Abstract
Pulmonary symptoms in cystic fibrosis (CF) begin in early life with chronic lung infections and concomitant airway inflammation leading to progressive loss of lung function. Gradual pulmonary function decline is interspersed with periods of acute worsening of respiratory symptoms known as CF pulmonary exacerbations (CFPEs). Cumulatively, CFPEs are associated with more rapid disease progression. In this study multiple sputum samples were collected from adult CF patients over the course of CFPEs to better understand how changes in microbiota are associated with CFPE onset and management. Data were divided into five clinical periods: pre-CFPE baseline, CFPE, antibiotic treatment, recovery, and post-CFPE baseline. Samples were treated with propidium monoazide prior to DNA extraction, to remove the impact of bacterial cell death artefacts following antibiotic treatment, and then characterised by 16S rRNA gene-targeted high-throughput sequencing. Partitioning CF microbiota into core and rare groups revealed compositional resistance to CFPE and resilience to antibiotics interventions. Mixed effects modelling of core microbiota members revealed no significant negative impact on the relative abundance of Pseudomonas aeruginosa across the exacerbation cycle. Our findings have implications for current CFPE management strategies, supporting reassessment of existing antimicrobial treatment regimens, as antimicrobial resistance by pathogens and other members of the microbiota may be significant contributing factors.
Introduction
Cystic fibrosis (CF) is a common recessive genetic disorder, primarily of Caucasians, affecting more than 8000 children and adults in the UK and an estimated 100 000 globally (Davies et al., 2014). Although this disorder is multisystemic, the leading cause of morbidity and mortality in an overwhelming majority (ca. 90%) of patients is chronic lung infections and concomitant airway inflammation (Foweraker, 2009). Until a treatment for the disorder can be found, for example, cystic fibrosis transmembrane conductance regulator gene therapy or therapies that improve cystic fibrosis transmembrane conductance regulator function, the best way to continue to raise life expectancy is to improve treatment of the chronic polymicrobial lung infections (LiPuma, 2014). Implicit within the aim of improving and directing treatment is to better understand the nature of those chronic polymicrobial infections, their response to standard treatments, and how they differ and change with disease progression.
Pulmonary symptoms in CF begin in early life and over time a combination of impaired mucociliary clearance, innate immune responses, and chronic infection lead to a progressive loss of lung function (Flume et al., 2009). This gradual decline in pulmonary function is interspersed with periods of acute worsening of respiratory symptoms known as CF pulmonary exacerbations (CFPEs) (Sibley et al., 2008). Cumulatively, these periods of CFPE are associated with more rapid disease progression and reduced survival as well as a recognised reduction in quality of life and an increase in overall healthcare costs (VanDevanter et al., 2013). There is currently no consensus for a generally applicable definition to describe when patients are experiencing a CFPE. This is mainly due to a lack of consistency in the symptoms experienced by CF patients of different ages and disease state, thus making it challenging for paediatric and adult physicians to come to an agreement (Smyth and Elborn, 2008). Despite this, clinical features can include decreased exercise tolerance, increased cough and sputum production, shortness of breath, chest pain, absence from school or work, increased adventitial sounds on lung examination, decreased appetite or weight loss, and a decline in lung function (Fuchs et al., 1994). Although lung function is still widely relied upon as an indicator of worsening symptoms, it has been revealed that short-term changes in lung function show little effect on the sensitivity or specificity of CFPE diagnosis (Rosenfeld et al., 2001).
As such, clinicians face the challenging role of determining if a patient is experiencing a CFPE and hence when intervention is appropriate. CFPEs are treated with high-dose intravenous antibiotics that are broad spectrum in their activity, but expected to have an impact on Pseudomonas aeruginosa (Smyth and Elborn, 2008). It is now recognised that immediate management of CFPE results in improved prognosis and increased likelihood of patient recovery, underlining the importance of CFPE diagnosis (Sanders et al., 2010). Therefore, there is an urgent need to identify and develop more reliable bioindicators of disease state change.
Studies using ecological analyses to aid in the interpretation of culture, independent data have shown associations between respiratory microbiota characteristics and clinical outcomes (LiPuma, 2014). For example, increasing age, disease progression, and reduced lung function have been associated with related changes in CF lung microbiota, summarised in reduced bacterial diversity and increasingly conserved community composition (e.g. Cox et al., 2010; Klepac-Ceraj et al., 2010; van der Gast et al., 2011; Zhao et al., 2012; van der Gast et al., 2014). However, a fundamental knowledge gap remains in relation to how the CF microbiota behaves and respond to CFPE and subsequent antibiotic interventions, largely due to a paucity of studies on this subject.
A useful framework proposed to better understand changes in microbiota associated with CFPE and antibiotic intervention has been to partition data into defined clinical periods: (B) baseline, (E) CFPE, (T) treatment, and (R) recovery (Zhao et al., 2012; Price et al., 2013). Set within this BETR classification, studies have provided useful insights including a clear demonstration that CFPE do not result from increased total bacterial density or density of a specific recognised CF pathogen, such as P. aeruginosa (Stressmann et al., 2011; Fodor et al., 2012; Carmody et al., 2013; Price et al., 2013). Conversely, how changes in microbiota composition and structure are associated with CFPE and intervention remain less clear; as different studies have shown either significant change or no change in CF bacterial communities, and in specific constituent species (Fodor et al., 2012; Stressmann et al., 2012; Daniels et al., 2013; Price et al., 2013; Smith et al., 2014). These contradictory findings may be attributed to differing experimental designs, patient cohorts, and sampling strategies. For example, studies have typically relied on single-point samples to be representative of a given disease state (e.g. baseline or exacerbation) which will not account for variance in either microbiota or clinical factors, such as lung function (commonly, FEV1, forced expiratory volume in 1 s), in a given clinical period (Carmody et al., 2013; Price et al., 2013; Zemanick et al., 2013). Studies have also been based in the main on point samples from subsets and different combinations of the clinical periods defined above (Fodor et al., 2012; Carmody et al., 2013; Price et al., 2013; Smith et al., 2014), making them difficult to compare, and as such the complete cycle of clinical periods from pre- to post-CFPE baseline, that is BETRB, has not been studied. However, in both instances this may be a reflection of the difficulty in obtaining respiratory samples and metadata over this type of longitudinal period.
