Abstract
Neuronal microtubules are subject to extensive posttranslational modifications and are bound by MAPs, tip-binding proteins, and other accessory proteins. All of these features, which are difficult to replicate in vitro, are likely to influence the translocation of kinesin motors. Here we describe assays for evaluating the translocation of a population of fluorescently labeled kinesin motor domains, based on their accumulation in regions of the cell enriched in microtubule plus ends. Neurons lend themselves to these experiments because of their microtubule organization. In axons, microtubules are oriented with their plus ends out; dendrites contain a mixed population of microtubules, but those near the tips are also plus end out. The assays involve the expression of constitutively active kinesins that can walk processively, but that lack the autoinhibitory domain in the tail that normally prevents their binding to microtubules until they attach to vesicles. The degree to which such motor domains accumulate at neurite tips serves as a measure of the efficiency of their translocation. Although these assays cannot provide the kind of quantitative kinetic information obtained from in vitro assays, they offer a simple way to examine kinesin translocation in living neurons. They can be used to compare the translocation efficiency of different kinesin motors and to evaluate how mutations or posttranslational modifications within the motor domain influence kinesin translocation. Changes to motor domain accumulation in these assays can also serve as readout for changes in the microtubule cytoskeleton that affect kinesin translocation.
INTRODUCTION
There are a number of assays to analyze the activity of kinesins translocating on reconstituted microtubules in vitro (Belyy & Yildiz, 2014; Cai, McEwen, Martens, Meyhofer, & Verhey, 2009; Friedman & Vale, 1999; Tomishige, Klopfenstein, & Vale, 2002; Vale et al., 1996; Yildiz, Tomishige, Vale, & Selvin, 2004). Such assays can provide detailed information about the kinetics of kinesin translocation, including the microtubule on-rate, velocity of translocation, and run length, but it can be difficult to translate this information into accurate predictions concerning the movements of these same kinesins in living cells. In cells, for example, the tubulin molecules incorporated into microtubules are subject to extensive posttranslational modifications (Garnham & Roll-Mecak, 2012; Janke, 2014; Magiera & Janke, 2014) and microtubules are bound by MAPs, tip-binding proteins, and other accessory proteins (Atherton, Houdusse, & Moores, 2013; Jiang & Akhmanova, 2011). While there has been some recent progress (Kaul, Soppina, & Verhey, 2014; Sirajuddin, Rice, & Vale, 2014), it has proven difficult to recapitulate the complexity of the cytoskeleton in vitro. One approach to investigate kinesin translocation in cells involves expressing kinesin motor domains, often tagged with multiple copies of a fluorescent protein, then analyzing the translocation of individual kinesins by TIRF microscopy (Cai et al., 2009; Cai, Verhey, & Meyhofer, 2007). While such assays can also provide valuable kinetic information, they are technically challenging and have only been applied in a limited number of cell types. To our knowledge, this approach has not yet been successfully applied in neurons.
Here we describe assays that evaluate the behavior of a population of fluorescently labeled kinesin motor domains, based on their accumulation in regions of the cell enriched in microtubule plus ends. The assay involves expressing constitutively active kinesin constructs that are capable of dimerizing, so that they can walk processively, but that lack the autoinhibitory domain in the tail that normally prevents them from translocating along microtubules until they attach to vesicles (Friedman & Vale, 1999; Hammond, Blasius, Soppina, Cai, & Verhey, 2010; Hammond et al., 2009; Kaan, Hackney, & Kozielski, 2011). While such population assays cannot provide the kind of quantitative kinetic information obtained from in vitro assays, they offer a simple way to examine kinesin translocation in living neurons. They can also serve as an indirect method to assess changes in microtubule properties that influence kinesin translocation, with a temporal resolution of tens of minutes. And most important, these experiments are easy to conduct and give rapid results that are straightforward to interpret.
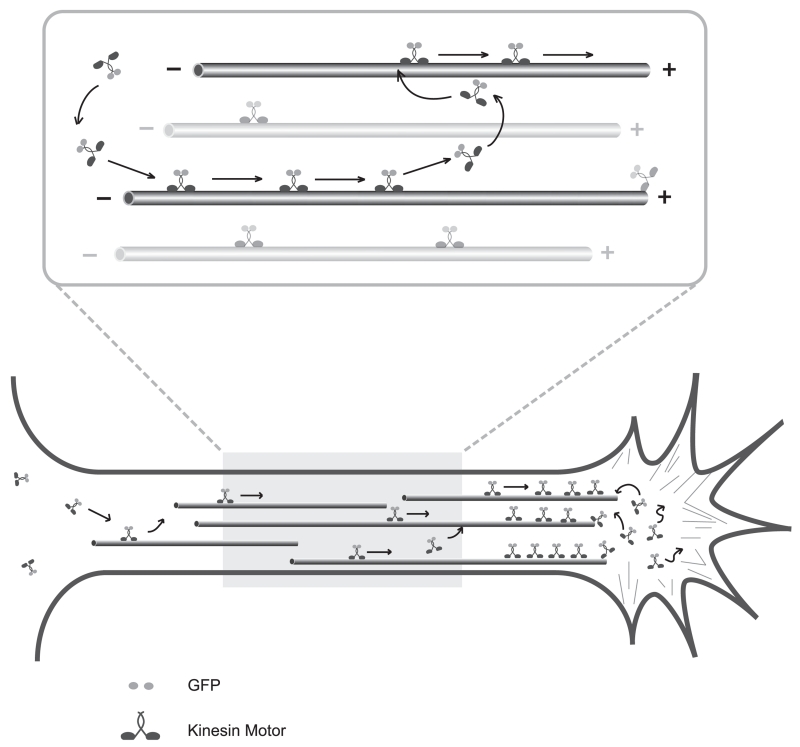
Figure 1 shows a diagram that illustrates how kinesin motor domains are thought to interact with microtubules in axons. Since these kinesins lack the tail domain and are unable to bind cargo, they walk as single molecules. Based on in vitro studies, each kinesin likely attaches to a microtubule, translocates for a few 100 steps (from 1 μm to perhaps as much as 10 μm), then detaches and becomes freely diffusible (Friedman & Vale, 1999; Soppina & Verhey, 2014). Before diffusing far, the released motor domain likely binds another microtubule and resumes its distal translocation. Thus over time, the distribution of individual kinesin molecules reflects two competing forces—active translocation directed predominantly toward axon tips and undirected, passive diffusion.
