Abstract
Since the sample size of a typical neuroimaging study lacks sufficient statistical power to explore unknown genomic associations with brain phenotypes, several international genetic imaging consortia have been organized in recent years to pool data across sites. The challenges and achievements of these consortia are considered here with the goal of leveraging these resources to study addiction.
The authors of this review have joined together to form an Addiction working group within the framework of the ENIGMA project, a meta-analytic approach to multisite genetic imaging data. Collectively, the Addiction working group possesses neuroimaging and genomic data obtained from over 10,000 subjects. The deadline for contributing data to the first round of analyses occurred at the beginning of May 2015. The studies performed on this data should significantly impact our understanding of the genetic and neurobiological basis of addiction.
Keywords: Addiction, Genetic imaging, ENIGMA, Neuroimaging
Introduction
The emergence of numerous large-scale international genetic imaging consortia in recent years is the product of several rapidly evolving factors. The maturing field of neuroimaging has made significant progress toward adopting a widely accepted set of best practices which have been incorporated into several competing software distributions (e.g., SPM, AFNI, FreeSurfer, FSL) that are free to download and relatively easy to install. Adaptation of imaging software to the developing needs of the neuroimaging community and greater automation have been accompanied by tremendous efforts to annotate the software and to educate a large cadre of scientists who are now able to apply these methods to studies with increasingly large sample sizes. Evidence of these efforts can readily be found on the busy message boards of any of the major neuroimaging platforms. Furthermore, with the development of standard anatomical templates and coordinate-based reference systems, researchers worldwide can now relate their findings to previous results in a consistent way. In combination, these factors have facilitated the formation of several large-scale collaborations to overcome the limitation of small sample sizes in typical genetic imaging studies.
The high dimensionality of genetic imaging datasets poses a difficult set of challenges. Human DNA consists of approximately 3 billion nucleotide base pairs. Variation in the population at any individual base is called a single-nucleotide polymorphism (SNP) and may contribute to the differential expression of phenotypic traits. Genomic studies have become a medical research priority because the identification of the genetic variation associated with a disease helps to clarify its molecular basis which, in turn, should lead to improved diagnostic categorization and more effective treatments (Sullivan et al., 2012). One way to proceed in identifying such associations is to investigate the relationship of traits of interest with candidate SNPs that are suggested on the basis of previous research (e.g., to examine the association of smoking behavior with SNPs related to the expression of nicotinic receptor subtypes). However, such a targeted approach is unlikely to expose the full range of SNPs involved in complex traits, such as addiction. To discover unknown trait-SNP associations, an unbiased search across the whole genome, known as a genome-wide association study (GWAS), is necessary. This latter strategy commonly involves testing hundreds of thousands to millions of SNPs and requires a strict multiple comparisons correction threshold, conventionally p ≤ 5 × 10−8, to avoid reporting spurious results. Furthermore, findings must be replicated in at least one independent cohort before they are considered credible or at least generalizable. To meet these stringent thresholds, sharing data across multiple sites has become necessary.
There are now many successful examples of genetic imaging consortia, including ADNI (Alzheimer's disease), IMAGEN (mental health and risk-taking behavior in teenagers), EPIGEN (epilepsy), the Saguenay Youth Study (development), fBIRN (schizophrenia), and CHARGE (heart and aging). These groups have pioneered the use of multisite data sharing protocols and have demonstrated that analyses using shared data produce meaningful findings. The purpose of this review is to discuss how these resources can be leveraged to study addiction.
2 Genetic Basis of Addiction
It is clear that addiction has a genetic component (Maes et al., 2004; Prescott and Kendler, 1999; Tsuang et al., 1998) although the specific set of genes involved remains obscure. Several GWAS of alcohol addiction have been published (Bierut et al., 2010; Edenberg et al., 2010; Heath et al., 2011; Treutlein et al., 2009) which have confirmed the risk of alcoholism associated with of a number of SNPs, such as the ADH and ALDH2 genes, previously identified through the candidate gene approach. These studies have also identified some additional but as yet unreplicated variants that may contribute to alcohol dependence (Rietschel and Treutlein, 2013). However, results have largely differed from one GWAS to another with later studies providing only modest evidence of replication of previous findings. A similar situation exists with regard to cannabis in that published GWAS have not reproduced previous findings (Agrawal et al., 2011; Han et al., 2012; Hopfer et al., 2007). The genetic basis of nicotine dependence has been more closely examined than other substance addictions although again only a handful of results have been replicated across studies (Berrettini et al., 2008; Drgon et al., 2009; Gelernter et al., 2015; Thorgeirsson et al., 2008; Uhl et al., 2008b; Wang et al., 2012a; Zuo et al., 2013). Only a few published GWAS have examined the genetic basis of other drug use (e.g., Uhl et al., 2008a). In summary, most of the genetic variation underlying addiction remains to be explained.
2.1 Brain Endophenotypes
The failure to identify a greater proportion of risk genes is disappointing given the high heritability of addiction. Recent estimates of the heritability of dependence on different addictive substances include: 56% for alcohol, 72% for cocaine, 40% for other stimulants, 48% for cannabis, and 51% for sedatives (Bienvenu et al., 2011). The intermediate “endophenotypes” approach may be a more sensitive way to determine how genes influence addiction vulnerability (Glahn et al., 2007, 2014). An intermediate endophenotype is a quantifiable biomarker (e.g., regional brain volume or activity) that is genetically correlated with disease liability and observed to a greater degree in affected individuals and their relatives than in unaffected nonrelatives. Since these biomarkers are arguably more proximal to the molecular expression of DNA than the related complex trait, it may be possible to generate simpler models of single aspects of the disorder to effectively bridge the gap in understanding between genotype and phenotype. In addition, the statistical power to detect genetic associations may be greater than using diagnostic categories because intermediate endophenotypes represent a continuous scale on which individuals can be ranked.
