Abstract
Identifying progressive mild cognitive impairment (pMCI) patients and predicting when they will convert to Alzheimer’s disease (AD) are important for early medical intervention. Multi-modality and longitudinal data provide a great amount of information for improving diagnosis and prognosis. But these data are often incomplete and noisy. To improve the utility of these data for prediction purposes, we propose an approach to denoise the data, impute missing values, and cluster the data into low-dimensional subspaces for pMCI prediction. We assume that the data reside in a space formed by a union of several low-dimensional subspaces and that similar MCI conditions reside in similar subspaces. Therefore, we first use incomplete low-rank representation (ILRR) and spectral clustering to cluster the data according to their representative low-rank subspaces. At the same time, we denoise the data and impute missing values. Then we utilize a low-rank matrix completion (LRMC) framework to identify pMCI patients and their time of conversion. Evaluations using the ADNI dataset indicate that our method outperforms conventional LRMC method.
1 Introduction
Alzheimer’s disease (AD) is the most prevalent dementia that is commonly associated with progressive memory loss and cognitive decline. It is incurable and requires attentive care, thus imposing significant socio-economic burden on many nations. It is thus vital to detect AD at its earliest stage or even before its onset, for possible therapeutic treatment. AD could be traced starting from its prodromal stage, called mild cognitive impairment (MCI), where there is mild but measurable memory and cognitive decline. Studies show that some MCI patients will recover over time, but more than half will progress to dementia within five years [2]. In this paper, we focus on distinguishing progressive MCI (pMCI) patients who will progress to AD from stable MCI (sMCI) patients who will not. We will at the same time predict when the conversion to AD will occur.
Biomarkers based on different modalities, such as magnetic resonance imaging (MRI), positron emission topography (PET), and cerebrospinal fluid (CSF), have been proposed to predict AD progression [15,4,12,14]. The Alzheimer’s disease neuroimaging initiative (ADNI) collects these data longitudinally from subjects ranging from cognitively normal elders to AD patients in an effort to use all these information to accurately predict AD progression. However, these data are incomplete because of dropouts and unavailability of a certain modality. The easiest and most popular way to deal with missing data is by discarding the samples with missing values [15]. But this will decrease the number of samples as well as the statistical power of analyses. One alternative is to impute the missing data, via methods like k-nearest neighbour (KNN), expectation maximization (EM), or low-rank matrix completion (LRMC) [10,16,1]. But these imputation methods do not often perform well on datasets with a large amount of values missing in blocks [9,13]. To avoid the need for imputation, Yuan et al. [13] divides the data into subsets of complete data, and then jointly learn the sparse classifiers for these subsets. Through joint feature learning, [13] enforces each subset classifier to use the same set of features for each modality. However, this will restrain samples with certain modality missing to use more features in available modality for prediction. Goldberg et al. [3], on the other hand, imputes the missing features and unknown targets simultaneously using a low-rank assumption. Thus, all the features are involved in the prediction of the target through rank minimization, while the propagation of the missing feature’s imputation errors to the target outputs is largely averted, as the target outputs are predicted directly and simultaneously. Thung et al. [9] improves the efficiency and effectiveness of [3] by performing feature and sample selection before matrix completion. However, by applying matrix completion on all the samples, the authors implicitly assumes that the data are from a single low-dimensional subspace. This assumption hardly holds for real and complex data.
To capture the complexity and heterogeneity of the pathology of AD progression, we assume that the longitudinal multi-modality data reside in a space that is formed by a union of several low-dimensional subspaces. Assuming that the data is low-rank as a whole is too optimistic, and missing values might not be imputed correctly. A better approach is to first cluster the data and then perform matrix completion on each cluster. In this paper, we propose a method, called low-rank subspace clustering and matrix completion (LRSC-MC), which will cluster the data into subspaces for improving prediction performance. More specifically, we first use incomplete low rank representation (ILRR) [5,8] to simultaneously determine a low-rank affinity matrix, which gives us an indication of the similarity between any pair of samples, estimate the noise, and obtain the denoised data. We then use spectral clustering [6] to split the data into several clusters and impute the output targets (status labels and conversion times) using low-rank matrix completion (LRMC) algorithm for each cluster. We tested our framework using longitudinal MRI data (and cross-sectional multi-modality data) for both pMCI identification and conversion time prediction, and found that the LRSC-MC outperforms LRMC. In addition, we also found that using denoised data will improve the performance of LRMC.