FIGURE 1. A schematic diagram illustrating the principles underlying the kinesin motor domain accumulation assay.
Axonal microtubules are oriented with their plus ends directed away from the cell body so that kinesins translocate toward the axon tip. As shown in the enlarged view, when an individual kinesin dimer binds to a microtubule, it walks processively toward the plus end. When it dissociates from the microtubule, it undergoes diffusion, but soon encounters another microtubule and resumes its plus end–directed translocation. The net result is that kinesins that translocate efficiently on axonal microtubules become highly concentrated at the tip of the axon.
The result is that motor domains that walk efficiently on axonal microtubules become highly concentrated at axon tips. Unlike axons, dendrites contain microtubules of mixed polarity, but near the distal tips of dendrites nearly all microtubules are plus end out (Baas, Deitch, Black, & Banker, 1988; Stepanova et al., 2003). Thus constitutively active kinesins can also accumulate at dendritic tips. The strong accumulation that occurs at neurite tips in this assay likely reflects the high efficiency of kinesin translocation in neurons and the dense packing of microtubules in axons and dendrites. When kinesin motor domains are expressed in fibroblasts, small spots of fluorescence can be detected near the cell periphery (presumably sites where plus-ends are concentrated), but the majority of the fluorescence is evenly distributed throughout the cell (Bentley, Decker, Luisi, & Banker, 2015).
The accumulation of kinesins at neurite tips observed in these assays requires active translocation along microtubules. Kinesins with mutations that interfere with ATPase activity fail to accumulate in these assays (Huang & Banker, 2012; Jacobson, Schnapp, & Banker, 2006; Nakata & Hirokawa, 2003). Dimerization is also required. For example, a Kinesin-1 construct truncated at amino acid 339 (after the neck linker but before the first coiled-coil) is uniformly distributed when expressed in hippocampal neurons, like a soluble protein (Jacobson et al., 2006). In most of our experiments, we have used constitutively active kinesin constructs generated by truncating the motor just beyond the first coiled-coil domain, which frequently is sufficient to ensure dimerization (Huang & Banker, 2012; Jacobson et al., 2006). Alternatively, constitutively active kinesins can be generated simply by deleting the autoinhibitory domain (Hammond, Blasius, et al., 2010; Nakata & Hirokawa, 2003); such constructs work equally well in this assay. If, for a particular kinesin, the domains that mediate dimerization have not been identified, the kinesin can still be investigated in the accumulation assay by including an artificial dimerization domain in the construct (Tomishige et al., 2002). For example, in the case of KIF13A and KIF13B, one can add an FKBP domain C-terminal to the motor domain, then induce homodimerization by adding an analog of rapamycin (Huang & Banker, 2012).
It should be noted that the motor domain constructs expressed in these assays could dimerize with endogenous kinesins, creating proteins that contain two motor domains but only a single tail. Such constructs would still be expected to walk processively, but could be deficient in cargo binding or in tail inhibition. Thus long-term expression of these constructs could disrupt aspects of vesicle transport and neural development. Although such effects have not been reported, it is probably desirable to minimize expression times in this assay insofar as possible.