At least three lines of evidence suggest that genetic neuroimaging may produce useful intermediate endophenotypes of addiction. First, 20 years of neuroimaging data amply demonstrate that brain structure and function interact with the use of addictive substances. For example, brain structure differences compared to healthy controls have been observed in cocaine-dependent individuals (Alia-Klein et al., 2011; Barros-Loscertales et al., 2011; Connolly et al., 2013; Hanlon et al., 2011; Ide et al., 2014; Mackey and Paulus, 2013; Matochik et al., 2003), cigarette smokers (Brener et al., 1995; Kuhn et al., 2010; Sutherland et al., 2013; Zhang et al., 2011), alcoholics (Cardenas et al., 2007; Jernigan et al., 1991; Rando et al., 2011), cannabis users (Batalla et al., 2013; Lorenzetti et al., 2014; Schacht et al., 2012; Yucel et al., 2008), and opiate users (Lyoo et al., 2006; Upadhyay et al., 2010; Wang et al., 2012b). These effects are widespread and likely reflect a mixture of preexisting differences that either confer vulnerability to addiction or are the cumulative effects of chronic exposure.
A second line of evidence suggesting that neuroimaging will generate useful intermediate phenotypes are twin- and SNP-based heritability studies which indicate a high heritability for structural brain measures, such as total amount of gray and white matter, overall brain volume, and addiction-relevant subcortical regions. Heritability estimates for brain measures (h2) are as high as 0.89 (Kremen et al., 2010) or even 0.96 (van Soelen et al., 2012) and subcortical regions appear to be moderately to highly heritable. One recent study reported high heritability estimates for the thalamus (0.80) and caudate nucleus (0.88) compared to a lower heritability for the left nucleus accumbens (0.44) (den Braber et al., 2013).
Third, biomarkers of addiction which are present to a greater degree in affected individuals and their relatives compared to unaffected nonrelatives have been reported. For example, a recent neuroimaging study acquired anatomical MRI and diffusion tensor image (DTI) scans in 50 biological sibling pairs and a group of non-related control subjects (Ersche et al., 2012). One sibling in each pair was dependent on cocaine or amphetamine. Fractional anisotropy in the DTI scans, an index of axonal integrity, was lower in dependent subjects and their nondependent siblings compared to the control subjects. Also, voxel-based morphometry indicated that gray matter volume in both dependent subjects and their siblings was lower in left posterior Sylvian fissure including parts of the postcentral gyrus, insula, and superior temporal gyrus and higher in the left putamen and left amygdala. The discovery of biomarkers that are quantifiably different in drug-dependent individuals and their siblings compared to nonrelated controls underscores the potential for neuroimaging to detect intermediate brain endophenotypes that will be useful in genomic research.
2.2 Challenges
The search for robust genetic and brain structural correlates of drug use and dependence faces a number of substantial challenges. The inability to find extensive significant genome-wide associations might be attributable to the large degree of heterogeneity due to polydrug use and the high incidence of mental health comorbidities among drug users. It will be necessary to disambiguate several sources of genetic variation. Epidemiological studies indicate that there will be genetic variation associated with a general vulnerability to addiction and to a lesser extent drug-specific associations as well as gene–environment interactions (Tsuang et al., 1998). Furthermore, lifetime drug use can be decomposed into a number of qualitatively different stages (e.g., initial experimentation, occasional use, transition to abuse and dependence, risk of relapse) that current research indicates will exhibit different sets of genetic associations (Belin and Deroche-Gamonet, 2012; Everitt and Robbins, 2013; Montigny et al., 2013). GWAS and candidate gene analyses also have their own unique shortcomings. While GWAS searches the whole genome for unknown associations, it will miss variants with small effect sizes that would pass the less stringent probability threshold of the candidate gene approach (Gizer and Ehlers, 2015). With the candidate gene approach, however, there is no way to verify whether published candidate gene studies are systematically biased toward reporting successes. To correct for this latter problem, it has even been suggested that candidate gene associations should be held to the same significance criterion as GWAS (Flint and Munafo, 2013). The solution will likely require a combination of the two search strategies to iteratively approximate the genetic polymorphisms involved in addiction using both intermediate endophenotypes and well-defined behavioral traits.
3 Enhancing Neuroimaging Genetics Through Meta-Analysis
In 2009, researchers from large-scale neuroimaging and genetics consortia, including IMAGEN, EPIGEN, SYS, FBIRN, and ADNI, formed the Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) project to work through the challenges of bringing together data from multiple samples and sites worldwide in a single meta-analytic framework (http://enigma.ini.usc.edu/) (Thompson et al., 2014). The first published ENIGMA meta-analysis reported that the mean bilateral volume of the hippocampus was significantly associated with the intergenetic variant rs7294919 (Stein et al., 2012). This proof-of-principle study established the feasibility of combining imaging and genomic data collected across multiple sites to investigate statistically significant effects of single-letter genomic differences in brain data. In addition, a follow-up study discovered eight genetic loci in which common variants were associated with the volumes of several subcortical structures, including the putamen, caudate, and hippocampus (Hibar et al., 2015). The SNPs associated with subcortical brain volumes were supported across 50 cohorts worldwide, suggesting the power to identify genetic effects that account for as little as 1% of the variance in regional brain volumes. Functional characterization of these genetic loci, in outbred mice, was consistent with possible effects on cell number and links to degenerative disease risk (Ashbrook et al., 2014). The protocol developed by the ENIGMA network to harmonize the data from multiple sites has been made freely available to collaborators and a support structure based in Dr. Paul Thompson's Imaging Genetics Center at the University of Southern California has been created to facilitate the application of the protocol to other projects.