In the current study, multiple sputum samples were collected from 10 CF adult patients over the course of a CFPE. The data were partitioned into five clinical periods: (B0) baseline pre-CFPE; (E) CFPE, 30 days prior to treatment; (T) treatment for CFPE; (R) recovery, 30 days post-CFPE; and (B1) baseline post-CFPE. In addition, multiple sputum samples were collected from two adult CF patients who were clinically stable over the same time period (B0 only). During periods B0, E, R, and B1, patients were receiving their standard doses of maintenance antibiotics. For T, patients were hospitalised and receiving increased antibiotic intervention, which was typically intravenous antibiotics (Table 1). To allow profiling of the extant CF lung microbiota, sputum samples were treated with propidium monoazide prior to DNA extraction, to remove the impact of extracellular DNA or DNA from dead or damaged bacterial cells, and then 16S rRNA gene-targeted high-throughput sequencing was carried out (Rogers et al., 2013). This allowed a detailed examination of (1) how changes in CF microbiota are associated with the onset of CFPEs, (2) the effect of antibiotic therapy administered for treatment of CFPE on the CF microbiota, and (3) identification of potential biomarkers of disease state change.
Table 1. Summary of clinical characteristics for individual patients.
| Patient | Age (years) | Gender | CFTR genotype | BMI | CF diabetes | CFPE antibioticsa |
|---|---|---|---|---|---|---|
| 1 | 30 | Male | ΔF508/NK | 29 | No | Ciprofloxacin p.o. |
| 2 | 45 | Female | ΔF508/NK | 18.2 | Yes | Colomycin i.v.+Tobramycin i.v. |
| 3 | 47 | Male | ΔF508/NK | 19.9 | Yes | |
| 4 | 22 | Female | ΔF508/ΔF508 | 18 | No | Cirprofloxacin p.o., then, Meropenem i.v.+Amakacin i.v. |
| 5 | 55 | Male | ΔF508/G58E | 23.9 | No | Ceftazidime i.v.+Gentamicin i.v. |
| 6 | 21 | Female | ΔF508/ΔF508 | 20.3 | No | Ciprofloxacin p.o. |
| 7 | 40 | Male | ΔF508/ΔF508 | 19.4 | Yes | |
| 8 | 22 | Male | ΔF508/ΔF508 | 18.4 | Yes | Meropenem i.v.+Colomycin i.v. |
| 9 | 17 | Female | ΔF508/ΔF508 | 22.5 | No | Ceftazidime i.v.+Gentamicin i.v. |
| 10 | 24 | Female | ΔF508/G542X | 21 | No | Clarithromycin p.o. |
| 11 | 20 | Male | ΔF508/ΔF508 | 20.4 | No | Ciprofloxacin p.o.+Metronidazole |
| 12 | 20 | Male | ΔF508/ΔF508 | 28.5 | No | Ceftazidime i.v.+Gentamicin i.v. |
Abbreviations: BMI, body mass index (kg m2); CFTR, cystic fibrosis transmembrane conductance regulator; NK, genotype not known (the clinical and functional translation of CFTR (CFTR2); http://cftr2.org).
Antibiotics administered as intervention for a clinically defined CFPE: p.o., oral; i.v., intravenous.
Materials and methods
Patients and clinical samples
This study was undertaken with the local ethical approval from Southampton and South West Hampshire Research Ethics Committee (06/Q1704/24). Sputum samples were collected from 12 adult CF patients attending the Southampton General Hospital, Southampton, UK (Table 1). All sputum samples were stored at 4oC immediately after expectoration and transferred to −80 °C storage within 12 h of expectoration. This timeframe was selected as recent evidence suggests that failure to stabilise samples at −80 °C within 12 h significantly impacts sequence-based microbiota profiling. (Cuthbertson et al., 2014). Subjects were selected due to their persistent production of sputum and history of CFPE. All patients were clinically defined as chronically colonised with P. aeruginosa. Samples were collected from patients during periods of baseline, defined as periods where patients were only receiving maintenance doses of antibiotics. During the study period 10 of the 12 patients were treated for a CFPE. The start and end of CFPE were identified by treating clinicians, and were defined for the purpose of this study as the period of time where patients received clinical intervention in the form of antibiotic treatment. The antibiotics administered to each patient are all broad spectrum in activity and are expected to have an impact on the predominant pathogen, P. aeruginosa (Table 1). Decisions to initiate treatment were based on worsening clinical symptoms (Fuchs et al., 1994). Stabilisation or improvement of these symptoms led to the termination of this treatment. Retrospectively samples were partitioned into five periods: (B0) baseline pre-CFPE; (E) CFPE, within 30 days prior to antibiotic treatment; (T) period of time patients were receiving treatment for pulmonary exacerbation; (R) recovery, within 30 days post-treatment for CFPE; and (B1) baseline post-CFPE. Clinically relevant symptoms were monitored throughout the sampling period. Lung function (FEV1) was assessed using a Koko PeakPro home spirometer (Ferraris Cardiorespiratory, Louisville, CO, USA) at the time of each sample collection. Patients were also required to assess respiratory symptoms: cough, breathlessness, sputum production, and general wellbeing, using a visual analogue scale scored 0–100, with 0 being the worst and 100 the best.