1. USING THIS ASSAY TO CHARACTERIZE KINESIN TRANSLOCATION IN NEURONS
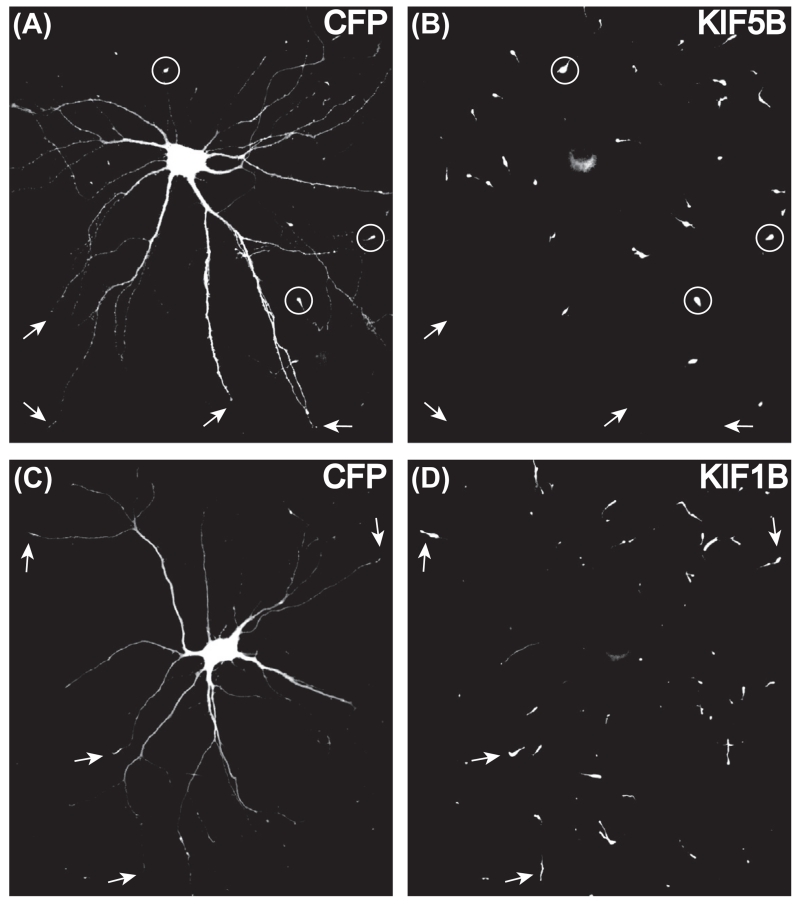
This assay was initially developed to investigate whether different kinesins walked preferentially on axonal or dendritic microtubules (Huang & Banker, 2012; Jacobson et al., 2006; Nakata & Hirokawa, 2003). Based on their patterns of accumulation, several kinesins were identified that preferentially accumulate at axon tips whereas other kinesins accumulate with equal efficiency at the tips of both axons and dendrites (Figure 2). No kinesins have yet been identified that accumulate preferentially at dendritic tips in this assay. Surprisingly, even closely related kinesins often exhibit quite different selectivity in this assay. For example, the Kinesin-3 family motor KIF13A accumulated selectively in the axon whereas the closely related KIF13B accumulated at both dendritic and axonal tips; likewise, the Kinesin-4 motor KIF21A was axon-selective, whereas KIF21B was not. The difference between axon-selective and nonselective motors, as revealed in this assay, is quite striking. For axon selective kinesins like KIF5C, it is difficult to find cells that have an accumulation of the motor domain in even a single dendritic tip. In contrast, in cells expressing nonselective motor domains like KIF1A and KIF1B, 80–90 % of all dendritic tips show an accumulation. More than 90% of all axonal tips are labeled by all of these motor domains (Jacobson et al., 2006). By creating chimeras of axon-selective and nonselective motors, it has been possible to identify elements within the microtubule binding loops that influence the selectivity of translocation (Huang & Banker, 2012; Nakata, Niwa, Okada, Perez, & Hirokawa, 2011). For example, replacing loop 12 of the motor domain of KIF5C with the corresponding residues from KIF1A resulted in a chimera that accumulated at the tips of both axons and dendrites (Huang & Banker, 2012).
FIGURE 2. Different kinesin motor domains exhibit different translocation selectivities in cultured hippocampal neurons.
(A,B) A hippocampal neuron expressing a truncated Kinesin-1 (KIF5B555-YFP), together with a soluble fluorescent protein (CFP) that fills the entire axonal and dendritic arbor. KIF5B555-YFP motors accumulated at the tips of most axonal branches, but did not label dendritic tips. (C, D) A neuron expressing truncated KIF1B (KIF1B386-tagRed) along with CFP. KIF1B accumulated at both axonal and dendritic tips. Circles show examples of labeled axonal tips and arrows indicate dendritic tips. Constructs were expressed by lipofectamine-mediated transfection in 7-day-old cultures and neurons were fixed and imaged after overnight expression.
This assay can also be used to assess the efficiency of kinesin translocation. In comparing the behavior of different kinesins in this expression assay, we observed that most motor domains were localized almost exclusively at neurite tips (Figure 2). In contrast, a few kinesins showed some tip accumulation, but a significant fraction of the label was distributed throughout the cell, in the pattern expected for a soluble protein. We interpret the presence of a soluble motor domain pool to indicate that these kinesins do not walk efficiently in neurons, presumably reflecting a low on-rate or reduced processivity. By calculating the ratio of kinesin motor domain fluorescence that accumulates in the tips of neurites to the total amount of fluorescence present throughout the cell, one can obtain an estimate of translocation efficiency.