The ENIGMA protocol contains several innovations to deal with special issues arising from multisite analyses, notably imputation of genomic data to a common reference panel and a pathway to harmonize neuroimaging data with standardized quality control procedures. For the initial ENIGMA study, all data were imputed to the HapMap3 reference panel because SNP data at the various sites were genotyped on different gene chips. The imputation protocol adds substantial power to the overall meta-analysis by creating a genomic dataset that is comparable across sites and by employing state-of-the-art approaches to account for hidden structure (e.g., ancestry) and relevant quality control variables. More recently, the ENIGMA imputation protocol implemented in MaCH (http://csg.sph.umich.edu/abecasis/MaCH/) has been updated to use the 1000 Genomes reference, a more in-depth analysis of the genome. To control for population stratification, multidimensional scaling (MDS) is applied to the genotyped data and the first four components are included as nuisance covariates in subsequent GWAS analyses (Hibar et al., 2015; Stein et al., 2012).
To process the neuroimaging data efficiently, one of two highly automated neuroimaging software packages (FSL's FIRST and FreeSurfer) was used for the initial ENIGMA publications although in future studies, including those undertaken by the Addiction working group, only FreeSurfer (Fischl et al., 2002) will be employed (Fig. 1). The use of these standard software programs ensures the comparability of neuroimaging results across sites. Despite the automation of FreeSurfer, considerable time is still required to test for statistical outliers, inspect distributions of brain structure volumes, genomic inflation factors, and other statistical summaries at each site.
Figure 1.
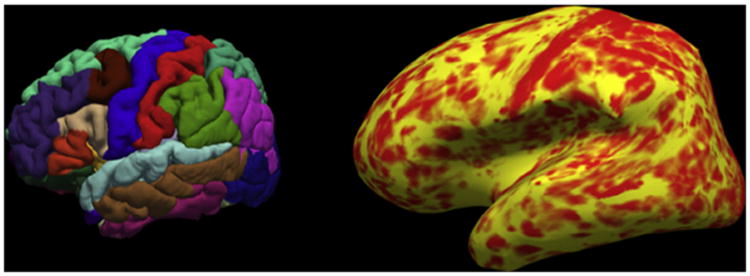
Illustration of a structural MRI brain scan processed with FreeSurfer. Left, example of automated parcellation of the cortex. Right, local cortical thickness projected onto inflated surface of the brain.
Rather than using an analysis strategy where all phenotypic and genotypic data are sent to one central site for processing, as for example in the Psychiatric Genomics Consortium (http://www.med.unc.edu/pgc), ENIGMA employs a meta-analytic strategy in which GWAS are computed locally using agreed upon covariates. The advantages of this approach include the active involvement in the analysis of the researchers who collect and curate the data, and the ability to draw upon local computer infrastructure at each site to ease demand on central data processing. Site-level GWAS are performed with Mach2qtl, a statistical genetics algorithm developed by Goncalo Abecasis and colleagues (Li et al., 2010). Multiple linear regression is performed on each SNP using trait as the dependent variable and allelic dosage (i.e., 0, 1, or 2 alleles) as the independent variable of interest. Sites control for a set of basic nuisance factors, namely the first four MDS components, age, sex, age × sex interaction effects, and nonlinear effects of age, including age2 and age2 × sex, by adding them as covariates in the regression model. Site-specific covariates may also be added (e.g., if data are acquired on two different scanners). Following quality control, the regression coefficient, standard error, and p-value for each SNP are forwarded to the coordinating site which conducts a unifying metaanalysis that weights the SNP coefficients by their standard error. This approach circumvents barriers associated with data sharing across sites and countries and allows sites to maintain responsibility for the integrity of their data. The meta-analysis is performed with an inverse standard error-weighted meta-analysis protocol implemented in METAL (Willer et al., 2010). Genomic control of p-values undertaken at the site level is repeated on the output of the meta-analysis to provide an additional control for population stratification or cryptic relatedness not accounted for by the MDS components (Devlin and Roeder, 1999). Additionally, associations are verified in replication samples that have been acquired independently of the discovery dataset.
3.1 Disease Working Groups
From the time that the pilot project by Stein et al. was published in April 2012, several ENIGMA working groups have been formed to focus more closely on applying the ENIGMA meta-analysis protocols to case–control differences in various brain-related diseases. With such large studies comes the ability to perform high power association studies to identify biomarkers for monitoring disease state and targets for drug therapies. ENIGMA working groups have been formed to study ADHD, schizophrenia, OCD, HIV, PTSD, major depressive disorder, and bipolar disorder (Jahanshad et al., 2013; Schmaal et al., 2015; van Erp et al., 2015).
4 Enigma Addiction Working Group
The authors of this review have joined together to leverage the structure of the ENIGMA project to study addiction. The international membership represents research laboratories from four continents and nine different time zones (Fig. 2). An initial site survey has identified datasets, including both case/control and cohort studies, that collectively contain neuroimaging and genomic data on over 10,000 subjects. Table 1 provides a summary of the Addiction working group datasets.
Figure 2.
World map of the current membership of the ENIGMA Addiction working group.