DNA extraction and pyrosequencing
Sputum samples were washed three times with 1 × phosphate-buffered saline to remove saliva as previously described (Rogers et al., 2006). Extracellular DNA and DNA from nonviable cells were excluded from analysis via cross-linking with propidium monoazide prior to DNA extraction, as previously described (Rogers et al., 2013). Barcode encoded FLX amplicon pyrosequencing was performed using the primer 338F (5′-ACTCCTACGGGAGGCAGCAG) and 926R (5′-CCGTCAATTCMTTTRAGT) as described previously (Cooper et al., 2013). Except, initial generation of 16S rRNA gene amplicons involved a one-step polymerase chain reaction of 25 cycles using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA). polymerase chain reaction-negative controls were included in each sequencing run (Salter et al., 2014). Four hundred and fifty-four pyrosequencing using the Lib-L kit was performed at the Wellcome Trust Sanger Institute (Hinxton, UK).
Sequence analysis
The mothur sequencing analysis platform was used to analyse the resulting data (Schloss et al., 2011). Sequence quality was ensured by trimming reads where the average quality score fell below 35 across a rolling window of 50 bases. Reads shorter than 400 bp in length and those that included mismatches in the barcode or 16S rRNA gene primer sequence, ambiguous base calls and homopolymeric stretches longer than 8 bases were subsequently removed. Chimeras were removed in mothur using the Perseus software program (Quince et al., 2011). Sequences were assembled into operational taxonomic units (OTUs) at 97% sequence identity to give an approximation of species (Schloss and Handelsman, 2006) and identified using the Ribosomal Database Project reference database provided in mothur. OTUs identified in negative controls were removed from further analysis (Salter et al., 2014).
As the Ribosomal Database Project classifier only provides classifications down to the genus level, representative sequences from each OTU were used to give appropriate species-level identifications using MegaBlast against the NCBI nucleotide archive. For some OTUs multiple, equally scoring, matches were obtained, As such, species-level designations should be considered putative. The raw sequence data reported in this study have been deposited in the European Nucleotide Archive under study accession numbers ERP005251 and ERP007059, and sample accession numbers ERS421603 and ERS551400. The relevant barcode information for each sample is shown in Supplementary Table S1.
Statistical analysis
Species were partitioned into core and rare species groups using a modification of the method previously described (van der Gast et al., 2011). Based on a significant positive distribution–abundance relationship, the persistent and abundant core species were defined as those in more than 75% of all samples, while all other species falling outside of the upper quartile were considered to be rare (Hedin et al., 2015). Species turnover between consecutive samples was measured using the method described by Brown and Kodric-Brown (1977). Turnover was defined as t=b+c/S1+S2. Where b=the number of species present only in the first sample; c=the number of species present only in the second sample; S1=the total number of species in the first sample; and S2 the total number of species in the second sample (Brown and Kodric-Brown, 1977). Species richness (S*) was used as previously described (Rogers et al., 2013). In brief, S* was calculated with a uniform re-sample size (to match the smallest sequence size in each group) following 1000 iterations in each instance and performed in R version 3.1.1 (Oksanen et al., 2013; The R Development Core Team, 2013). Two sample t-tests, regression analysis, coefficients of determination (r2), residuals and significance (P) were calculated using Minitab software (version 16; Minitab, University Park, PA, UK). Analysis of similarity (ANOSIM) and similarity of percentages analysis (SIMPER) were performed using the PAST (Paleontological Statistics, version 3.01) program available from the University of Oslo (http://folk.uio.no/ohammer/past). The Bray–Curtis quantitative index of similarity was used as the underpinning community similarity measure for both ANOSIM and SIMPER analyses.
Mixed-effect models were fitted in R version 3.1.1 using the GLMMADMB package version 0.8.0 (The R Development Core Team, 2013). Values of r2 were calculated using the R package MuMIN. To analyse species-level changes over the disease periods, mixed-effect models (GLMMADMB) with negative bionomial errors were used as data were over dispersed (greater variability than would be expected based on Poisson distribution). For each species, the change in abundance across all patients was measured using the disease period as the fixed effect and variation between patients was accounted for by including patient as a random effect. The model fits changes in abundance on the logit scale. The null hypothesis for each species was there would be no change in species abundance between periods.
Results
An expectation for patterns of lung function (FEV1) within individual patients was that it would entail a consistent decline in FEV1 from baseline (B0) through the CFPE and into the treatment periods, followed by an improvement in the subsequent periods. However, no common pattern in lung function by clinical period was observed across the 10 patients that experienced CFPE (Supplementary Figure S1). Therefore, within the current study, FEV1 was found to be a poor indicator of short-term disease state.
Microbiota diversity and composition from respiratory samples was assessed using 16S rRNA gene-targeted pyrosequencing. From 237 samples, a total of 386 002 bacterial sequence reads (mean±standard deviation (s.d.) per sample, 1628±84) were included in the final analysis, identifying 103 genera and 183 distinct OTUs classified to the species level (Supplementary Table S2); however, given the relative length of the ribosomal sequences analysed, these identities should be considered putative. The average numbers of bacterial sequence reads per sample were similar among the five disease states: (B0) n=56, 1845±247 (mean±s.d.); (E) n=41, 1357±136; (T) n=67, 1643±127; (R) n=32, 1845±151; and (B1) n=41, 1449±212 (Supplementary Table S3).
