Abstract
PTEN hamartoma tumour syndrome (PHTS) is caused by heterozygous variants in PTEN and is characterised by tumour predisposition, macrocephaly, and cognition impairment. Bi-allelic loss of PTEN activity has not been reported so far and animal models suggest that bi-allelic loss of PTEN activity is embryonically lethal. Here, we report the identification of a novel homozygous variant in PTEN, NM_000314.4; c.545T>C; p.Leu182Ser, in two adolescent siblings with severe macrocephaly and mild intellectual disability. The variant is predicted to be damaging and is associated with significantly increased phospho-S6 downstream of PTEN. The absence of tumours in the two homozygous siblings as well as lack of symptoms of PHTS in the heterozygous carriers of the family suggest that this particular variant is functionally hypomorphic rather than deleterious.
INTRODUCTION
Dominant pathogenic variants in PTEN cause a broad spectrum of symptoms and overlapping phenotypes that include Cowden syndrome and its subphenotype Lhermitte–Duclos disease (cerebelloparenchymal disorder VI), Bannayan–Riley–Ruvalcaba syndrome as well as autism spectrum disorders and macrocephaly.1, 2 Due to the substantial phenotypic overlap and due to the lack of distinct genotype phenotype associations, all phenotypes of individuals with germline PTEN variants are summarised as PTEN hamartoma tumour syndrome (PHTS).3 The spectrum of PHTS associated findings includes macrocephaly in nearly all individuals, multiple tumours, mucocutaneous lesions, as well as autism and intellectual disability in about 15–20% of the affected individuals.2, 4, 5, 6
PTEN (phosphatase and tensin homologue) acts as a dual-specificity phosphatase of proteins and of lipids.7, 8, 9 In the nervous system, PTEN has a role in axon regeneration and elongation10 and is a key modulator of the AKT-mTOR signalling pathway controlling neurogenesis, neuron positioning, dendrite development, and synapse formation.11 In mice, homozygous deletion of Pten activity leads to abnormal organ development and embryonic lethality.12 Bi-allelic, tissue-specific inactivation of Pten in cerebral cortex and hippocampus leads to macrocephaly, neuronal hypertrophy, and abnormal dendritic arborisation, with increased dendritic thickness and spine density.13 In humans, no homozygous or compound heterozygous variants in PTEN have been reported to date. In the context of a systematic study of the genetic causes of autosomal recessive intellectual disability at the Institute of Human Genetics in Erlangen, we examined a series of affected families. Here, we describe in one of those families a homozygous pathogenic variant in PTEN and discuss the potential functional implications.
PATIENTS AND METHODS
Patients
This study was approved by the Ethics Committees of the Universities of Erlangen-Nürnberg, Leipzig, and Oxford. Informed consent of patient 1 (P1) and both parents (including their consent for P2) was obtained.
Whole exome sequencing
After genome-wide autozygosity mapping (HomozygosityMapper, http://www.homozygositymapper.org), we performed exome sequencing of the index patient (P1) as described previously.14, 15 To validate the results, we repeated the sequencing in P1 and performed exome sequencing in P2. DNA was enriched using the SureSelect Human All Exon Kit version 5 (Agilent, Santa Clara, CA, USA), and paired-end sequenced (100 bp, 100 bp) on a HiSeq2500 instrument (Illumina, San Diego, CA, USA). Analysis, base calling, and variant annotation were performed according to standard methods.15 We achieved an average coverage of 145 × ; 95% of the target sequence was covered at least 20 × and 96% were covered at least 5 ×.