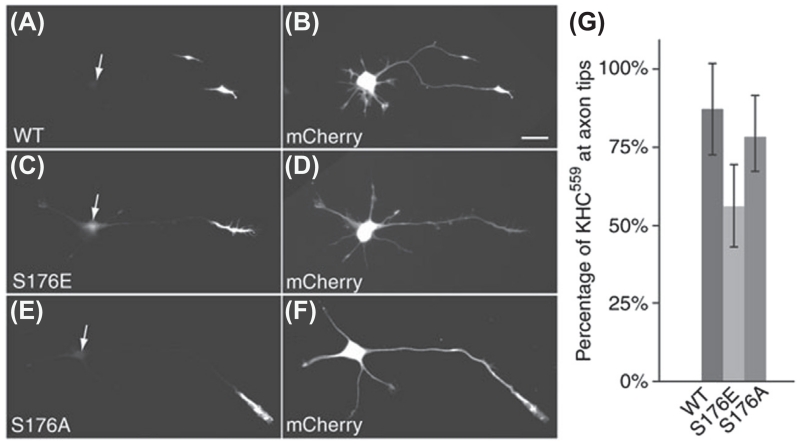
Using this approach, it is possible to determine how modifications in the motor domain influence the efficiency of translocation. For example, cJun N-terminal kinase 3 (JNK3 kinase) phosphorylates KIF5C on serine 176, which lies within loop 8, one of 3 microtubule-binding loops present in this kinesin. By expressing phosphomimetic mutants in which this serine was replaced by a glutamate residue, it was possible to investigate whether phosphorylation at this site influenced motor domain translocation (Morfini et al., 2009). As shown in Figure 3, about 85% of expressed wild-type KIF5C motor domain accumulates in the tip of the axon in this assay. Only about 50% of the phosphomimetic construct localized to the axon tip, with the remainder being diffusely distributed throughout the cell. Mutating the same residue to an alanine, thus preventing phosphorylation, had no effects on motor domain accumulation. Thus JNK3 phosphorylation appears to significantly reduce the translocation efficiency of Kinesin-1. This same approach has been used to evaluate how mutations affect motor domain accumulation. Mutations in the Kinesin-4 motor KIF21A are associated with congenital fibrosis of the extraocular muscles (Cheng et al., 2014; Nugent, Kolpak, & Engle, 2012). Niwa, Takahashi, and Hirokawa (2013) demonstrated that one of these mutations greatly reduced the ability of the motor to accumulate at axon tips when expressed in cultured hippocampal neurons (Niwa et al., 2013). Finally, in some kinesins, sequences outside the motor domain are capable of interacting with microtubules and modifying motor domain translocation (Friedman & Vale, 1999; Soppina et al., 2014). By preparing constitutively active constructs that include different elements within the tail, this assay can be adapted to identify tail domains that affect the pattern or efficiency of accumulation (Huang & Banker, 2012).
FIGURE 3. A mutation mimicking phosphorylation at Serine 176 markedly reduces the translocation efficiency of constitutively active Kinesin-1 in cultured hippocampal neurons.
(A–F) Stage 3 neurons were transfected with GFP-tagged constructs expressing KIF5C559 (A,C,E) and mCherry, a soluble marker protein that diffuses throughout the cell (B,D,F). Wild-type KIF5C and the nonphosphorylatable mutant were localized almost exclusively at axon tips. In contrast, a substantial fraction of the pseudophosphorylated S176E mutant was diffusely distributed throughout the cell. Arrows in A, C, and E indicate the location of the neuronal cell bodies shown in B, D, and F. Cells were analyzed 5 h after transfection. Scale bar: 20 μm. (G) The percentage of total KIF5C-GFP fluorescence localized at the axon tip was measured in cells expressing wild-type KIF5C, KIF5CS176E or KIF5CS176A. Significantly less KIF5CS176E accumulated at axon tips than the wild-type construct (T test, p < 0.0001). Error bars indicate ± s.d.
Reproduced from Morfini et al. (2009).
2. USING THIS ASSAY TO DETECT CHANGES IN MICROTUBULES THAT INFLUENCE KINESIN TRANSLOCATION
Changes in the pattern or efficiency of kinesin motor domain accumulation in this assay can also serve as readout for changes in the microtubule cytoskeleton. For example, this strategy has been useful in elucidating the role of posttranslational modifications of tubulin in modulating kinesin translocation (Hammond, Huang, et al., 2010; Konishi & Setou, 2009; Nakata et al., 2011). In neurons, axonal microtubules are characterized by significantly higher levels of acetylation, detyrosination, and glutamylation than dendritic microtubules and the KIF5C motor domain accumulates only in the tips of axons (Hammond, Huang, et al., 2010). Treatment with a deacetylase inhibitor resulted in increased acetylation of microtubules in both axons and dendrites. However, this treatment did not change the accumulation pattern of KIF5C, suggesting that acetylation alone cannot account for axon selectivity. In contrast, treating cells with paclitaxel, which stabilizes microtubules and induces an increase in acetylation, detyrosination, and glutamylation, resulted in a profound change in the pattern of KIF5C accumulation (Hammond, Huang, et al., 2010; Huang & Banker, 2012). Within 10–20 min of paclitaxel treatment, KIF5C began to accumulate at dendritic tips, a pattern never observed in control neurons. This strategy has been used to investigate how mutations in tubulin associated with genetic diseases that lead to axonal degeneration influence kinesin translocation (Niwa et al., 2013). When these mutant tubulins were expressed in cultured neurons, some of the mutations severely impaired the ability of KIF5, KIF1B, and KIF21A motor domains to accumulate at axon tips.