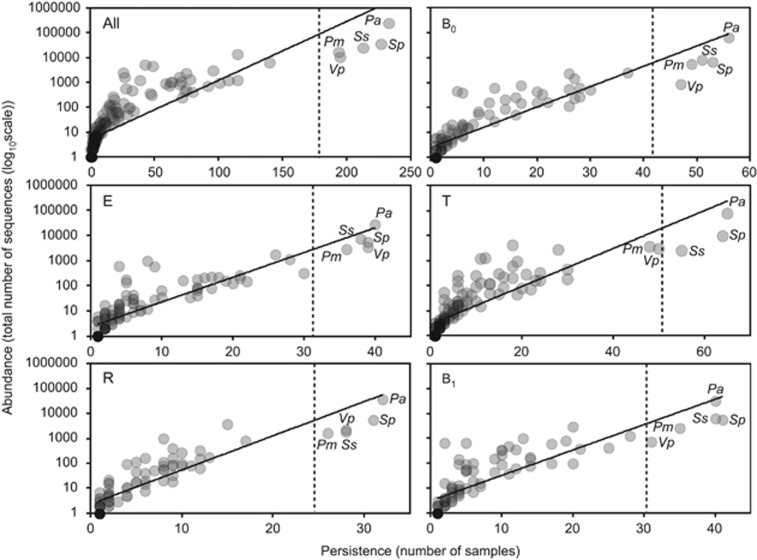
We have previously established that categorisation of component species in CF microbiota into core and rare species revealed important aspects of species-abundance distributions with a metacommunity that would be neglected without such a distinction (van der Gast et al., 2011, 2014). A coherent metacommunity could be expected to exhibit a direct relationship between prevalence and proportional abundance of individual species within the constituent communities. Consistent with this prediction, the abundance of OTUs, across all samples and within specific disease periods (Figure 1), was significantly correlated with the number of individual respiratory sample communities that those OTUs occupied. On that basis, OTUs present in more than 75% of all samples, and hence highly abundant, were classified as core (Hedin et al., 2015). These were derived from five common taxa, P. aeruginosa, Streptococcus pneumoniae group, Streptococcus sanguinis group, Prevotella melaninogenica/veroralis/histicola (abbreviated hereafter as P. melaninogenica), and Veillonella parvula/rogosae (abbreviated as P. parvula), and accounted for over 84% of the total sequence abundance. Conversely, the rare OTU group accounted for the majority of the diversity, 178 OTUs.
Figure 1.
The persistence and abundance (total number of sequence reads) of bacteria taxa present in all longitudinal samples and within each of the five partitioned periods. (All) All samples collected from the 12 CF patients, irrespective of disease state, within the study (n=237, r2=0.7, F1,182=425.7, P<0.001); (B0) baseline pre-CFPE (n=56, r2=0.8, F1,106=429.3, P<0.001); (E) CFPE, 30 days prior to treatment (n=41, r2=0.8, F1,121=415.1, P<0.001); (T) CFPE treatment period (n=67, r2=0.7, F1,140=363.3, P<0.001); (R) recovery, 30 days post-CFPE treatment (n=32, r2=0.8, F1,86=316.7, P<0.001); and (B1) baseline post-CFPE (n=41, r2=0.7, F1,92=221.1, P<0.001). Core OTUs were defined as those that fell within the upper quartile (dashed lines), and rare OTUs defined as those that did not. Taxa defined as core across all samples are labelled in each panel: Prevotella melaninogenica (Pm), Pseudomonas aeruginosa (Pa), Streptococcus pneumoniae group (Sp), Streptococcus sanguinis group (Ss), and Veillonella parvula (Vp).
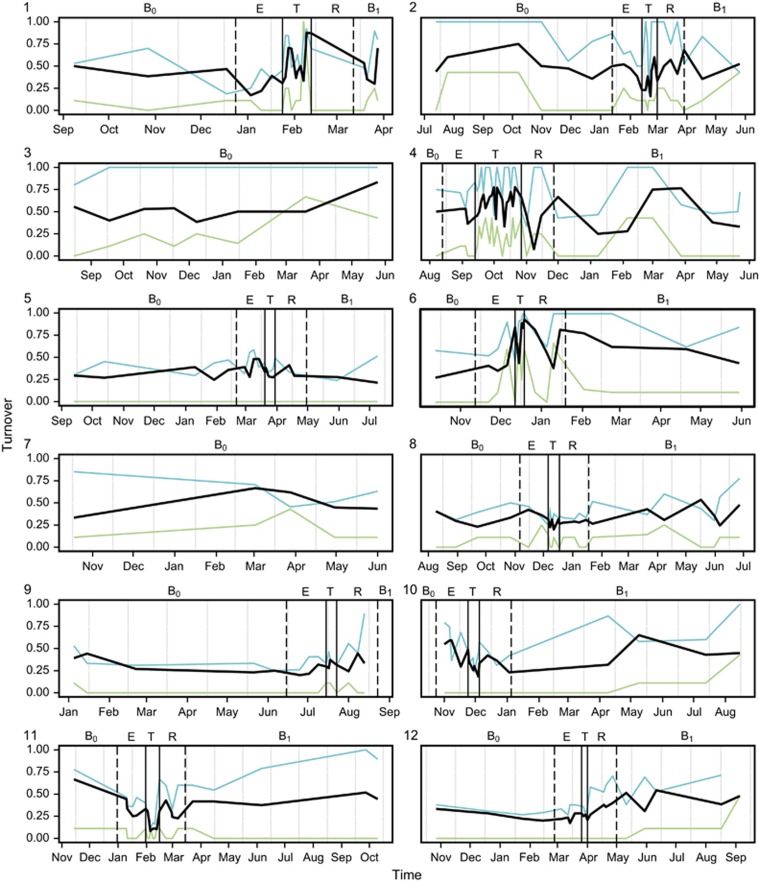
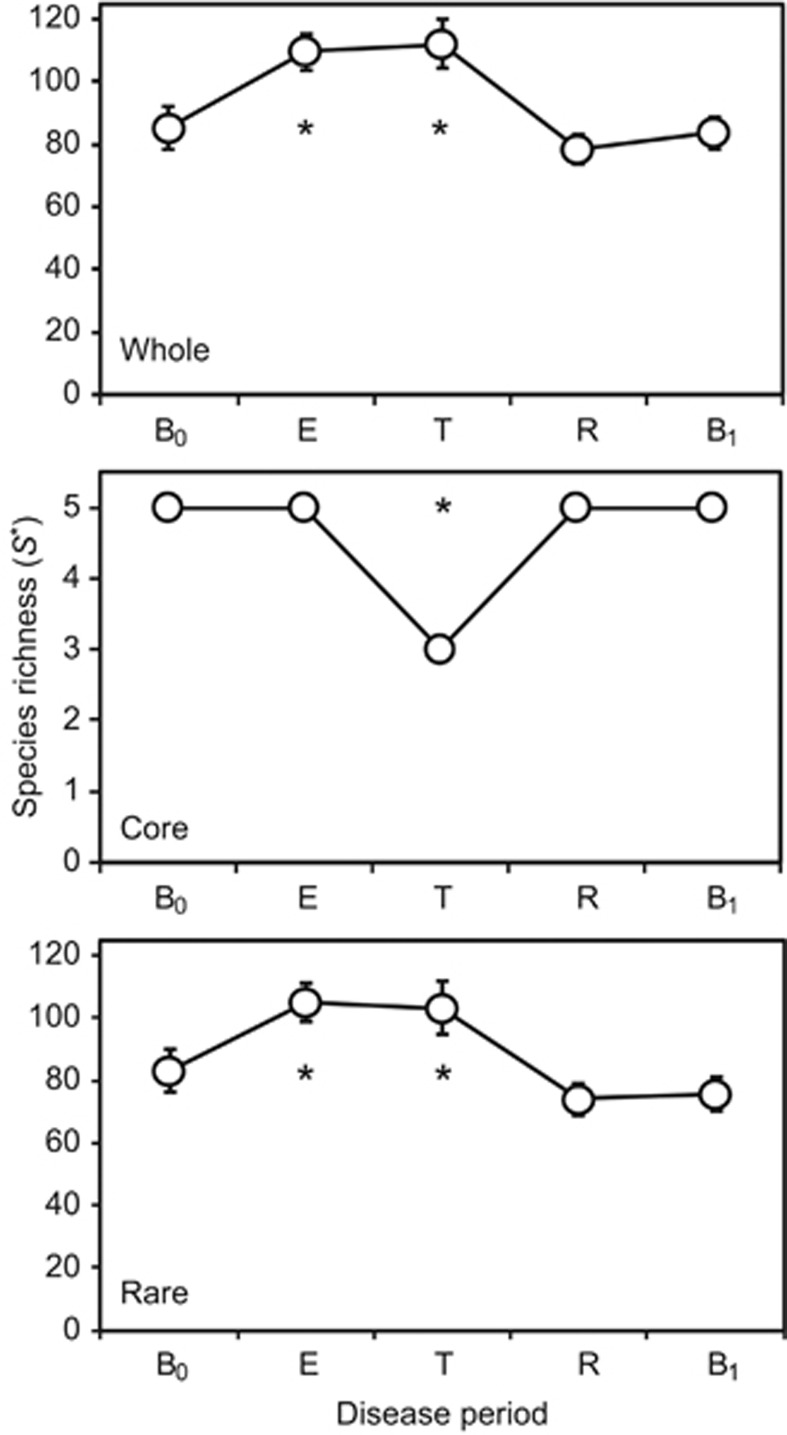
The rate of species turnover, the number of OTUs eliminated and replaced over time, was assessed for each patient over the study period (Figure 2). The rate of OTU turnover showed more variation in the periods surrounding and during treatment for CFPE. Whereas pre- and post-baseline periods had relatively consistent OTU turnover rates, particularly evident for patients 3 and 7 who did not experience a CFPE over the course of the study (Figure 2). The rate of turnover in the rare microbiota group was greater and more variable than that observed for the core microbiota group, suggesting that the more diverse rare microbiota was driving the observed changes in the whole microbiota. Species richness (S*) within the whole microbiota increased significantly from baseline (B0) within the CFPE and treatment periods, returning to B0 levels in the subsequent recovery and post-CFPE baseline (B1) periods. Again, these patterns of diversity were driven by changes within the rare microbiota (Figure 3). Diversity within the core microbiota significantly decreased within the treatment period, with Prevotella melaninogenica and Veillonella parvula moving into the rare group for that period (Figures 1 and 3).