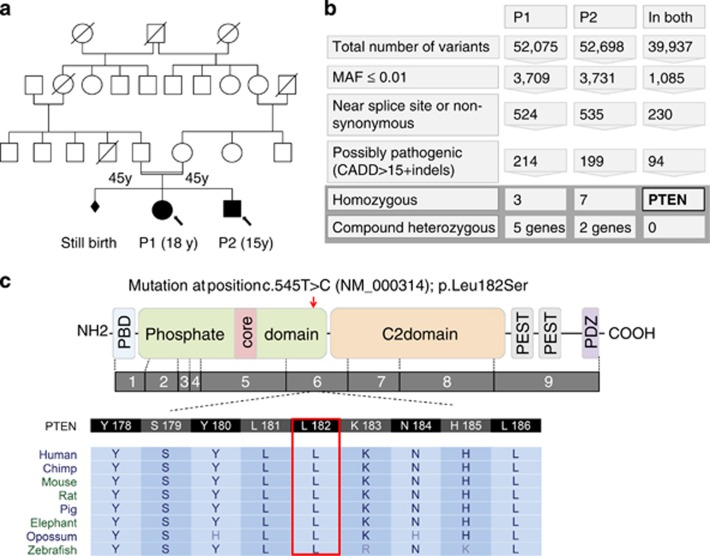
Only variants with coverage of 5 × or more were analysed. Variants with minor allele frequency of less than 0.01 in exome variant sever (6500 individuals), 1000 genomes project (version 2012 with 1200 individuals), and in our in house exomes (728 individuals) were considered. We then repeated filtering steps based on conservation, pathogenicity prediction, and mutation scoring using several sources such as SIFT, PhyloP, PolyPhen2, LRT, MutationTaster, MutationAssessor, GERP++, and CADD. We present in Figure 1 variant numbers obtained after filtering based on CADD values. We compared our findings within the PTEN gene to the NCBI reference sequence NG_007466.2 (Homo sapiens phosphatase and tensin homolog, PTEN). We submitted the PTEN variant to the public database of ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) with the accession number SCV000212235.
Figure 1.
Family tree, filtering, and variant characteristics. (a) Pedigree of the family. The first pregnancy in the family was a still birth 3 weeks before delivery possibly due to gestational diabetes. No malformation was detected. P1 and P2 carry the homozygous variant. Both have mild intellectual disability and macrocephaly that, especially in P2, is reminiscent of Bannayan–Riley–Ruvalcaba syndrome. (b) Filtering. We followed several filtering strategies based on different in silico parameters and inheritance mode. All filtering strategies suggested one variant in PTEN. (c) PTEN gene structure. Protein domain structure showing phosphatase domain as well as C2 domain with a p.Leu182Ser alteration. Below genomic structure of PTEN (NM_000314.4) and evolutionary conservation of the Leu182 residue.
Cell lines and phosflow
Lymphoblastoid cells lines (LCL) were generated from PBMCs with supernatant from the Epstein-Barr virus (EBV)-producing marmoset cell line B95-8 according to standard protocols. For analysis of intracellular phosphoproteins, EBV cell lines were cultured at similar cell density in complete medium, fixed with BD cytofix, and were stained with anti-pS6-AF488 (pS235/pS236) by BD Phosflow according to the manufacturer's protocol (BD Biosciences, Oxford, UK). Signals were acquired on a BD LSRFortessa (BD Biosciences) with FACS-Diva software (BD Biosciences) and analysed with FlowJo (Tree Star Inc., Ashland, OR, USA).
Mitochondrial depolarisation assay
EBV-transformed LCLs were seeded in wells of a 48-well plate at 750 000 cells/well in RPMI supplemented with 10% fetal calf serum and culture overnight. Depolarisation in the mitochondrial membrane potential in live cells was measured by the TMRE (tetramethylrhodamine, ethyl ester) assay (TMRE Mitochondrial Membrane Potential Assay Kit, Abcam, Cambridge, UK). Cells were incubated with the TMRE dye (400 nM, 25 min), washed in PBS and stained with a cell viability dye to exclude dead cells from subsequent FACS analyses. Small molecule inhibition of PTEN was performed using increasing concentrations of the PTEN inhibitor, SF1670 (Echelon Biosciences Inc., Salt Lake City, UT, USA). Cells were incubated for 20 h in the presence of varying concentrations of the inhibitor before FACS detection of TMRE signal.