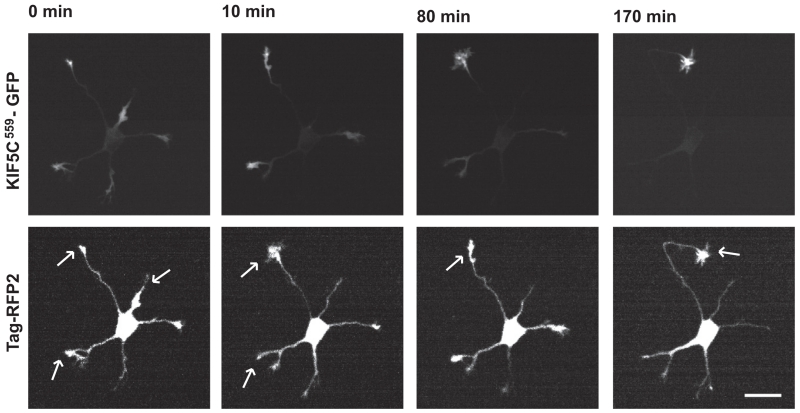
This assay has also been useful for studying how the efficiency of kinesin translocation changes during normal development. Most nonselective kinesins accumulate efficiently at neurite tips at all stages of development. In contrast, axon-preferring members of the Kinesin-3 and Kinesin-4 families fail to accumulate at stage 2, but accumulate efficiently at stage 4. This change roughly correlates with the increases in posttranslational modification of axonal tubulin, especially increases in glutamylation. Members of the Kinesin-1 family, which accumulate preferentially at axon tips in mature neurons, display a particularly interesting change in their pattern of accumulation during development (Figure 4). During stage 2, before axon specification, the pattern of accumulation of KIF5C changes continually, accumulating first in one neurite, then another. Coincident with the spurt of growth that marks the beginning of axonal specification, KIF5C accumulates exclusively at the tip of the emerging axon and remains there. A similar series of dynamic changes in the accumulation of KIF5C have been observed during axon specification in cultured retinal ganglion cells (Randlett, Poggi, Zolessi, & Harris, 2011). The stable accumulation of KIF5C motor domain in a single neurite is among the earliest indicators of axon specification, both in cell culture and in vivo (Distel, Hocking, Volkmann, & Köster, 2010; Jacobson et al., 2006; Randlett et al., 2011; Yamamoto et al., 2012).
FIGURE 4. The Kinesin-1 motor domain translocates into different neurites until the axon is specified.
Selected frames from a time-lapse recording of a hippocampal neuron expressing constitutively active Kinesin-1 (KIF5C559-GFP) along with a soluble protein (Tag-RFP2) to visualize the cell’s geometry. Upper panels show the accumulation of the kinesin motor domain while lower panels illustrate the cell’s morphology; arrows denote neurites with significant kinesin accumulation. Prior to axon specification, the motor domain accumulates in two or three of the cell’s five neurites. As one neurite begins the period of extended growth that marks it as the axon, Kinesin-1 accumulates exclusively in this neurite, losing its ability to accumulate at the tips of any other neurites. Scale bar: 20 μm.
3. PROTOCOLS
In the following sections we provide two protocols, one for assessing the accumulation of kinesin motor domains by fluorescence imaging of fixed cells, the second for live-cell fluorescence imaging to analyze the dynamics of kinesin accumulation. The first assay can be used to ask whether a given kinesin translocates preferentially on axonal or dendritic microtubules, to compare the efficiency of accumulation of different kinesins, or to examine how alterations in the motor domain influence translocation. The second assay is intended to detect temporal changes in the pattern or efficiency of kinesin accumulation, such as those that precede axon specification during development (Jacobson et al., 2006) or after experimental manipulations that alter neuronal microtubules (Hammond, Huang, et al., 2010). The specific parameters described in these protocols (DNA concentrations, transfection methods, expression times, etc.) are based on our experience in applying these methods to rat hippocampal neurons, cultured as previously described (Kaech & Banker, 2006). This same approach can also be used with other types of cultured neurons, or even in intact animals, but of course preliminary experiments may be needed to optimize these parameters.
3.1 PROTOCOL 1: ANALYZING THE PATTERN AND EFFICIENCY OF KINESIN MOTOR DOMAIN ACCUMULATION IN CULTURED NEURONS
In this assay, constructs are expressed in neurons at the desired stage of development and the extent of accumulation is quantified by comparing the localization of the expressed motor domain with that of a coexpressed soluble protein, such as GFP. Details concerning the kinesin constructs that can be used in this assay are provided in Huang and Banker (2012). All were expressed using vectors containing the CAG β-actin promoter (Niwa, Yamamura, & Miyazaki, 1991). Several of these constructs are available from Addgene.
Step 1: Prepare neuronal cultures on glass coverslips. For hippocampal neurons, we plate cells onto polylysine-treated 18 mm coverslips (Fisher Scientific, catalog #12-545-84-1D) at a density of 150,000–250,00 cells per 6 cm dish (~7–12,000 cells/cm2) (Kaech & Banker, 2006).