Figure 2.
Changes in the rate of OTU turnover for each patient over time. Solid vertical lines indicate the start and end of treatment for CFPE, and dashed lines indicate the start and end of the 30-day period either side of the treatment period. Disease states are denoted by (B0) baseline pre-CFPE; (E) CFPE, 30 days prior to treatment; (T) treatment for clinical exacerbation; (R) recovery, 30 days post-CFPE treatment; and (B1) stable post-CFPE. Black lines represent turnover in the whole microbiota, while green and blue lines represent turnover within the core and rare OTUs groups, respectively.
Figure 3.
Changes in metacommunity diversity across the disease periods. Given are the whole microbiota, core and rare OTUs groups. Disease periods are denoted by (B0) baseline pre-CFPE; (E) CFPE, 30 days prior to treatment; (T) treatment for clinical exacerbation; (R) recovery, 30 days post-CFPE treatment; and (B1) baseline post-CFPE. Richness (S*) was calculated with a uniform re-sample size following 1000 iterations in each instance; re-sample sizes by group were n=55 656 (whole), n=47 092 (core), and n=8712 (rare) equating to the lowest number of sequences by disease period within each group. Error bars represent the standard deviation of the mean. Asterisks denote significant differences between B0 and subsequent disease periods at the P<0.05 level and determined by two sample t-tests.
ANOSIM, using the Bray–Curtis quantitative index of similarity, was used to determine how composition within the whole, core, and rare microbiota changed by disease period (Table 2). No significant change was observed between disease periods within the whole microbiota. Conversely, significant differences within the core microbiota were observed between pre-CFPE baseline and treatment periods (Table 2). Additionally, significant differences within the rare microbiota were observed between pre-CFPE baseline and the treatment and recovery periods. No significant compositional differences were observed between pre- and post-CFPE baseline periods for both the core and rare OTU groups (Table 2). The switching of the two core OTUs, P. melaninogenica and Veillonella parvula, to the rare group in the treatment period will have likely contributed to the significant differences for the core and rare microbiota between pre-CFPE baseline and treatment periods but not for the whole microbiota, where group membership is not accounted for. SIMPER analysis was employed to assess the contribution of individual OTUs to differences in similarity within the whole microbiota, core, and rare OTU groups between disease periods (Supplementary Tables S4–S6). P. aeruginosa, followed by S. pneumoniae group, contributed most to dissimilarity in the whole microbiota and core OTU group between all disease periods (Supplementary Tables S4 and S5). Within the rare OTU group, Porphyromonas catoniae was observed to provide the greatest contribution to dissimilarity between all disease periods, with the exception of the comparison between the CFPE treatment and recovery periods, where V. parvula provided the largest contribution.