RESULTS
Clinical report
We reinvestigated two adolescent siblings (P1 and P2, Figure 1) with a mild intellectual disability and substantial macrocephaly. The family has been described in a former report as having a likely recessive trait of ‘progressive megalencephaly and dilated periventricular cystic Virchow-Robin spaces'.16
Birth measurements of the affected female (P1) were normal (Table 1), but growth of head circumference was accelerated and crossed the percentiles within the first year of life. The girl showed clinical symptoms due to adenoid hypertrophy. The developmental milestones were delayed, with walking at the age of 19 months and first words at the age of 24 months. IQ testing with a Kaufman Assessment Battery test for Children (K-ABC) at the age of 7 years did show variable results but substantially reduced visual short-term-memory, spatial-constructive abilities, and motoric reproduction of gesticulations. A Beery-Buktenica Developmental Test of Visual-Motor Integration indicated delayed integration and coordination of visual perceptual and motoric skills. At age of 15 years, scores in the Hamburg–Wechsler Intelligence test for children (HAWIK) were 63 points in speech comprehension, 81 points in processing speed, 49 points in logical reasoning, and 74 points in working memory. Taken together, HAWIK resulted in an average IQ of 57 (normal range is 85–115), which suggests a mild intellectual disability. She attended kindergarten and a school for children with special needs and started to train for a simple profession. She can read and write in two languages but has remarkable deficits in abstract logic thinking such as estimating time and in mathematical skills. She is an interactive and friendly woman without autistic features.
Table 1. Clinical signs and anthropometric characteristics.
|
Birth parameters |
Parameters at examination |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| OFCa (cm) | Length (cm) | Weight (g) | OFCa (cm) | Height (cm) | Weight (kg) | Tumours | IDb | Further clinical aspects | |
| P1, female | Born after 37 weeks of gestation | Examination at age of 18 years | |||||||
| 35.3 | 52 | 3.5 | 63 | 166.2 | 60 | No | Mild | ||
| (+1.3 SD) | (+0.6 SD) | (+1.5 SD) | (+5.9 SD) | (+0.3 SD) | (+0.2 SD) | Neonatally: several absence seizures; childhood: adenoid hypertrophy, serotympanon; at examination: dolichocephalic head, prominent front is reminiscent of Bannayan–Riley–Ruvalcaba syndrome | |||
| P2, male | Born after 35 weeks of gestation | Examination at age of 15 years | |||||||
| Unremarkablec | 49 | 2.5 | 64 | 180.5 | 80 | No | Mild | ||
| (+0.6 SD) | (−0.1 SD) | (+5.5 SD) | (+0.3 SD) | Childhood: several grand mal seizures, muscular hypotonia, reduced proprioceptive reflexes, penile freckling | |||||
| Mother | NA | NA | NA | 54 | 159.2 | 61 | No | No | |
| (−0.1 SD) | (−0.5 SD) | (+0.2 SD) | |||||||
| Father | NA | NA | NA | 58 | 174.8 | 84 | No | No | |
| (+0.8 SD) | (+0.4 SD) | (+0.8 SD) | |||||||
NA, data not available.
Occipito-frontal head circumference.
ID is intellectual disability.
At age of 2 months.
Birth measurements of the affected boy (P2) were in the normal range but showed a similar accelerated growth of head circumference like in P1. He started walking at age of 19 months and spoke first words at age of 27 months. At the age of about 3 years, two episodes of seizures were observed within 3 months and waking EEG revealed pathological intermittent slow waves on the right central side but no evidence for epileptic periods. At the age of 4 years, a lumbering and broad-based gait, a crude pinch grip, reduced spontaneous movements, and periods of diminished concentration and disorientation for time and space were noted. In the HAWIK at age of 12 years he scored 67 points in speech comprehension, 71 points in processing speed, but much lower values in logical reasoning and working memory. Summarised IQ value was 54, that is, mild intellectual disability. The boy attends the same schools as his sister. He also has significant problems in abstract logic thinking such as estimating time and in calculating. He is socially well integrated and does not show autistic features. The father reported that the boy has penile freckling.
The parents were first-degree cousins of Turkish origin. Both are completely healthy, socially integrated, and intellectually normal. None of them underwent formal intellectual testing. Parents were examined for features of PHTS such as head circumference (Table 1) and cancer awareness was discussed. The parents did not meet the PHTS diagnostic criteria and no formal cancer screening was initiated.
