samples and that is due to an incomplete cleavage of the N-terminus by the 3C protein. For samples used in the analysis of ligand and Nb effects, we almost eliminate this peak by optimizing the 3C cleavage step as described in the method section (treatment with 100 µM TCEP). The # sign in the panel i between WT (No TCEP) and K271R (100 µM TCEP) is an example of the improvement of 3C cleavage in TCEP treated samples as indicated by the almost complete disappearance of the peak in both R and LRNb conditions (K271R, red line).
The K98 peak was assigned by deduction (see Fig 1a). Indeed, we have generated and recorded NMR spectra of eleven mutants of the twelve endogenous lysines and the only peaks that were never affected in all the mutant spectra and left unassigned necessarily correspond to the K98 residue. (l) The assignments of D1 (N-terminus full-length) and G52 (cleaved N-terminus) were inferred from the spectra obtained before (red line) and after cleavage with 3C (black line). The G52 loss of signal is better observed in the LRNb situation (G52* is almost completely disappearing). Asterisks indicate the chemical shifts of methyl probes in the ternary complex condition.
Extended data Figure 3.
Comparison of Nb33 and Nb39 effects on NMR spectra. HMQC spectrum of 13C-dimethylated μOR in the ternary complex situation (R: 30 µM, BU72: 150 µM and Nb 60 µM) for Nb33 acquired with a 700 MHz spectrometer (blue trace) and for Nb39 acquired with a 500 MHz spectrometer (red trace). Both spectra are very similar and are characteristic to the fully agonist-induced state of μOR. 1D slice of HMQC spectra in the 1H dimension (right panels) highlighting the similar effect of Nb33 and Nb39 in the presence of BU72 on the indicated set of dimethyllysines and compared to the untreated sample (black dotted line).
Extended data Figure 4.
Partial reversal of agonist effects by treatment with the antagonist naloxone. HMQC spectra of (a) unliganded 13C-dimethylated μOR (30 µM, R, black), (b) the same sample bound to a saturating concentration of ligand (150 µM, LR, [Dmt1]-DALDA (blue) or naloxone (red) and (c) the same sample treated with a saturating concentration of Nb33 (60 µM, LRNb, [Dmt1]-DALDA (green) or naloxone (light pink). (d) Spectra of the [Dmt1]-DALDA-RNb sample treated with naloxone (900 µM) (L2RNb, magenta). Green arrows in 1D traces highlight the peaks movement in the activation process (c) and for the partial reversal effect of naloxone (d) for K209 and K344/K98 peaks.
Extended data Figure 5.
(a) HMQC spectra of unliganded 13C-dimethylated μOR (30 µM, R, black), 13C-dimethylated μOR treated with a saturating concentration of [Dmt1]-DALDA (150 µM, LR, blue), 13C-dimethylated μOR treated with [Dmt1]-DALDA and a saturating concentration of Nb33 (60 µM, LRNb, green) and of unliganded 13C-dimethylated μOR treated with a saturating concentration of Nb33 (60 µM, RNb, orange). The small crosses indicate the position of peaks visible in the apo-state that are reduced in intensity or no longer visible in the other spectra. Arrows indicate the position of the 1D slice represented in (b). (b) 1D slice of HMQC spectra in the 1H dimension highlighting the effect of [Dmt1]-DALDA (blue) and [Dmt1]-DALDA + Nb33 (green) on the N-terminus (G52) and on indicated set of dimethyllysines (ECL2, TM5, TM6, ICL1 and H8 lysines). The yellow and blue boxes indicate the positions in the extracellular and intracellular domains respectively. The zoom level for each peak and the 13C chemical shifts are indicated in the top left of each panel. Asterisks indicate the chemical shifts of methyl probes in the ternary complex condition.
Extended data Figure 6.