2 Materials and Preprocessing
2.1 Materials
We used both longitudinal and cross-sectional data from ADNI1 for our study. We used MRI data of MCI subjects that were scanned at baseline, 6th, 12th and 18th months to predict who progressed to AD in the monitoring period from 18th to 60th month. MCI subjects that progressed to AD within this monitoring period were retrospectively labeled as pMCI, whereas those that remained stable were labeled as sMCI. MCI subjects that progressed to AD on and before 18th month were not used in this study. There are two target outputs in this study – class label and conversion month. For estimating these outputs, we used the MRI data, PET data, and clinical scores (e.g., Mini-Mental State Exam (MMSE), Clinical Dementia Rating (CDR), and Alzheimer’s Disease Assessment Scale (ADAS)). Table 1 shows the demographic information of the subjects used in this study.
Table 1.
Demographic information of subjects involved in this study. (Edu.: Education; std.: Standard Deviation)
| No. of subjects | Gender (M/F) | Age (years) | Edu. (years) | |
|---|---|---|---|---|
| pMCI | 65 | 49/16 | 75.3 ± 6.7 | 15.6 ± 3.0 |
| sMCI | 53 | 37/16 | 76.0 ± 7.9 | 15.5 ± 3.0 |
| Total | 118 | 86/32 | - | - |
2.2 Preprocessing and Feature Extraction
We use region-of-interest (ROI)-based features from the MRI and PET images in this study. The processing steps involved, for each MRI image, are described as follows. Each MRI image was AC-PC corrected using MIPAV2, corrected for intensity inhomogeneity using the N3 algorithm, skull stripped, tissue segmented, and registered to a common space [11,7]. Gray matter (GM) volumes, normalized by the total intracranial volume, were extracted as features from 93 ROIs [11]. We also linearly aligned each PET image to its corresponding MRI image, and used the mean intensity value at each ROI as feature.
3 Method
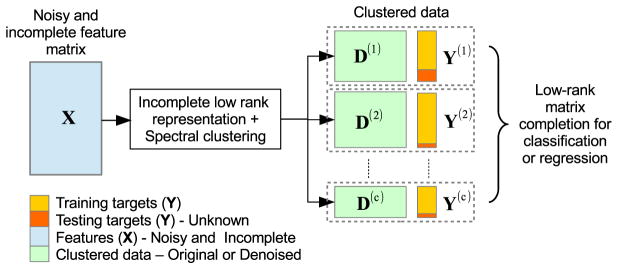
Figure 1 shows an overview of the proposed low-dimensional subspace clustering and matrix completion framework. The three main components in this framework are 1) incomplete low-rank representation, which computes the low-rank affinity matrix, 2) spectral clustering, which clusters data using the affinity matrix acquired from the previous step, and 3) low-rank matrix completion, which predicts the target outputs using the clustered data. In the following subsections, each of these steps will be explained in detail.
Fig. 1.
Low-rank subspace clustering and matrix completion for pMCI diagnosis and prognosis.
3.1 Notation
We use X ∈ ℝn×m to denote the feature matrix with n samples of m features. n depends on the number of time points and the number of modalities used. Each sample (i.e., row) in X is a concatenation of features from different time points and different modalities (MRI, PET and clinical scores). Note that X can be incomplete due to missing data. The corresponding target matrix is denoted as Y ∈ ℝn×2, where the first column is a vector of labels (1 for pMCI, −1 for sMCI), and the second column is a vector of conversion times (e.g., the number of months to AD conversion). The conversion times associated with the sMCI samples should ideally be set to infinity. But to make the problem feasible, we set the conversion times to a large value computed as 12 months in addition to the maximum time of conversion computed for all pMCI samples. For any matrix M, Mj,k denotes its element indexed by (j, k), whereas Mj,: and M:,k denote its j-th row and k-th column, respectively. We denote ||M||* = Σσi(M) as the nuclear norm (i.e., sum of the singular values {σi} of M), ||M||1 = Σ|Mj,k| as l1 norm, and M′ as transpose of M. I is the identity matrix.
3.2 Low-Rank Matrix Completion (LRMC)
Assuming linear relationship between X and Y, we let for the k-th target Y:,k = Xak + bk = [X1] × [ak; bk], where 1 is a column vector of 1’s, ak is the weight vector, and bk is the offset. Assuming a low rank X, then the concatenated matrix M= [X 1 Y] is also low-rank [3], i.e., each column of M can be linearly represented by other columns, or each row of M can be linearly represented by other rows. Based on this assumption, low-rank matrix completion (LRMC) can be applied on M to impute the missing feature values and the output target simultaneously by solving , where Ω is the index set of known values in M. In the presence of noise, the problem can be relaxed as [3]
| (1) |
where Ll(·, ·) and Ls(·, ·) are the logistic loss function and mean square loss function, respectively. Ωl and Ωs are the index sets of the known target labels and conversion times, respectively. The nuclear norm || · ||* in Eq. (1) is used as a convex surrogate for matrix rank. Parameters μ and λ are the tuning parameters that control the effect of each term.