Table 1. Summary of ENIGMA Addiction Working Group Datasets as of February 2015.
| Substance | Pattern of Use | Cases | Female Cases | Cases and Controls |
|---|---|---|---|---|
|
| ||||
| Alcohol | Occasional | 150 | 75 | 150 |
| Dependent | 1695 | 560 | 2124 | |
| Abstinent | 61 | 24 | 177 | |
| Nicotine | Occasional | 0 | 0 | 0 |
| Current | 1132 | 385 | 1797 | |
| Abstinent | 10 | 0 | 10 | |
| Cannabis | Occasional | 91 | 30 | 213 |
| Dependent | 238 | 33 | 348 | |
| Abstinent | 17 | 7 | 34 | |
| Stimulants | Occasional | 175 | 69 | 228 |
| Dependent | 906 | 182 | 1408 | |
| Abstinent | 68 | 9 | 108 | |
| Gambling | Occasional | 0 | 0 | 0 |
| Dependent | 59 | 0 | 187 | |
| Abstinent | 0 | 0 | 0 | |
| Heroin | Occasional | 0 | 0 | 0 |
| Dependent | 0 | 0 | 0 | |
| Abstinent | 38 | 15 | 70 | |
| Cohort | – | – | 6445 | |
| Totals | 4640 | 1389 | 13,299 | |
The strengths of the working group are currently found in several large developmental cohorts as well as case/control studies of dependent users of alcohol, nicotine, stimulants (cocaine and methamphetamine), and cannabis.
Each site has committed to completing the site-level analyses through local personnel. ENIGMA provides detailed image analysis protocols that will be adopted at all sites (http://enigma.ini.usc.edu/protocols/imaging-protocols/). The fact that many sites already employ these protocols or very similar processing pipelines will minimize the time required for data preprocessing. Data analysis support will be provided by a postdoctoral associate (S.M.) at the University of Vermont and will also be available from the engineers and analysts in Dr. Thompson's ENIGMA support team. Easy-to-use instructions on how to preprocess the neuroimaging and genomic data and check for data quality have also been prepared.
A multisite genetic neuroimaging meta-analysis will only be successful if careful attention is paid to the assessment of behavioral variables. The experience of the ENIGMA research consortium shows that the pooling of neuroimaging data requires the evaluation, and where possible standardization, of site effects on phenotypic characterizations and brain measures. The chosen phenotypes and brain measures must offer optimal sensitivity to disease effects, clinically relevant modulators of disease, and treatment effects. While each site possesses extensive phenotyping on its research participants, there are important differences across sites in the instruments and questionnaires used. The Addiction working group will develop common measures of quantity and frequency of use derived from the different instruments and assessments obtained at each site. Standardized addiction scores will be generated across the varying developmental and clinical profiles. This approach has been effective in harmonizing measures of alcohol consumption for the purpose of large genetics studies, such as the Gene–Environment Association Studies (GENEVA) consortium. For example, the GENEVA consortium was able to convert disparate alcohol measures into useful categories representing onset and safe compared with unsafe consumption (Holman and English, 1995; Holman et al., 1996). There are many methodological problems associated with measurement heterogeneity for alcohol consumption in the context of genomic studies. These include questions with regard to how abstention should be interpreted, the episodic nature of alcohol consumption, the coding of current drug use state at the time of scanning, the quantity and frequency of substance use across reference periods, differences in cultural norms, the standardization of drinking units, as well as recall and other respondent biases (see review, Agrawal et al., 2012). As recommended by Agrawal et al., the Addiction working group will use the guidelines and where possible attempt to align the addiction-related phenotypes with the NIH PhenX toolkit measures for alcohol and drug consumption (e.g., lifetime use, age at first use, and symptoms of dependence).
4.1 Initial Project
The first analysis will examine the structural correlates of four simple drug use categories, no lifetime use, occasional use, abuse, and dependence. Data related to four substances, i.e., alcohol, nicotine, stimulants (cocaine and methamphetamine), and cannabis, will be used to identify the neural substrates of core addiction processes as well as substance-specific factors. Performing GWAS on the identified brain regions will significantly reduce the dimensionality of the brain imaging data. This first analysis will establish relationships between the sites and bring to light any major difficulties that need to be addressed. Analysis will begin after the first data freeze which will occur at the beginning of May 2015. Likely, the most important early challenge for the group will be the development of an assessment instrument that harmonizes the different drug use measures at the various sites.
5 Summary and Future Directions
Several international consortia have been organized in recent years to improve the statistical power of genetic imaging association analyses by pooling data from multiple sites. The authors of this review have formed an Addiction working group within the framework of the ENIGMA project to leverage the acquired knowledge about data sharing across multiple sites to study the genetic and neurobiological mechanisms underlying addiction. The ENIGMA Addiction working group will attempt to identify brain endophenotypes starting with a volumetric investigation of the core neural substrates of addiction. The identification of core brain regions using structural MRI will reduce the number of dimensions in subsequent genomic analyses of problematic substance use. The Addiction working group will adopt the meta-analytic methods used successfully by the ENIGMA project. However, a mega-analysis approach, i.e., analysis of all pooled raw data at one location, may offer opportunities to conduct in-depth examinations of the neurobiology of drug use that are not possible in a meta-analysis. While practical concerns about sharing data were part of the motivation for the meta-analysis approach used by ENIGMA, more sensitive analyses may be possible by going beyond the pooling of effect sizes and the sharing of summary statistics (e.g., volume measurements of specific cortical and subcortical structures) to the sharing of complete, fully anonymized datasets, where available. We believe that the obstacles to this level of data sharing are surmountable. Depending on how the consortium grows (i.e., the addition of new members and of new datasets from current members), the Addiction working may in also decide to include multi-modal assessments of brain function including task-related and resting-state fMRI, DTI, and EEG (e.g., Jahanshad et al., 2015; Kochunov et al., 2015). At the present time, the working group is focused on resolving problems related to multisite data pooling with a manageable number of 18 sites.