Step 2: Transfect cultured neurons with cDNAs encoding the desired kinesin motor domains. For hippocampal cultures, we typically transfect between day 6 and day 9 using Lipofectamine 2000 (Kaech & Banker, 2006). Include DNA for the fluorescently tagged kinesin motor domain (0.4–1 μg) and for a soluble fluorescent protein in a different color to label the entire cell arborization (0.1–0.4 μg). Because soluble fluorescent proteins are very stable and express efficiently, a smaller amount of DNA is needed.
Step 3: Return the cells to the incubator for a suitable period to allow expression of the transfected constructs. For cultured hippocampal neurons, we routinely allow constructs to express for 6–24 h. For other cell types, expression times should be worked out in pilot experiments.
Step 4: Fix the cultures and mount the coverslips on slides using an appropriate mounting medium for fluorescence imaging. For hippocampal cultures we fix in 4% paraformaldehyde (in PBS containing 4% sucrose) and mount with polyvinyl alcohol (Banker & Goslin, 1998; p. 128), Prolong Diamond antifade mountant (Life Technologies), or other suitable mounting medium.
Step 5: Scan the coverslips to locate cells expressing the soluble marker protein and record images of both the kinesin motor domain and the marker protein (to capture the cell’s morphology). High NA oil immersion objectives are needed to adequately capture the kinesin fluorescence of transfected neurons. We typically use 40×/1.3NA or 60×/1.4NA plan-apochromat objectives. Because neurons have a sprawling morphology, especially after several days of growth in culture, it will be necessary to take several images and stitch them together to get coverage of the entire cell.
Step 6: Quantify the accumulation of motor domains in neurite tips.
To determine the accumulation pattern of a motor domain, one simply needs to compare the fraction of all axonal and dendritic tips that show a clear accumulation of the kinesin motor. We score tips as positive if the average fluorescence intensity ratio of tip to neurite shaft is three times that of the GFP fill. Soluble GFP has a tip to shaft ratio somewhat greater than 1, reflecting the greater volume of the growth cone compared with the neurite (Jacobson et al., 2006). In most cases, levels of accumulation are far greater than this minimum, so accumulation can be scored without actually measuring fluorescence intensity.
As an estimate of the efficiency of motor translocation, we determine the fraction of expressed motor domain that has accumulated at neurite tips. This can be quantified by measuring the total intensity values for the cell (after background subtraction) and the total intensity localized in the tips of neurites. As an example, about 85% of all wild-type Kinesin-1 fluorescence intensity accumulates in the tips of axons (Morfini et al., 2009). If desired, the fraction of kinesin fluorescence present at tips can be normalized to the fraction of soluble protein present at tips.
It is important not to mistake diffusively distributed soluble motor domain for motor domain that has accumulated at neurite tips. Since dendrites are short and thick compared with axons, motor domains that do not accumulate efficiently may appear to preferentially label the cell body and dendrites, but this should not be mistaken for accumulation due to translocation, which will be restricted to dendritic tips.
3.2 PROTOCOL 2: ASSESSING DYNAMIC CHANGES IN KINESIN MOTOR DOMAIN ACCUMULATION BY TIME-LAPSE IMAGING
This protocol describes the approach we use to image changes in the pattern of kinesin motor domain accumulation that occur in developing neurons during the transitioning from stage 2 to stage 3 (Jacobson et al., 2006).
In principle, any fluorescence microscope setup that allows the cultured neurons to be maintained at an appropriate temperature could be used for such an experiment. In practice, several other factors greatly increase the probability of obtaining useful data in a time-lapse experiment such as this (Kaech, Huang, & Banker, 2012). First, it is extremely useful to use a microscope equipped with an automated, programmable stage, since this allows multiple cells to be recorded in a single experiment. Second, it is very helpful to have a means to compensate for focal drift, using either a hardware device that measures the distance between the objective front lens or software that maximizes image contrast. Finally, to reduce the chance of photo-damage due to repeated imaging, it is advantageous to use a spinning disk confocal microscope rather than a laser scanning confocal microscope or a conventional wide-field microscope. The images shown in Figure 4 were obtained using a 40×/1.3 Plan Fluor objective on a Nikon Ti-E microscope equipped with a Yokogawa CSU-W1 spinning disk attachment, hardware-based drift compensation to maintain focus, and an Andor Zyla sCMOS camera. Coverslips are mounted in a chamber containing imaging medium. The chamber is surrounded by an environmental enclosure that maintains temperature, humidity, and CO2 concentration throughout imaging.
Long-term time-lapse experiments can be challenging. Cells die or suffer damage from light exposure, they grow out of the field of view or their neurites become entangled with those from a neighboring cell, focus is inexplicably lost, or the software crashes. If, in a typical time-lapse experiment, we select 5–10 cells to follow, we consider ourselves lucky to obtain useful data from one or two cells.