If the samples come from different low-dimensional subspaces instead of one single subspace, applying (1) to the whole dataset is inappropriate and might cause inaccurate imputation of missing data. We thus propose instead to apply LRMC to the subspaces, where samples are more similar and hence the low-rank constraint is more reasonable. The problem now becomes
| (2) |
where Qc ∈ ℝn×n is a diagonal matrix with values {0, 1}, C is the number of clusters, and . The effect of QcZ is to select from the rows of Z the samples that are associated with each cluster. For example, if the value of Q(i, i) is one, the i-th sample (i.e., the i-th row) in Z will be selected. Qc is estimated from the feature matrix X by performing low-rank representation subspace clustering [5] and spectral clustering [6], which will be discussed next.
3.3 Incomplete Low-Rank Representation (ILRR)
Low-rank representation (LRR) [5] is widely used for subspace clustering. LRR assumes that data samples are approximately (i.e., the data are noisy) drawn from a union of multiple subspaces and aims to cluster the samples into their respective subspaces and at the same time remove possible outliers. However, LRR cannot be applied directly to our data because they are incomplete. We therefore apply instead the method reported in [8], which is called incomplete low-rank representation (ILRR). For an incomplete matrix X, the ILRR problem is given by
| (3) |
where 𝒳 is the completed version of X, which is self-represented by A𝒳, A ∈ ℝn×n is the low-rank affinity matrix, E is the error matrix, and α is the regularizing parameter. Each element of A indexed by (i, j) is an indicator of the similarity between the i-th sample and the j-th sample. Eq. (3) is solved using inexact augmented Lagragrian multiplier (ALM), as described in [8]. ILRR 1) denoises the data through prediction of E, providing us with clean data D = A𝒳, and 2) predicts the affinity matrix A, which is used next to cluster the data.
3.4 Spectral Clustering
After acquiring the affinity matrix A between all the subjects, we perform spectral clustering [6] on X (or D). The clustering procedures are:
Symmetrize the affinity matrix, A ← A + A′ and set diag(A) = 0.
Compute Laplacian matrix, L = S−1/2AS−1/2, where S is a diagonal matrix consisting of the row sums of A.
Find k largest eigenvectors of L and stack them as columns of matrix V ∈ ℝn×k.
Renormalize each row of V to have unit length.
Treat each row of V as a point in ℝk and cluster them into k clusters using the k-means algorithm.
Based on the clustering indices, we now have clustered data X(c) (or D(c)), which together with the corresponding target Y(c), can be used for classification. As can be seen in Fig. 1, we now have multiple classification problems, one for each subspace. Thus, in practice, we implement (2) by completing concatenated matrix [X(c)1Y(c)] (or [D(c)1Y(c)]) using LRMC, and combine the outputs predicted in all subspaces to obtain a single classification or regression outcome.
4 Results and Discussions
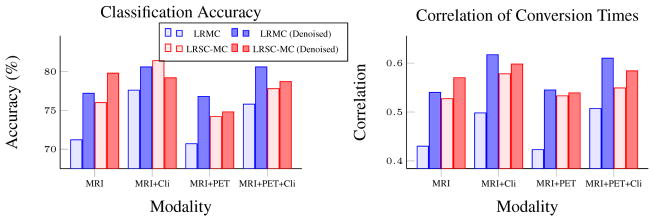
We evaluated the proposed method, LRSC-MC, using both longitudinal and multi-modality data. In both Fig. 3 and Fig. 2, the baseline method – LRMC using original data, is denoted by light blue bar, while the rests either using part(s) or full version of the proposed method – LRMC using the denoised data, and LRSC-MC using the original and the denoised data. Two clusters (C = 2) were used for LRSC-MC. Parameter α = 0.005 is used for LRSC, and parameters μ and λ of LRMC are searched through the range {10−n, n = 6, …, 3} and {10−3 – 0.5}, respectively. All reported results are the average of the 10 repetitions of a 10-fold cross validation.
Fig. 3.
Average pMCI/sMCI classification accuracy and the correlation of conversion month predictions, using data from different number of time points.
Fig. 2.
Average pMCI/sMCI classification accuracy and the correlation of conversion month predictions using multi-modality data.
Fig. 2 shows the comparison result between the conventional LRMC and LRSC-MC, using different combinations of MRI, PET, and clinical scores, at first time point. The result shows that LRSC-MC always outperforms the conventional LRMC, using either original or denoised data, both in terms of pMCI identification and conversion time prediction (i.e., red bar is higher than light blue bar). This confirms the effectiveness of our method for heterogeneous data that exhibit multiple low dimensional subspaces. In addition, we also found that LRMC is improved using the denoised data from the first component of our method (ILRR) (i.e., dark blue bar is higher than light blue bar). This is probably because denoising the data using ILRR is similar to smoothing the data to a low-dimensional subspace, (when affinity matrix A is low rank, A𝒳 is also low rank), which fulfills the assumption of LRMC. Sometimes, the improvement due to the use of denoised data is more than the use of clustering. Thus, there are 2 ways to improve LRMC on data that consists of multiple subspaces, i.e., by first denoise/smooth the data, or by clustering the data. Nevertheless, the best pMCI classification is still achieved by LRSC-MC using modality combination of MRI+Cli, at around 81%, using the original data.