Table 2. Analysis of similarities (ANOSIM) of whole, common, and rare microbiota between disease periods.
| B0 | E | T | R | B1 | |
| Whole | |||||
| B0 | — | 0.210 | 0.092 | 0.413 | 0.562 |
| E | 0.012 | — | 0.276 | 0.177 | 0.342 |
| T | 0.019 | 0.012 | — | 0.778 | 0.396 |
| R | 0.001 | 0.017 | −0.034 | — | 0.199 |
| B1 | −0.007 | 0.001 | 0.003 | 0.014 | — |
| Common | |||||
| B0 | — | 0.197 | 0.01* | 0.477 | 0.533 |
| E | 0.011 | — | 0.04* | 0.208 | 0.488 |
| T | 0.145 | 0.157 | — | 0.518 | 0.152 |
| R | −0.005 | 0.013 | −0.008 | — | 0.140 |
| B1 | −0.006 | −0.004 | 0.029 | 0.022 | — |
| Rare | |||||
| B0 | — | 0.123 | 0.0001* | 0.017* | 0.574 |
| E | 0.027 | — | 0.0002* | 0.024* | 0.005* |
| T | 0.195 | 0.152 | — | 0.0002* | 0.0001* |
| R | 0.194 | 0.156 | 0.206 | — | 0.008* |
| B1 | −0.007 | 0.149 | 0.194 | 0.071 | — |
ANOSIM test statistic (R) and probability (P) that two compared groups are significantly different at the P<0.05 level (denoted with asterisks) are given in the lower and upper triangles, respectively. ANOSIM R and P values were generated using the Bray–Curtis measure of similarity. Disease states are denoted by (B0) stable pre-CFPE; (E) CFPE, 30 days prior to CFPE treatment; (T) treatment for clinical exacerbation; (R) 30 days post-CFPE treatment; and (B1) stable post-CFPE.
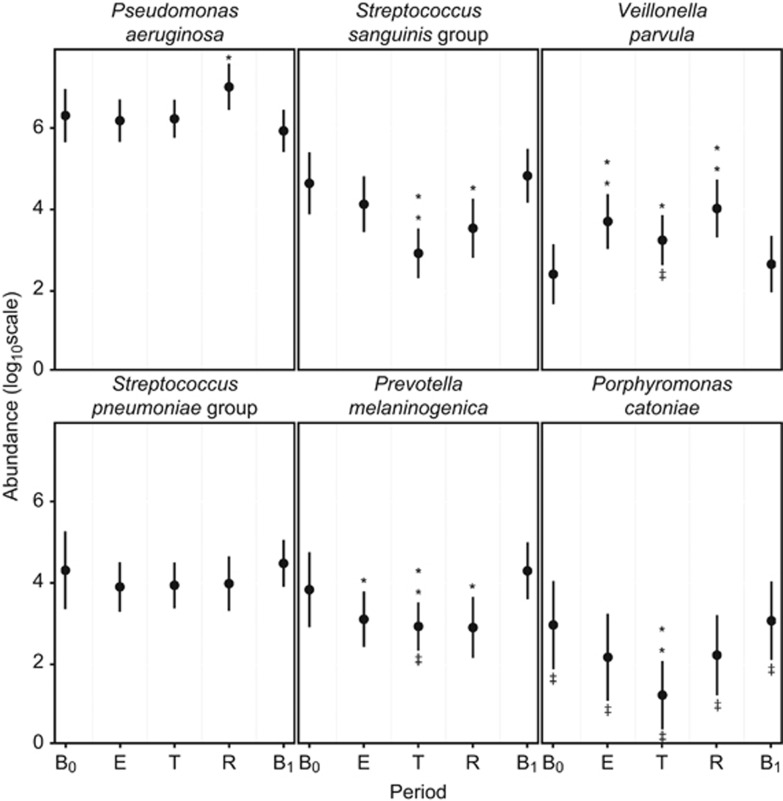
SIMPER analysis also revealed changes in the percentage contribution of individual OTUs within the core microbiota by disease period. Mixed effects models were used to further investigate how individual core OTUs changed between disease periods (Figure 4). P. catoniae, the most abundant of the rare OTU group, was also included in this analysis. These models allowed for the inclusion of both fixed (disease period) and random effects (patient), and therefore variation between patients was accounted for in each model. For P. aeruginosa, no significant change in relative abundance from pre-CFPE baseline (B0) was observed in the CFPE and treatment periods. However, a significant increase in proportional abundance was observed within the recovery period returning to B0 abundance levels in the post-CFPE baseline period (B1). No significant changes in relative abundance from B0 were observed for the S. pneumoniae group OTU in any of the disease periods. S. sanguinis group and P. catoniae both had significantly reduced abundance within the treatment period, with the former OTU also significantly lower in the recovery period. P. melaninogenica proportional abundance significantly reduced from baseline (B0) over the CFPE, treatment, and recovery periods before returning to pre-CFPE baseline (B0) levels in period B1. Conversely, the proportional abundance of V. parvula significantly increased over the same periods (Figure 4).
Figure 4.
Trends in the relative abundance of bacterial species with change in disease period. Parameters are extracted from mixed-effects models based on 237 samples from 12 patients. (B0) baseline pre-CFPE, (E) CFPE, 30 days prior to antibiotic treatment, (T) period of time patients were receiving treatment for pulmonary exacerbation, (R) recovery, 30 days post-treatment, (B1) baseline post-CFPE. **P<0.001, *P<0.05. ‡ indicates rare group membership.