(a) HMQC spectra of unliganded 13C-dimethylated μOR (30 µM, R, black), 13C-dimethylated μOR treated with a saturating concentration of DAMGO (150 µM, LR, blue), 13C-dimethylated μOR treated with DAMGO and a saturating concentration of Nb33 (60 µM, LRNb, green) and of unliganded 13C-dimethylated μOR treated with a saturating concentration of Nb33 (60 µM, RNb, orange). The small crosses indicate the position of peaks visible in the apo-state that are reduced in intensity or no longer visible in the other spectra. Arrows indicate the position of the 1D slice represented in (b). (b) 1D slice of HMQC spectra in the 1H dimension highlighting the effect of DAMGO alone (blue) and DAMGO + Nb33 (green) on the N-terminus (G52) and on indicated set of dimethyllysines (ECL2, TM5, TM6, ICL1 and H8 lysines). The yellow and blue boxes indicate the positions in the extracellular and intracellular domains respectively. The zoom level for each peak and the 13C chemical shifts are indicated in the top left of each panel. Asterisks indicate the chemical shifts of methyl probes in the ternary complex condition.
Acknowledgments
We acknowledge support from INSERM (S.G.) and CNRS (H.D.) and from the National Institutes of Health Grant (NIDA-DA036246 to B.K.K and S.G). We also acknowledge the National Institute of Drug Abuse Drug Supply Program for providing [Dmt1]-DALDA.
A.M., T.L., J.S., and B.K.K. have filed a patent for active-state stabilizing nanobodies for opioid receptors.
Footnotes
AUTHOR CONTRIBUTIONS
R.S, H.D., A.M., W.H., B.K and S.G. designed experiments, performed research and analysed data. C.M. expressed, purified and characterized receptor and nanobody preparations. J.S and T.L developed the G protein mimetic nanobodies. H.D supervised NMR data analysis. S.G. and R.S prepared the manuscript with the help of H.D and B.K.. S.G. supervised the overall project.
REFERENCES
- 1.Manglik A, et al. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthes HW, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 4.Macht DI, Herman NB, Levy CS. A Quantitative Study of Cutaneous Analgesia Produced by Various Opium Alkaloids. Proc Natl Acad Sci U S A. 1915;1:582–585. doi: 10.1073/pnas.1.12.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasternak GW, Pan YX. Mu opioids and their receptors: evolution of a concept. Pharmacol Rev. 2013;65:1257–1317. doi: 10.1124/pr.112.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raehal KM, Schmid CL, Groer CE, Bohn LM. Functional selectivity at the mu-opioid receptor: implications for understanding opioid analgesia and tolerance. Pharmacol Rev. 2011;63:1001–1019. doi: 10.1124/pr.111.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPherson J, et al. mu-opioid receptors: correlation of agonist efficacy for signalling with ability to activate internalization. Mol Pharmacol. 2010;78:756–766. doi: 10.1124/mol.110.066613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephenson RP. A modification of receptor theory. British journal of pharmacology and chemotherapy. 1956;11:379–393. doi: 10.1111/j.1476-5381.1956.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kofuku Y, et al. Efficacy of the beta(2)-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat Commun. 2012;3:1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kofuku Y, et al. Functional dynamics of deuterated beta2 -adrenergic receptor in lipid bilayers revealed by NMR spectroscopy. Angewandte Chemie. 2014;53:13376–13379. doi: 10.1002/anie.201406603. [DOI] [PubMed] [Google Scholar]
- 16.Kim TH, et al. The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J Am Chem Soc. 2013;135:9465–9474. doi: 10.1021/ja404305k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokoch MP, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 20.Neilan CL, et al. Characterization of the complex morphinan derivative BU72 as a high efficacy, long-lasting mu-opioid receptor agonist. Eur J Pharmacol. 2004;499:107–116. doi: 10.1016/j.ejphar.2004.07.097. [DOI] [PubMed] [Google Scholar]
- 21.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Sequence of late molecular events in the activation of rhodopsin. Proc Natl Acad Sci U S A. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Ernst OP, et al. Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin alpha and gamma subunits. J Biol Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 24.Kaye RG, Saldanha JW, Lu ZL, Hulme EC. Helix 8 of the M1 muscarinic acetylcholine receptor: scanning mutagenesis delineates a G protein recognition site. Mol Pharmacol. 2011;79:701–709. doi: 10.1124/mol.110.070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, et al. Structural basis of G protein-coupled receptor-G protein interactions. Nat Chem Biol. 2010;6:541–548. doi: 10.1038/nchembio.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]