In Fig. 3, we show the results of using only MRI data at 1 to 4 time points for pMCI identification and conversion time prediction. The results indicate that LRSC-MC outperforms LRMC using either data from 1, 2 and 3 time points. The improvement of the LRSC-MC is more significant for the case of 1 time point (at 18th month), where the pMCI identification is close to 80% of accuracy using the denoised data, while LRMC on the original data is only about 72%. In general, when using the original data, LRSC-MC outperforms LRMC, while when using the denoised data, LRSC-MC is either better or on par with LRMC. This is probably because the denoised data is closer to the assumption of single low-dimensional subspace, opposing the assumption made by the LRSC-MC – the data is a union of several low-dimensional subspaces. The only case where LRSC-MC performs worse than LRMC is when data from 4 time points are used. This is probably because when 2 or more TPs are used, the earlier data (e.g., 12th, 6th month) are added. These added data is farther away from the conversion time and could be noisier and less reliable for prediction. Since in our current model, we assume equal reliability of features, when the noisy features started to dominate the feature matrix using 4 TPs, LRSC is unable to give us a good affinity matrix and thus poorer performance.
In addition, we also compare our method with [9,13] in Table 2. However, since both methods rely on the subset of complete data for feature selection and learning, they fail in cases when the subset contains too little samples for training (those cases are left blank in the table). The results show that the proposed method outperforms or on par with these state-of-the-art methods.
Table 2.
Comparison with other methods.
5 Conclusion
We have demonstrated that the proposed method, LRSC-MC, outperforms conventional LRMC using longitudinal and multi-modality data in many situations. This is in line with our hypothesis that, for data that reside in a union of low-dimensional subspaces, subspace low-rank imputation is better than whole-space low-rank imputation.
Acknowledgments
This work was supported in part by NIH grants EB009634, AG041721, and AG042599.
Footnotes
References
- 1.Candès EJ, et al. Exact matrix completion via convex optimization. Foundations of Computational Mathematics. 2009;9(6):717–772. [Google Scholar]
- 2.Gauthier S, et al. Mild cognitive impairment. The Lancet. 2006;367(9518):1262–1270. doi: 10.1016/S0140-6736(06)68542-5. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg A, et al. Transduction with matrix completion: three birds with one stone. Advances in Neural Information Processing Systems. 2010;23:757–765. [Google Scholar]
- 4.Li F, et al. A robust deep model for improved classification of AD/MCI patients. IEEE Journal of Biomedical and Health Informatics. 2015;99 doi: 10.1109/JBHI.2015.2429556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G, et al. Robust recovery of subspace structures by low-rank representation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2013;35(1):171–184. doi: 10.1109/TPAMI.2012.88. [DOI] [PubMed] [Google Scholar]
- 6.Ng AY, et al. On spectral clustering: Analysis and an algorithm. Advances in Neural Information Processing Systems. 2002;2:849–856. [Google Scholar]
- 7.Shen D, et al. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 8.Shi J, et al. Low-rank representation for incomplete data. Mathematical Problems in Engineering 2014. 2014 [Google Scholar]
- 9.Thung KH, et al. Neurodegenerative disease diagnosis using incomplete multi-modality data via matrix shrinkage and completion. Neuroimage. 2014;91:386–400. doi: 10.1016/j.neuroimage.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Nie J, Yap P-T, Shi F, Guo L, Shen D. Robust deformable-surface-based skull-stripping for large-scale studies. In: Fichtinger G, Martel A, Peters T, editors. MICCAI 2011, Part III. LNCS. Vol. 6893. Springer; Heidelberg: 2011. pp. 635–642. [DOI] [PubMed] [Google Scholar]
- 12.Weiner MW, et al. The Alzheimer’s disease neuroimaging initiative: A review of papers published since its inception. Alzheimer’s & Dementia. 2013;9(5):e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan L, et al. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. NeuroImage. 2012;61(3):622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan L, et al. Comparison of nine tractography algorithms for detecting abnormal structural brain networks in Alzheimers disease. Frontiers in Aging Neuroscience. 2015:7. doi: 10.3389/fnagi.2015.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, et al. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage. 2012;59(2):895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, et al. Missing value estimation for mixed-attribute data sets. IEEE Transactions on Knowledge and Data Engineering. 2011;23(1):110–121. [Google Scholar]