Discussion
Cross-sectional studies have informed our understanding of CF chronic lung infections, revealing a complex and highly variable microbiota in a constantly perturbed ecosystem (van der Gast et al., 2011, 2014). However, in order to better understand the progressive nature of chronic lung infection in CF, it is important to investigate how the bacterial communities change through time in relation to disease state and progression. A small number of studies have attempted to understand bacterial community dynamics within the CF lung (Stressmann et al., 2012; Zhao et al., 2012; Carmody et al., 2013; Price et al., 2013). A common limitation, as stated by Carmody and colleagues (2013) for their own study, has been narrow frequency of sampling, which would not allow fluctuations in airway microbiota to be accounted for, within or between clinical periods. Indeed this within clinical period fluctuation was particularly highlighted in the current study, where species turnover was more variable in the CFPE, treatment, and recovery periods (Figure 2). By collecting samples and associated metadata at more frequent intervals the current study was able to not only account for variation within the microbiota over time but also investigate how this related to changes in disease state through the full cycle of an exacerbation event.
Resistance can be defined as ‘the degree to which microbial composition remains unchanged in the face of a disturbance' and resilience as ‘the rate at which microbial composition returns to its original composition after being disturbed' regardless of the system studied (Allison and Martiny, 2008). Previously, Price et al. (2013) and Stressmann et al. (2011) reported no significant change in community composition through CFPE and treatment. Congruent with those studies we found that the whole microbiota did not significantly change in composition across the five clinical periods studied, indicating resistance to perturbations within the lung (Table 2). However, partitioning the whole chronic lung infection microbiota revealed there was resilience to intervention with antibiotics within both the core and rare OTU groups (Table 2), a pattern also reflected within the diversity based analysis (Figure 3).
Infection with P. aeruginosa has been highlighted as a key factor in poor clinical outcomes and is typically one of the main targets for treatment. It was therefore surprising that its proportional abundance remained high across all clinical periods, even exhibiting a significant increase in abundance in the recovery period (Figure 4). While the relative abundance from high-throughput sequencing data of a given species may not be considered strictly quantitative, Price et al. (2013) demonstrated a highly significant correlation between the results of P. aeruginosa-targeted quantitative polymerase chain reaction and sequencing data. One possible explanation for the increase in P. aeruginosa proportional abundances observed after the conclusion of treatment is that antibiotic therapy led to a reduction in the abundance of other species, thereby allowing P. aeruginosa to take advantage of newly available physical niche space. Even so, the proportional abundance of P. aeruginosa returned to baseline after the 30-day post-treatment recovery period, which may be due to the resilience of the members of the rare species group.
The lack of a generally applicable definition of CFPE and a robust general marker of disease state represents a major challenge for the timely treatment of exacerbations. As stated by Rosenfeld et al. (2001) and confirmed from the FEV1 data (Supplementary Figure S1), changes in lung function demonstrates little sensitivity or specificity of CFPE diagnosis. Molecular diagnostic tests are being increasingly employed in diagnosis of infectious diseases, allowing disease-associated biomarkers to be used to identify, predict, and monitor changes in infections (Pattison et al., 2013). The identification of biomarkers in CF could potentially provide important tools to improve CFPE identification and therefore improve outcomes for CF patients. Within the current study, Veillonella species related to V. parvula/rogosae and Prevotella species related to P. melaninogenica/veroralis/histicola represent potential biomarkers of disease state due to their significant changes in abundance from baseline prior to the start of antibiotic treatment (Figure 4). Development of targeted quantitative polymerase chain reaction based analysis is required to test the efficacy of these two OTUs as molecular diagnostic tools for the onset of CFPE.
While our study was limited to a detailed examination of a relatively small number of patients, our findings have relevance to current management strategies for CFPEs. CF patients receive frequent and prolonged courses of antibiotics, often at high doses, especially for CFPE treatment. Smyth and Elborn (2008) stated there is no comparable medical condition where so many individuals are exposed to antibiotics and risk of cumulative antibiotic side effects in this way. While the resilience within the core and rare species groups will be of concern with regard to treatment, of particular interest to clinicians will be the lack of substantial negative impact on P. aeruginosa. Antibiotics administered in response to the worsening symptoms associated with CFPE are expected to target P. aeruginosa, which is the emblematic species of chronic lung infections in CF patients. Antimicrobial resistance by recognised respiratory pathogens and other members of the microbiota may be a significant contributing factor underpinning the findings of this study (Sherrard et al., 2014). Therefore this study strongly highlights the merits of continual reassessment of existing antimicrobial treatment regimens and antibiotic delivery strategies (Sherrard et al., 2014).
From a more fundamental perspective, the composition of the bacterial microbiota is most likely not the whole story. At the population level, studies have shown a variety of co-infection interactions between P. aeruginosa and other co-colonizing microbes, increasing the virulence of this core CF pathogen. For example, P. aeruginosa can use peptidoglycan shed by Gram-positive bacteria as a cue for enhanced virulence (Korgaonkar et al., 2013), metabolites are transferred (e.g. 2,3-butanediol) between fermenting bacteria and P. aeruginosa (Venkataraman et al., 2014; Whiteson et al., 2014), and, similarly, ethanol produced by the fungus Candida albicans can influence P. aeruginosa biofilm formation and phenazine production (Chen et al., 2014). Therefore the wider microbial community and the myriad of interactions therein need to be considered through, for example, metagenomic, metatranscriptomic, and metabolomic approaches to elucidate the roles of bacteria, fungi, and respiratory viruses in tandem, alongside host immune responses. Studies of this nature could reveal the underlying cause or causes of CFPE and it may well be the case that there is no consistent factor (infection or host) that trigger exacerbations, either within or between patients.
Data deposition
The sequence data reported in this paper have been deposited in the European Nucleotide Archive under study accession numbers ERP005251 and ERP007059, and sample accession numbers ERS421603 and ERS551400.