PTEN variant identification
Due to the occurrence of two affected siblings we assumed recessive inheritance in the family and, because of the consanguinity of the parents, a homozygous variant. Autozygosity mapping identified only one candidate region on chromosome 10 between 87.99 and 91.02 Mb. First exome sequencing using DNA from P1 revealed a total of 53 474 SNVs and 3206 indels. Only one variant in PTEN (NM_000314.4: chr10:g.89711927T>C; c.545T>C; p.Leu182Ser) was located in the candidate region, affecting an evolutionary highly conserved amino acid position, that was predicted to be pathogenic by in silico programs (Figures 1c and 2a). We validated the identified variant by Sanger sequencing and confirmed familial segregation with the macrocephaly phenotype. To exclude other variants, we repeated the sequencing of P1 and we also sequenced P2 using the new version of Agilent exome enriching kit and Illumina HiSeq2500 (see Patients and methods). We considered variants that have a minor allele frequency of ≤0.01 in 1000 Genomes, Exome Variant Server (ESP), and in 728 in-house exomes, that may affect the protein sequence (splice site or non-synonymous), and that are possibly pathogenic (Figure 1b for details on filtering). Only the homozygous c.545T>C variant in PTEN was identified in both affected individuals, and it was confirmed by Sanger sequencing using genomic DNA of both patients. Sequencing of cDNA obtained from LCL of the index patient confirmed the results on RNA level, NM_000314.4; r.1576T>C. We also evaluated the exomes for compound-heterozygous variants and found no combination that is shared by both siblings (Figure 1b).
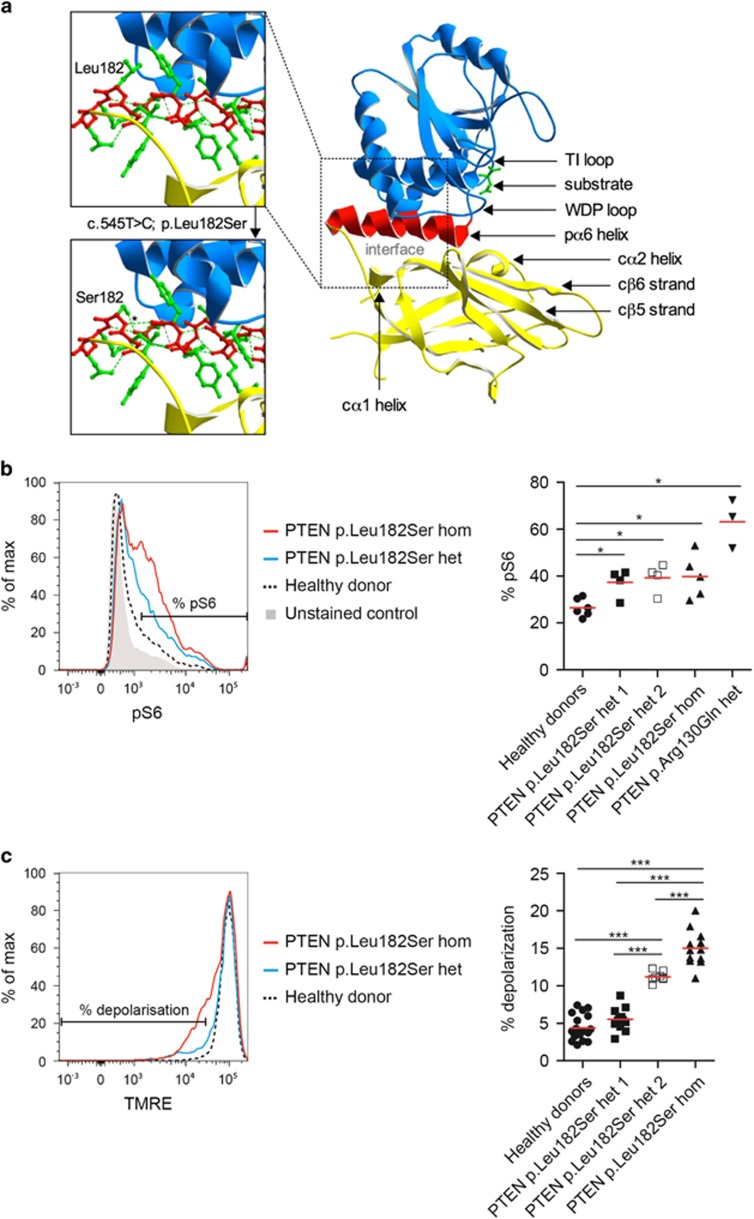
Figure 2.