Recent advances in the statistical analysis of genomic data present several promising new ways to investigate the combined datasets. We will explore the application of genome-wide complex trait analyses (Yang et al., 2011) to assess the heritability and genetic correlations among brain regions and phenotypic measures associated with alcohol and drug use. This method produces estimates of the variance explained by all SNPs over the whole genome for a complex trait and is suitable for large samples of nonrelated subjects. The working group will also investigate emerging statistical methods to detect significant associations in high dimensional data, such as the parallel independent components analysis with a reference mask (Liu et al., 2012), meta-analysis of voxel-based data (Jahanshad et al., 2015), and novel applications of correspondence analysis (Cioli et al., 2014).
5.1 Addiction Medicine
There are multiple ways in which the progress of the working group could impact the practice of addiction medicine. Since there is strong evidence that addiction has a genetic component (Maes et al., 2004; Prescott and Kendler, 1999; Tsuang et al., 1998), a GWAS with sufficient power, such as the one envisaged by the working group, will likely detect novel genetic associations with behavioral features of addiction or with intermediate brain phenotypes. Not only will these novel associations drive future research aimed at understanding the neural processes involved in problematic substance use and potentially provide novel targets for pharmacological intervention, but they could also lead to the development of predictive genetic and neuroimaging biomarkers. Addiction medicine would benefit enormously from a set of predictive tools that could be used to estimate risk at various stages of the disorder, e.g., risk of transition from healthy to problematic patterns of use or risk of relapse after treatment (Paulus, 2015). Current research also points toward a heterogeneity of causes (Tsuang et al., 1998). If addictive behavior can be attributed to many small effects in a range of brain systems, it is possible that combined neuroimaging and genetic testing could identify differential vulnerabilities which could be used to customize treatment to address the specific challenges of the individual patient.
Acknowledgments
This work was supported by a National Institute on Drug Abuse (NIDA) Grant 1R21DA038381 and by a National Institutes of Health (NIH) Grant U54 EB 020403 with funds provided for the trans-NIH Big Data to Knowledge (BD2K) initiative. Support was also provided by an NIH Grant 1P20GM103644-01A1 awarded to the Vermont Center on Behavior and Health.
References
- Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, Neuman R, Howells W, Fisher S, Fox L, Cloninger R, Dick DM, Doheny KF, Edenberg HJ, Goate AM, Hesselbrock V, Johnson E, Kramer J, Kuperman S, Nurnberger JI, Jr, Pugh E, Schuckit M, Tischfield J, GENEVA Consortium. Rice JP, Bucholz KK, Bierut LJ. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol. 2011;16:514–518. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Freedman ND, Cheng YC, Lin P, Shaffer JR, Sun Q, Taylor K, Yaspan B, Cole JW, Cornelis MC, DeSensi RS, Fitzpatrick A, Heiss G, Kang JH, O'Connell J, Bennett S, Bookman E, Bucholz KK, Caporaso N, Crout R, Dick DM, Edenberg HJ, Goate A, Hesselbrock V, Kittner S, Kramer J, Nurnberger JI, Jr, Qi L, Rice JP, Schuckit M, van Dam RM, Boerwinkle E, Hu F, Levy S, Marazita M, Mitchell BD, Pasquale LR, Bierut LJ GENEVA Consortium. Measuring alcohol consumption for genomic meta-analyses of alcohol intake: opportunities and challenges. Am J Clin Nutr. 2012;95:539–547. doi: 10.3945/ajcn.111.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alia-Klein N, Parvaz MA, Woicik PA, Konova AB, Maloney T, Shumay E, Wang R, Telang F, Biegon A, Wang GJ, Fowler JS, Tomasi D, Volkow ND, Goldstein RZ. Gene x disease interaction on orbitofrontal gray matter in cocaine addiction. Arch Gen Psychiatry. 2011;68:283–294. doi: 10.1001/archgenpsychiatry.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbrook DG, Williams RW, Lu L, Stein JL, Hibar DP, Nichols TE, Medland SE, Thompson PM, Hager R. Joint genetic analysis of hippocampal size in mouse and human identifies a novel gene linked to neurodegenerative disease. BMC Genomics. 2014;15:850. doi: 10.1186/1471-2164-15-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros-Loscertales A, Garavan H, Bustamante JC, Ventura-Campos N, Llopis JJ, Belloch V, Parcet MA, Avila C. Reduced striatal volume in cocaine-dependent patients. Neuroimage. 2011;56:1021–1026. doi: 10.1016/j.neuroimage.2011.02.035. [DOI] [PubMed] [Google Scholar]