Step 1: Express kinesin motor domain constructs by electroporation of dissociated neurons prior to plating. For hippocampal cultures, we typically electroporate 500,000 hippocampal neurons with an Amaxa rat neuron nucleofector device and the corresponding kit (Kaech & Banker, 2006). Include DNA for the fluorescent kinesin motor domain (0.4–1 μg) of interest, as well as a soluble fill in a different color (0.1–0.4 μg). The soluble fill is important for visualizing cell morphology. To decrease the concentration of transfected cells on each coverslip, electroporated cells can be diluted with untreated cells prior to plating. We recommend diluting cells by about one in three before plating (e.g., in a typical experiment we plate 60,000 electroporated cells together with 120,000 unlabeled cells in a 60-cm dish), but this may need to be adjusted depending on experimental details (cell type, time in culture, etc.).
Step 2: After plating cells, place the cultures in a CO2 incubator and allow them to develop to the desired stage for imaging. For example, to assess changes in motor accumulation during the process of axon specification in cultured hippocampal neurons, we allow cultures to develop for 24–36 h.
Step 3: Before imaging, equilibrate environmental enclosure and coverslip chamber to 37 °C.
Step 4: Mount a coverslip in a suitable chamber for live-cell imaging. For long-term recording (greater than 1 h), we use glia-conditioned medium lacking buffered phenol red and maintain the chamber at 5% CO2. For shorter periods of time, buffered imaging medium (Hibernate E) can be used. During all live-cell imaging, cells should be maintained between 34 °C and 37 °C.
Step 5: Place the imaging chamber on the microscope and choose the set of cells to be followed by time-lapse imaging. Locate a transfected cell at the desired developmental stage and record its stage coordinates (X, Y, and Z). Continue until you have obtained a set of 5–10 cells. Ensure that automated focus control is engaged. With a 40× high NA objective, it should be possible to get entire stage 3 cells in the field of view.
Step 6: Program the image acquisition software to capture an image of each cell in each channel at the desired recording interval and begin recording. To observe changes in kinesin motor domain localization in neurons at stage 2 of development, we typically obtain an image every 5–10 min and plan to maintain the recording for at least 10 h.
Step 7: After the recording session is complete, prepare movies by deinterlacing the stacks, so all the images from one cell are arranged together. Eliminate cells whose development is impaired by monitoring growth cone movements and the rate of neurite extension. A reduction in cell dynamics over time is a warning that the cell’s health is deteriorating, making subsequent data on kinesin accumulation suspect.
4. MATERIALS AND EQUIPMENT
Lipofectamine 2000 (Life Technologies, cat. no. 11668027)
Polyvinyl alcohol mounting medium (Banker & Goslin, 1998; p. 128)
Prolong Diamond antifade mountant (Life Technologies, cat. no. 36961)
Lonza neuron electroporation kit (Lonza, cat. no. VPG-1003)
Hibernate E Low Fluorescence (BrainBits, SKU: HE-Lf)
MEM, no glutamine, no phenol red, for preparing conditioned medium (Life Technologies 51200-038)
Electroporator: Nucleofector II (Lonza, cat. no. AAD-1001)
Imaging chamber: Chamlide CMB for 18 mm coverslips (Quorum Technologies, cat. no. CM-B18-1)
ACKNOWLEDGMENTS
This work was supported by NIH grant MH066179. We thank Dr. Stefanie Kaech for her helpful advice on imaging and her comments on the manuscript. We thank Julie Luisi and Barbara Smoody for their outstanding technical assistance.
REFERENCES
- Atherton J, Houdusse A, Moores C. MAPping out distribution routes for kinesin couriers. Biology of the Cell. 2013;105(10):465–487. doi: 10.1111/boc.201300012. http://dx.doi.org/10.1111/boc.201300012. [DOI] [PubMed] [Google Scholar]
- Baas PW, Deitch JS, Black MM, Banker G. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(21):8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K. Culturing nerve cells. MIT Press; 1998. [Google Scholar]
- Belyy V, Yildiz A. Processive cytoskeletal motors studied with single-molecule fluorescence techniques. FEBS Letters. 2014;588(19):3520–3525. doi: 10.1016/j.febslet.2014.05.040. http://dx.doi.org/10.1016/j.febslet.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M, Decker H, Luisi J, Banker G. A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. The Journal of Cell Biology. 2015;93(10):4604. doi: 10.1083/jcb.201408056. http://dx.doi.org/10.1083/jcb.201408056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biology. 2009;7(10):e1000216. doi: 10.1371/journal.pbio.1000216. http://dx.doi.org/10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Verhey KJ, Meyhofer E. Tracking single kinesin molecules in the cytoplasm of mammalian cells. Biophysical Journal. 2007;92(12):4137–4144. doi: 10.1529/biophysj.106.100206. http://dx.doi.org/10.1529/biophysj.106.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Desai J, Miranda CJ, Duncan JS, Qiu W, Nugent AA, et al. Human CFEOM1 mutations attenuate KIF21A autoinhibition and cause oculomotor axon stalling. Neuron. 2014;82(2):334–349. doi: 10.1016/j.neuron.2014.02.038. http://dx.doi.org/10.1016/j.neuron.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Hocking JC, Volkmann K, Köster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. The Journal of Cell Biology. 2010;191(4):875–890. doi: 10.1083/jcb.201004154. http://dx.doi.org/10.1083/jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nature Cell Biology. 1999;1(5):293–297. doi: 10.1038/13008. http://dx.doi.org/10.1038/13008. [DOI] [PubMed] [Google Scholar]