Molecular modelling and functional impact. (a) Molecular modelling. Position of the p.Leu182Ser alteration is shown in relation to the catalytic core (substrate in green). PTEN crystal structural (Rontgen diffractometry, 2.1 Å) was taken from http://www.rcsb.org/pdb/ (#1d5r) based on Lee et al.27 For visualising the catalytic core of PTEN, the domains as well as the affected amino acid, data from Swiss-Pdb viewer were used. The substrate L(+)-tartrate and the critical PTEN pα6 helix that is likely altered by the variant is highlighted. (b) Functional analysis. Impact of allele number of the p.Leu182Ser variant on S6 phosphorylation in EBV-transformed LCLs. These cells were generated from PTEN c.545T>C homozygotes, PTEN c.545T>C heterozygous parents, and controls. S6 phosphorylation was assessed by intracellular staining and fluorescence intensity determined by FACS analysis. Significance of differences was assessed on pooled data sets for healthy controls, p.Leu182Ser heterozygous as well as p.Leu182Ser homozygous patients and compared with a PHTS p.Arg130Gln patient line. Differences were compared using Mann–Whitney U test. (c) Mitochondrial fitness. LCLs were assessed by the TMRE assay to measure relative mitochondrial depolarisation. Gates for depolarised cell populations were defined by comparison with LCLs generated from healthy controls. Significance of differences was assessed on pooled data sets and analysed using Mann–Whitney U test (*P<0.05, ***P<0.001).
The c.545T>C variant was not previously reported in the exome variant server (6500 individuals), the 1000 genomes project (version 2012 with 1200 individuals), the ClinVar database nor in the in house generated exome database (728 individuals). We have further investigated the Exome Aggregation Consortium (ExAC) database that includes 60 706 unrelated individuals sequenced as part of various disease-specific and population genetic studies using whole exome sequencing (http://exac.broadinstitute.org/gene/ENSG00000171862). The p.Leu182Ser variant was not present in the heterozygous nor in the homozygous state. Indeed among 42 likely protein affecting variants in PTEN (frameshift, splice region, missense, stop-gain), found in 188 individuals, there was no homozygous PTEN variant. Finally, the p.Leu182Ser variant was also not found in >200 DNA samples of PHTS patients (Charis Eng, Cleveland; Stefan Aretz, Bonn; personal communications).
The missense substitution c.545T>C causes p.Leu182Ser amino acid exchange localised within the pα6 helix (Figure 2a) which is relevant for the interface between phosphatase and the C2 domain of PTEN. The phosphatase C2 interface has seven hydrophobic and aromatic residues forming a buried core, and nine residues participating in hydrogen bond networks. In particular, the WDP, TI loop, and pα6 helix from the phosphatase domain and cβ5, cβ6 strands, and cα1, cα2 helices of C2 domain form a spacious domain–domain interface that has relevance for both phosphatase and C2 domain but also offers access sites for binding proteins. Several heterozygous variants of PTEN have been described in the ClinVar database that likely alter the phosphatase C2 interface, suggesting that functional alterations can cause PHTS.
Functional analysis of PI3K signalling in p.Leu128Ser PTEN cell lines
To determine whether the p.Leu182Ser variant has a functional impact on PI3K signalling downstream of PTEN, we performed intracellular staining of phospho-S6 in EBV-transformed LCL at steady state. Compared with healthy donor LCL, significantly increased levels of phosho-S6 (pS235/pS236) were observed in heterozygous and homozygous p.Leu182Ser variant PTEN lines (Figure 2b). However, phosphorylation levels were not as high as in LCL derived from a PHTS patient with a heterozygous PTEN missense mutation p.Arg130Gln that affects phosphatase catalytic core activity.
As PTEN-mediated negative regulation of PI3K signalling is important for the maintenance of mitochondrial fitness, we investigated whether the heterozygous and homozygous variants resulted in compromised mitochondrial function in LCLs. We found a gene-dose effect in the loss of mitochondrial membrane potential (Figure 2c), with the homozygous PTEN variant LCL showing the highest proportion of depolarised fraction. To validate the experimental setting, we treated LCLs generated from a healthy donor with increasing concentrations of a small molecule PTEN inhibitor SF1670 (IC50 2μM according to supplier) to mimic a gene-dose effect of PTEN loss-of-function (Supplementary Figure 1). SF1670 has been described as a PTEN inhibitor in cell-free as well as cellular assays but the mode of activity of SF1670 is not resolved.17, 18, 19 Most recent data suggest there might be an irreversible interaction dependent on reducing conditions.19 Our findings support the observation that the hypomorphic p.Leu182Ser variant of PTEN results in compromised mitochondrial fitness in a gene dose-dependent manner.