- Batalla A, Bhattacharyya S, Yucel M, Fusar-Poli P, Crippa JA, Nogue S, Torrens M, Pujol J, Farre M, Martin-Santos R. Structural and functional imaging studies in chronic cannabis users: a systematic review of adolescent and adult findings. PLoS One. 2013;8:e55821. doi: 10.1371/journal.pone.0055821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin D, Deroche-Gamonet V. Responses to novelty and vulnerability to cocaine addiction: contribution of a multi-symptomatic animal model. Cold Spring Harb Perspect Med. 2012;2:a011940. doi: 10.1101/cshperspect.a011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenu OJ, Davydow DS, Kendler KS. Psychiatric ‘diseases’ versus behavioral disorders and degree of genetic influence. Psychol Med. 2011;41:33–40. doi: 10.1017/S003329171000084X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP Gene Environment Association Studies Consortium. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brener ND, Collins JL, Kann L, Warren CW, Williams BI. Reliability of the Youth Risk Behavior Survey Questionnaire. Am J Epidemiol. 1995;141:575–580. doi: 10.1093/oxfordjournals.aje.a117473. [DOI] [PubMed] [Google Scholar]
- Cardenas VA, Studholme C, Gazdzinski S, Durazzo TC, Meyerhoff DJ. Deformation-based morphometry of brain changes in alcohol dependence and abstinence. Neuroimage. 2007;34:879–887. doi: 10.1016/j.neuroimage.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioli C, Abdi H, Beaton D, Burnod Y, Mesmoudi S. Differences in human cortical gene expression match the temporal properties of large-scale functional networks. PLoS One. 2014;9:e115913. doi: 10.1371/journal.pone.0115913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly CG, Bell RP, Foxe JJ, Garavan H. Dissociated grey matter changes with prolonged addiction and extended abstinence in cocaine users. PLoS One. 2013;8:e59645. doi: 10.1371/journal.pone.0059645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Braber A, Bohlken MM, Brouwer RM, van't Ent D, Kanai R, Kahn RS, de Geus EJ, Hulshoff Pol HE, Boomsma DI. Heritability of subcortical brain measures: a perspective for future genome-wide association studies. Neuroimage. 2013;83:98–102. doi: 10.1016/j.neuroimage.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Drgon T, Montoya I, Johnson C, Liu QR, Walther D, Hamer D, Uhl GR. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Mol Med. 2009;15:21–27. doi: 10.2119/molmed.2008.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersche KD, Jones PS, Williams GB, Turton AJ, Robbins TW, Bullmore ET. Abnormal brain structure implicated in stimulant drug addiction. Science. 2012;335:601–604. doi: 10.1126/science.1214463. [DOI] [PubMed] [Google Scholar]
- Everitt BJ, Robbins TW. From the ventral to the dorsal striatum: devolving views of their roles in drug addiction. Neurosci Biobehav Rev. 2013;37:1946–1954. doi: 10.1016/j.neubiorev.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafo MR. Candidate and non-candidate genes in behavior genetics. Curr Opin Neurobiol. 2013;23:57–61. doi: 10.1016/j.conb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, Zhao H, Farrer LA. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77:493–503. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Ehlers CL. Genome-wide association studies of substance use: considerations regarding populations and phenotypes. Biol Psychiatry. 2015;77:423–424. doi: 10.1016/j.biopsych.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007;28:488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventos H, Blangero J, Gottesman II, Almasy L. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:122–130. doi: 10.1002/ajmg.b.32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Yang BZ, Kranzler HR, Oslin D, Anton R, Farrer LA, Gelernter J. Linkage analysis followed by association show NRG1 associated with cannabis dependence in African Americans. Biol Psychiatry. 2012;72:637–644. doi: 10.1016/j.biopsych.2012.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon CA, Dufault DL, Wesley MJ, Porrino LJ. Elevated gray and white matter densities in cocaine abstainers compared to current users. Psychopharmacology (Berl) 2011;218:681–692. doi: 10.1007/s00213-011-2360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, Aribisala BS, Armstrong NJ, Bernard M, Bohlken MM, Boks MP, Bralten J, Brown AA, Mallar Chakravarty M, Chen Q, Ching CR, Cuellar-Partida G, den Braber A, Giddaluru S, Goldman AL, Grimm O, Guadalupe T, Hass J, Woldehawariat G, Holmes AJ, Hoogman M, Janowitz D, Jia T, Kim S, Klein M, Kraemer B, Lee PH, Olde Loohuis LM, Luciano M, Macare C, Mather KA, Mattheisen M, Milaneschi Y, Nho K, Papmeyer M, Ramasamy A, Risacher SL, Roiz-Santianez R, Rose EJ, Salami A, Samann PG, Schmaal L, Schork AJ, Shin J, Strike LT, Teumer A, van Donkelaar MM, van Eijk KR, Walters RK, Westlye LT, Whelan CD, Winkler AM, Zwiers MP, Alhusaini S, Athanasiu L, Ehrlich S, Hakobjan MM, Hartberg CB, Haukvik UK, Heister AJ, Hoehn D, Kasperaviciute D, Liewald DC, Lopez LM, Makkinje RR, Matarin M, Naber MA, Reese McKay D, Needham M, Nugent AC, Putz B, Royle NA, Shen L, Sprooten E, Trabzuni D, van der Marel