- Garnham CP, Roll-Mecak A. The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton (Hoboken, N.J.) 2012;69(7):442–463. doi: 10.1002/cm.21027. http://dx.doi.org/10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. The Journal of Cell Biology. 2010;189(6):1013–1025. doi: 10.1083/jcb.201001057. http://dx.doi.org/10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, et al. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biology. 2009;7(3):e1000072. doi: 10.1371/journal.pbio.1000072. http://dx.doi.org/10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Huang C-F, Kaech S, Jacobson C, Banker G, Verhey KJ. Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Molecular Biology of the Cell. 2010;21(4):572–583. doi: 10.1091/mbc.E09-01-0044. http://dx.doi.org/10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-F, Banker G. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 2012;13(4):549–564. doi: 10.1111/j.1600-0854.2011.01325.x. http://dx.doi.org/10.1111/j.1600-0854.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson C, Schnapp B, Banker GA. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49(6):797–804. doi: 10.1016/j.neuron.2006.02.005. http://dx.doi.org/10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Janke C. The tubulin code: molecular components, readout mechanisms, and functions. The Journal of Cell Biology. 2014;206(4):461–472. doi: 10.1083/jcb.201406055. http://dx.doi.org/10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Akhmanova A. Microtubule tip-interacting proteins: a view from both ends. Current Opinion in Cell Biology. 2011;23(1):94–101. doi: 10.1016/j.ceb.2010.08.008. http://dx.doi.org/10.1016/j.ceb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Kaan HYK, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333(6044):883–885. doi: 10.1126/science.1204824. http://dx.doi.org/10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nature Protocols. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. http://dx.doi.org/10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kaech S, Huang C-F, Banker G. Long-term time-lapse imaging of developing hippocampal neurons in culture. Cold Spring Harbor Protocols. 2012;2012(3) doi: 10.1101/pdb.prot068239. http://dx.doi.org/10.1101/pdb.prot068239.pdb.prot068239–pdb.prot068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul N, Soppina V, Verhey KJ. Effects of α-tubulin K40 acetylation and detyrosination on Kinesin-1 motility in a purified system. Biophysical Journal. 2014;106(12):2636–2643. doi: 10.1016/j.bpj.2014.05.008. http://dx.doi.org/10.1016/j.bpj.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nature Neuroscience. 2009;12(5):559–567. doi: 10.1038/nn.2314. http://dx.doi.org/10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Magiera MM, Janke C. Post-translational modifications of tubulin. Current Biology. 2014;24(9):R351–R354. doi: 10.1016/j.cub.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Morfini GA, You Y-M, Pollema SL, Kaminska A, Liu K, Yoshioka K, et al. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nature Neuroscience. 2009;12(7):864–871. doi: 10.1038/nn.2346. http://dx.doi.org/10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. The Journal of Cell Biology. 2003;162(6):1045–1055. doi: 10.1083/jcb.200302175. http://dx.doi.org/10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Niwa S, Okada Y, Perez F, Hirokawa N. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. The Journal of Cell Biology. 2011;194(2):245–255. doi: 10.1083/jcb.201104034. http://dx.doi.org/10.1083/jcb.201104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Niwa S, Takahashi H, Hirokawa N. ß-Tubulin mutations that cause severe neuropathies disrupt axonal transport. EMBO Journal. 2013;32(10):1352–1364. doi: 10.1038/emboj.2013.59. http://dx.doi.org/10.1038/emboj.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent AA, Kolpak AL, Engle EC. Human disorders of axon guidance. Current Opinion in Neurobiology. 2012;22(5):837–843. doi: 10.1016/j.conb.2012.02.006. http://dx.doi.org/10.1016/j.conb.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randlett O, Poggi L, Zolessi FR, Harris WA. The oriented emergence of axons from retinal ganglion cells is directed by laminin contact in vivo. Neuron. 2011;70(2):266–280. doi: 10.1016/j.neuron.2011.03.013. http://dx.doi.org/10.1016/j.neuron.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nature Cell Biology. 2014;16:335–344. doi: 10.1038/ncb2920. http://dx.doi.org/10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, et al. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(15):5562–5567. doi: 10.1073/pnas.1400759111. http://dx.doi.org/10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Verhey KJ. The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors. Molecular Biology of the Cell. 2014;25:2161–2170. doi: 10.1091/mbc.E14-01-0696. http://dx.doi.org/10.1091/mbc.E14-01-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, et al. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) Journal of Neuroscience. 2003;23(7):2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297(5590):2263–2267. doi: 10.1126/science.1073386. http://dx.doi.org/10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380(6573):451–453. doi: 10.1038/380451a0. http://dx.doi.org/10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Demura T, Morita M, Banker GA, Tanii T, Nakamura S. Differential neurite outgrowth is required for axon specification by cultured hippocampal neurons. Journal of Neurochemistry. 2012;123(6):904–910. doi: 10.1111/jnc.12001. http://dx.doi.org/10.1111/jnc.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303(5658):676–678. doi: 10.1126/science.1093753. http://dx.doi.org/10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]