DISCUSSION
Both clinical and functional studies suggest that the homozygous PTEN c.545T>C variant is an extremely rare and unusual causative genetic variant. Macrocephaly, mild intellectual disability, penile freckling, and adenoid hypertrophy seen in the patients with the homozygous PTEN c.545T>C variant are typical and common symptoms of PTEN pathogenic variants.20 Indeed, the risk calculator for estimating a patient's risk for PTEN pathogenic variant in the adult patient P1 (http://www.lerner.ccf.org/gmi/ccscore/) as well as the Cleveland Clinic Paediatric Clinical Criteria for PTEN testing in patient P2 revealed scores justifying genetic testing for PTEN pathogenic variants just due to clinical appearance. Interestingly, Hartel et al,16 who previously reported on the family, had already considered clinical Bannayan–Riley–Ruvalcaba syndrome, but did not perform PTEN molecular analysis due to the seemingly discrepant inheritance mode. Second, our functional experiments suggest increased phospho-S6 downstream of the PTEN variant and a potential effect on mitochondrial fitness.
The PTEN c.545T>C variant has several unusual features. In contrast to the classical autosomal dominant occurrence of PHTS, the heterozygous variant in the healthy parents (both 45 years old) and in further obligate carriers in the family did not show any symptoms characteristic of the PHTS spectrum. This supports a threshold model for the phosphatase activity, in which a hypomorphic protein alteration is functionally compensated if the variant is heterozygous, but pathogenic in a homozygous state. In contrast to the classical form of PHTS, the homozygous c.545T>C variant is associated with macrocephaly and intellectual disability but not with early-onset tumours. We can only speculate that this might be explained by a differential impact of the p.Leu182Ser alteration on protein versus lipid phosphatase activity. There is increasing evidence that the protein phosphatase activity of PTEN has an essential role for neuron activity. Zhang et al21 demonstrated that overexpression of wild-type PTEN leads to a decrease in spine density in neurons and that it was the protein phosphatase activity, but not the lipid phosphatase activity of PTEN that was essential for this effect. In addition, there is increasing recognition that PTEN function is linked to its spatial distribution within cells.22 Busa et al23 recently described a pathogenic dominant p.Met134Ile alteration in PTEN that leads to a phenotype with macrocephaly and learning difficulties but the absence of early tumour formation.
To our knowledge we present the first case that suggests that hypomorphic PTEN variants can follow a recessive inheritance pattern.
In a recent study, it has been shown that PTEN does affect mitochondrial metabolism.24 PTEN regulates the activity of mitochondrial energy metabolism and fitness via direct and indirect mechanisms, such as regulation of mTOR and downstream signalling as well as direct localisation of PTEN within mitochondria.24, 25 PTEN-dependent mitochondrial function was assessed by measuring mitochondrial depolarisation in T cells.26 Here, we establish this assay in EBV-transformed LCLs from patients with different PTEN status and show dose-dependent effects of the PTEN p.Leu182Ser variant. Further functional data are required to clarify whether the effect of the p.Leu182Ser variant is due to impairment of protein phosphatase activity causing neurodevelopmental problems but allowing sufficient lipid phosphatase activity to maintain tumor suppressor function.