Acknowledgments
This study was supported by the UK Natural Environment Research Council (NE/H019456/1) to CJvdG, by the Wellcome Trust (WT 098051) to AWW and JP for sequencing costs, and by The Anna Trust (KB2008) to KDB. AWW and The Rowett Institute of Nutrition and Health, University of Aberdeen, receive core funding support from the Scottish Government Rural and Environmental Science and Analysis Service (RESAS). We thank Paul Scott, Richard Rance and the Wellcome Trust Sanger Institute's sequencing team for generating 16S rRNA gene sequence data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SD, Martiny JBH. (2008). Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA 105: 11512–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Kodric-Brown A. (1977). Turnover rates in insular biogeography: effect of immigration and extinction. Ecology 58: 445–449. [Google Scholar]
- Carmody LA, Zhao J, Schloss PD, Petrosino JF, Murray S, Young VB et al. (2013). Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc 10: 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S et al. (2014). Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10: e1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M et al. (2013). Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS ONE 8: e76573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA et al. (2010). Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE 5: e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Rogers GB, Walker AW, Oliver A, Hafiz T, Hoffman LR et al. (2014). Time between collection and storage significantly influences bacterial sequence composition in sputum samples from cystic fibrosis respiratory infections. J Clin Microbiol 52: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels TWV, Rogers GB, Stressmann FA, van der Gast CJ, Bruce KD, Jones GR et al. (2013). Impact of antibiotic treatment for pulmonary exacerbations on bacterial diversity in cystic fibrosis. J Cyst Fibros 12: 22–28. [DOI] [PubMed] [Google Scholar]
- Davies JC, Ebdon A-M, Orchard C. (2014). Recent advances in the management of cystic fibrosis. Arch Dis Child 11: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Flume PA, Mogayzel PJ, Robinson KA, Goss CH, Rosenblatt RL, Kuhn RJ et al. (2009). Cystic fibrosis pulmonary guidelines. Am J Respir Crit Care Med 180: 802–808. [DOI] [PubMed] [Google Scholar]
- Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM et al. (2012). The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS ONE 7: e45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foweraker J. (2009). Recent advances in the microbiology of respiratory tract infection in cystic fibrosis. Br Med Bull 89: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs HJ, Borowitz DS, Christiansen DH, Morris EM, Nash ML, Ramsey BW et al. (1994). Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. New Engl J Med 331: 637–642. [DOI] [PubMed] [Google Scholar]
- Hedin C, van der Gast CJ, Rogers GB, Cuthbertson L, McCartney S, Stagg AJ et al. (2015). Siblings of patients with Crohn's disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut e-pub ahead of print 8 April 2015 doi:10.1136/gutjnl-2014-308896. [DOI] [PubMed]
- Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA et al. (2010). Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 12: 1293–1303. [DOI] [PubMed] [Google Scholar]
- Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA 110: 1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiPuma JJ. (2014). Expanding our understanding of respiratory microbiota in cystic fibrosis. Ann Am Thorac Soc 11: 1084–1085. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2013) Package Vegan: Community Ecology Package. R package version 2.0-7. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Pattison SH, Rogers GB, Crockard M, Elborn JS, Tunney MM. (2013). Molecular detection of CF lung pathogens: current status and future potential. J Cyst Fibros 12: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price K, Hampton T, Gifford A, Dolben E, Hogan D, Morrison H et al. (2013). Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome 1: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport R, Turnbaugh P. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Kehagia V et al. (2006). Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J Clin Microbiol 44: 2601–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Cuthbertson L, Hoffman LR, Wing PAC, Pope C, Hooftman DAP et al. (2013). Reducing bias in bacterial community analysis of lower respiratory infections. ISME J 7: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M, Emerson J, Williams-Warren J, Pepe M, Smith A, Montgomery AB et al. (2001). Defining a pulmonary exacerbation in cystic fibrosis. J Pediatr 139: 359–365. [DOI] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DB, Bittner RCL, Rosenfeld M, Hoffman LR, Redding GJ, Goss CH. (2010). Failure to recover to baseline pulmonary function after cystic fibrosis pulmonary exacerbation. Am J Respir Crit Care Med 182: 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. (2006). Toward a census of bacteria in soil. PLoS Comput Biol 2: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard LJ, Tunney MM, Elborn JS. (2014). Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384: 703–713. [DOI] [PubMed] [Google Scholar]
- Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. (2008). A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA 105: 15070–15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Badrick AC, Zakrzewski M, Krause L, Bell SC, Anderson GJ et al. (2014). Pyrosequencing reveals transient cystic fibrosis lung microbiome changes with intravenous antibiotics. Eur Respir J 44: 922–930. [DOI] [PubMed] [Google Scholar]
- Smyth A, Elborn JS. (2008). Exacerbations in cystic fibrosis: 3. Management. Thorax 63: 180–184. [DOI] [PubMed] [Google Scholar]
- Stressmann FA, Rogers GB, van der Gast CJ, Marsh P, Vermeer LS, Carroll MP et al. (2012). Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax 67: 867–873. [DOI] [PubMed] [Google Scholar]
- Stressmann FA, Rogers GB, Marsh P, Lilley AK, Daniels TWV, Carroll MP et al. (2011). Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J Cyst Fibros 10: 357–365. [DOI] [PubMed] [Google Scholar]
- The R Development Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW et al. (2011). Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Gast CJ, Cuthbertson L, Rogers GB, Pope C, Marsh RL, Redding GJ et al. (2014). Three clinically distinct chronic pediatric airway infections share a common core microbiota. Ann Am Thorac Soc 11: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDevanter DR, Elkin EP, Pasta DJ, Morgan WJ, Konstan MW. (2013). Changing thresholds and incidence of antibiotic treatment of cystic fibrosis pulmonary exacerbations, 1995–2005. J Cyst Fibros 12: 332–337. [DOI] [PubMed] [Google Scholar]
- Venkataraman A, Rosenbaum MA, Werner JJ, Winans SC, Angenent LT. (2014). Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa. ISME J 8: 1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteson KL, Meinardi S, Lim YW, Schmieder R, Maughan H, Quinn R et al. (2014). Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. ISME J 8: 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemanick ET, Harris JK, Wagner BD, Robertson CE, Sagel SD, Stevens MJ et al. (2013). Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS ONE 8: e62917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF et al. (2012). Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci USA 109: 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.