Acknowledgments
We thank the patients and their parents for participating in this study. We are grateful to Arif Ekici, Steffen Uebe, and Mandy Krumbiegel for support with the SNP arrays and whole exome sequencing, Farah Radwan, Angelika Diem, and Petra Rothe for excellent technical assistance, as well as Oliver Rompel for examining the MRI images. We thank Charis Eng and Stefan Aretz for helpful discussions and sharing unpublished results. This study was supported by grants from the Deutsche Forschungs-gemeinschaft to RAJ (AB393/2-2) and by Cancer Research UK grant number C38302/A12278, through the Oxford Cancer Research Centre Development Fund (OCRC0612-HU) to HHU. HHU and HC declare industry collaboration via the Oxford UCB target program.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Teresi RE, Zbuk KM, Pezzolesi MG, Waite KA, Eng C: Cowden syndrome-affected patients with PTEN promoter mutations demonstrate abnormal protein translation. Am J Hum Genet 2007; 81: 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Mester J, Peterson C et al: A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet 2011; 88: 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester J, Eng C: When overgrowth bumps into cancer: the PTEN-opathies. Am J Med Genet 2013; 163C: 114–121. [DOI] [PubMed] [Google Scholar]
- Hanssen AM, Fryns JP: Cowden syndrome. J Med Genet 1995; 32: 117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA, Dinulos MB, Leppig KA, Sybert VP, Eng C, Hudgins L: The spectrum and evolution of phenotypic findings in PTEN mutation positive cases of Bannayan-Riley-Ruvalcaba syndrome. J Med Genet 2001; 38: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarski R, Burt R, Kohlman W, Pho L, Shannon KM, Swisher E: Cowden syndrome and the PTEN hamartoma tumor syndrome: systematic review and revised diagnostic criteria. J Natl Cancer Inst 2013; 105: 1607–1616. [DOI] [PubMed] [Google Scholar]
- Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW: PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci 2010; 30: 9306–9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE: The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 1998; 273: 13375–13378. [DOI] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C et al: P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA 1997; 94: 9052–9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Park KK: Neuron-intrinsic inhibitors of axon regeneration: PTEN and SOCS3. Int Rev Neurobiol 2012; 105: 141–173. [DOI] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY et al: DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 2009; 63: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles B, Groszer M, Wang S, Jiao J, Wu H: PTENless means more. Dev Biol 2004; 273: 175–184. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM et al: Pten regulates neuronal arborization and social interaction in mice. Neuron 2006; 50: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Jamra R, Wohlfart S, Zweier M et al: Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur J Hum Genet 2011; 19: 1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Tawamie H, Maeda Y et al: Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy. PLoS Genet 2014; 10: e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartel C, Bachmann S, Bonnemann C, Meinecke P, Sperner J: Familial megalencephaly with dilated Virchow-Robin spaces in magnetic resonance imaging: an autosomal recessive trait? Clin Dysmorphol 2005; 14: 31–34. [PubMed] [Google Scholar]
- Rosivatz E, Matthews JG, McDonald NQ et al: A small molecule inhibitor for phosphatase and tensin homologue deleted on chromosome 10 (PTEN). ACS Chem Biol 2006; 1: 780–790. [DOI] [PubMed] [Google Scholar]
- Li Y, Prasad A, Jia Y et al: Pretreatment with phosphatase and tensin homolog deleted on chromosome 10 (PTEN) inhibitor SF1670 augments the efficacy of granulocyte transfusion in a clinically relevant mouse model. Blood 2011; 117: 6702–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli L, Lindsay YE, Leslie NR: PTEN inhibitors: an evaluation of current compounds. Adv Biol Regul 2015; 57: 102–111. [DOI] [PubMed] [Google Scholar]
- Heindl M, Händel N, Ngeow J et al: Autoimmunity, intestinal lymphoid hyperplasia, and defects in mucosal B-cell homeostasis in patients with PTEN hamartoma tumor syndrome. Gastroenterology 2012; 142: 1093–1096.e1096. [DOI] [PubMed] [Google Scholar]
- Zhang XC, Piccini A, Myers MP, Van Aelst L, Tonks NK: Functional analysis of the protein phosphatase activity of PTEN. Biochem J 2012; 444: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis P, Leondaritis G, Lieberam I, Eickholt BJ: Subcellular targeting and dynamic regulation of PTEN: implications for neuronal cells and neurological disorders. Front Mol Neurosci 2014; 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busa T, Chabrol B, Perret O, Longy M, Philip N: Novel PTEN germline mutation in a family with mild phenotype: difficulties in genetic counseling. Gene 2013; 512: 194–197. [DOI] [PubMed] [Google Scholar]
- Liang H, He S, Yang J et al: PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 2014; 19: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, He L, Zeng N et al: Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) signaling regulates mitochondrial biogenesis and respiration via estrogen-related receptor alpha (ERRalpha). J Biol Chem 2013; 288: 25007–25024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H: Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol 2015; 16: 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM et al: Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 1999; 99: 323–334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.