SS, van Hulzen KJ, Walton E, Wolf C, Almasy L, Ames D, Arepalli S, Assareh AA, Bastin ME, Brodaty H, Bulayeva KB, Carless MA, Cichon S, Corvin A, Curran JE, Czisch M, de Zubicaray GI, Dillman A, Duggirala R, Dyer TD, Erk S, Fedko IO, Ferrucci L, Foroud TM, Fox PT, Fukunaga M, Raphael Gibbs J, Goring HH, Green RC, Guelfi S, Hansell NK, Hartman CA, Hegenscheid K, Heinz A, Hernandez DG, Heslenfeld DJ, Hoekstra PJ, Holsboer F, Homuth G, Hottenga JJ, Ikeda M, Jack CR, Jr, Jenkinson M, Johnson R, Kanai R, Keil M, Kent JW, Jr, Kochunov P, Kwok JB, Lawrie SM, Liu X, Longo DL, McMahon KL, Meisenzahl E, Melle I, Mohnke S, Montgomery GW, Mostert JC, Muhleisen TW, Nalls MA, Nichols TE, Nilsson LG, Nothen MM, Ohi K, Olvera RL, Perez-Iglesias R, Bruce Pike G, Potkin SG, Reinvang I, Reppermund S, Rietschel M, Romanczuk-Seiferth N, Rosen GD, Rujescu D, Schnell K, Schofield PR, Smith C, Steen VM, Sussmann JE, Thalamuthu A, Toga AW, Traynor BJ, Troncoso J, Turner JA, Valdes Hernandez MC, van't Ent D, van der Brug M, van der Wee NJ, van Tol MJ, Veltman DJ, Wassink TH, Westman E, Zielke RH, Zonderman AB, Ashbrook DG, Hager R, Lu L, McMahon FJ, Morris DW, Williams RW, Brunner HG, Buckner RL, Buitelaar JK, Cahn W, Calhoun VD, Cavalleri GL, Crespo-Facorro B, Dale AM, Davies GE, Delanty N, Depondt C, Djurovic S, Drevets WC, Espeseth T, Gollub RL, Ho BC, Hoffmann W, Hosten N, Kahn RS, Le Hellard S, Meyer-Lindenberg A, Muller-Myhsok B, Nauck M, Nyberg L, Pandolfo M, Penninx BW, Roffman JL, Sisodiya SM, Smoller JW, van Bokhoven H, van Haren NE, Volzke H, Walter H, Weiner MW, Wen W, White T, Agartz I, Andreassen OA, Blangero J, Boomsma DI, Brouwer RM, Cannon DM, Cookson MR, de Geus EJ, Deary IJ, Donohoe G, Fernandez G, Fisher SE, Francks C, Glahn DC, Grabe HJ, Gruber O, Hardy J, Hashimoto R, Hulshoff Pol HE, Jonsson EG, Kloszewska I, Lovestone S, Mattay VS, Mecocci P, McDonald C, McIntosh AM, Ophoff RA, Paus T, Pausova Z, Ryten M, Sachdev PS, Saykin AJ, Simmons A, Singleton A, Soininen H, Wardlaw JM, Weale ME, Weinberger DR, Adams HH, Launer LJ, Seiler S, Schmidt R, Chauhan G, Satizabal CL, Becker JT, Yanek L, van der Lee SJ, Ebling M, Fischl B, Longstreth WT, Jr, Greve D, Schmidt H, Nyquist P, Vinke LN, van Duijn CM, Xue L, Mazoyer B, Bis JC, Gudnason V, Seshadri S, Ikram MA, Alzheimer's Disease Neuroimaging Initiative, CHARGE Consortium, EPI-GEN, IMAGEN, SYS. Martin NG, Wright MJ, Schumann G, Franke B, Thompson PM, Medland SE. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman CD, English DR. An improved aetiologic fraction for alcohol-caused mortality. Aust J Public Health. 1995;19:138–141. doi: 10.1111/j.1753-6405.1995.tb00363.x. [DOI] [PubMed] [Google Scholar]
- Holman CD, English DR, Milne E, Winter MG. Meta-analysis of alcohol and all-cause mortality: a validation of NHMRC recommendations. Med J Aust. 1996;164:141–145. doi: 10.5694/j.1326-5377.1996.tb122011.x. [DOI] [PubMed] [Google Scholar]
- Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, Corley RP, Hewitt JK, Krauter KS, Mikulich-Gilbertson SK, Rhee SH, Smolen A, Young SE, Crowley TJ. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend. 2007;89:34–41. doi: 10.1016/j.drugalcdep.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide JS, Zhang S, Hu S, Sinha R, Mazure CM, Li CS. Cerebral gray matter volumes and low-frequency fluctuation of BOLD signals in cocaine dependence: duration of use and gender difference. Drug Alcohol Depend. 2014;134:51–62. doi: 10.1016/j.drugalcdep.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov P, Armstrong N, Bastin M, Bearden C, Brouwer R, Deary I, Fears S, Franke B, Fullerton J, Hariri A, Hashimoto R, Hellard S, Hibar D, Kelly S, Knickmeyer R, Lemaitre H, McIntosh A, Schumann G, Sprooten E, Roberts G, Pol H, Nyberg L, Wen W, van Hulzen K, Zwiers M, Szeszko P, Nichols T, Alzheimer's Disease Neuroimaging Initiative. Wright M, Haberg A, Thompson P, Glahn D. Organization for Human Brain Mapping (OHBM) Honolulu, Hawaii: 2015. Meta-Analyzing Genome-Wide Associations with White Matter Microstructure—The ENIMGA-DTI Group. [Google Scholar]
- Jahanshad N, Kochunov PV, Sprooten E, Mandl RC, Nichols TE, Almasy L, Blangero J, Brouwer RM, Curran JE, de Zubicaray GI, Duggirala R, Fox PT, Hong LE, Landman BA, Martin NG, McMahon KL, Medland SE, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Hulshoff Pol HE, Bastin ME, McIntosh AM, Deary IJ, Thompson PM, Glahn DC. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. Neuroimage. 2013;81:455–469. doi: 10.1016/j.neuroimage.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan TL, Butters N, DiTraglia G, Schafer K, Smith T, Irwin M, Grant I, Schuckit M, Cermak LS. Reduced cerebral grey matter observed in alcoholics using magnetic resonance imaging. Alcohol Clin Exp Res. 1991;15:418–427. doi: 10.1111/j.1530-0277.1991.tb00540.x. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Marcus D, Winkler A, Sprooten E, Nichols TE, Wright SN, Hong LE, Patel B, Behrens T, Jbabdi S, Andersson J, Lenglet C, Yacoub E, Moeller S, Auerbach E, Ugurbil K, Sotiropoulos SN, Brouwer RM, Landman B, Lemaitre H, den Braber A, Zwiers MP, Ritchie S, van Hulzen K, Almasy L, Curran J, de Zubicaray GI, Duggirala R, Fox P, Martin NG, McMahon KL, Mitchell B, Olvera RL, Peterson C, Starr J, Sussmann J, Wardlaw J, Wright M, Boomsma DI, Kahn R, de Geus EJ, Williamson DE, Hariri A, van't Ent D, Bastin ME, McIntosh A, Deary IJ, Hulshoff Pol HE, Blangero J, Thompson PM, Glahn DC, Van Essen DC. Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage. 2015;111:300–311. doi: 10.1016/j.neuroimage.2015.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ, Pacheco J, Perry ME, Stevens A, Schmitt JE, Grant MD, Seidman LJ, Thermenos HW, Tsuang MT, Eisen SA, Dale AM, Fennema-Notestine C. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Schubert F, Gallinat J. Reduced thickness of medial orbitofrontal cortex in smokers. Biol Psychiatry. 2010;68:1061–1065. doi: 10.1016/j.biopsych.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ghassemi MM, Michael AM, Boutte D, Wells W, Perrone-Bizzozero N, Macciardi F, Mathalon DH, Ford JM, Potkin SG, Turner JA, Calhoun VD. An ICA with reference approach in identification of genetic variation and associated brain networks. Front Hum Neurosci. 2012;6:21. doi: 10.3389/fnhum.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti V, Solowij N, Fornito A, Lubman DI, Yucel M. The association between regular cannabis exposure and alterations of human brain morphology: an updated review of the literature. Curr Pharm Des. 2014;20:2138–2167. doi: 10.2174/13816128113199990435. [DOI] [PubMed] [Google Scholar]
- Lyoo IK, Pollack MH, Silveri MM, Ahn KH, Diaz CI, Hwang J, Kim SJ, Yurgelun-Todd DA, Kaufman MJ, Renshaw PF. Prefrontal and temporal gray matter density decreases in opiate dependence. Psychopharmacology (Berl) 2006;184:139–144. doi: 10.1007/s00213-005-0198-x. [DOI] [PubMed] [Google Scholar]
- Mackey S, Paulus M. Are there volumetric brain differences associated with the use of cocaine and amphetamine-type stimulants? Neurosci Biobehav Rev. 2013;37:300–316. doi: 10.1016/j.neubiorev.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, Kendler KS. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34:1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Matochik JA, London ED, Eldreth DA, Cadet JL, Bolla KI. Frontal cortical tissue composition in abstinent cocaine abusers: a magnetic resonance imaging study. Neuroimage. 2003;19:1095–1102. doi: 10.1016/s1053-8119(03)00244-1. [DOI] [PubMed] [Google Scholar]
- Montigny C, Castellanos-Ryan N, Whelan R, Banaschewski T, Barker GJ, Buchel C, Gallinat J, Flor H, Mann K, Paillere-Martinot ML, Nees F, Lathrop M, Loth E, Paus T, Pausova Z, Rietschel M, Schumann G, Smolka MN, Struve M, Robbins TW, Garavan H, Conrod PJ, Consortium I. A phenotypic structure and neural correlates of compulsive behaviors in adolescents. PLoS One. 2013;8:e80151. doi: 10.1371/journal.pone.0080151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus MP. Pragmatism instead of mechanism: a call for impactful biological psychiatry. JAMA Psychiatry. 2015;72:631–632. doi: 10.1001/jamapsychiatry.2015.0497. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Rando K, Hong KI, Bhagwagar Z, Li CS, Bergquist K, Guarnaccia J, Sinha R. Association of frontal and posterior cortical gray matter volume with time to alcohol relapse: a prospective study. Am J Psychiatry. 2011;168:183–192. doi: 10.1176/appi.ajp.2010.10020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J. The genetics of alcohol dependence. Ann N Y Acad Sci. 2013;1282:39–70. doi: 10.1111/j.1749-6632.2012.06794.x. [DOI] [PubMed] [Google Scholar]
- Schacht JP, Hutchison KE, Filbey FM. Associations between cannabinoid receptor-1 (CNR1) variation and hippocampus and amygdala volumes in heavy cannabis users. Neuropsychopharmacology. 2012;37:2368–2376. doi: 10.1038/npp.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L, Veltman DJ, van Erp TG, Samann PG, Frodl T, Jahanshad N, Loehrer E, Tiemeier H, Hofman A, Niessen WJ, Vernooij MW, Ikram MA, Wittfeld K, Grabe HJ, Block A, Hegenscheid K, Volzke H, Hoehn D, Czisch M, Lagopoulos J, Hatton SN, Hickie IB, Goya-Maldonado R, Kramer B, Gruber O, Couvy-Duchesne B, Renteria ME, Strike LT, Mills NT, de Zubicaray GI, McMahon KL, Medland SE, Martin NG, Gillespie NA, Wright MJ, Hall GB, MacQueen GM, Frey EM, Carballedo A, van Velzen LS, van Tol MJ, van der Wee NJ, Veer IM, Walter H, Schnell K, Schramm E, Normann C, Schoepf D, Konrad C, Zurowski B, Nickson T, McIntosh AM, Papmeyer M, Whalley HC, Sussmann JE, Godlewska BR, Cowen PJ, Fischer FH, Rose M, Penninx BW, Thompson PM, Hibar DP. Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol psychiatry. 2015 doi: 10.1038/mp.2015.69. http://dx.doi.org/10.1038/mp.2015.69. [DOI] [PMC free article] [PubMed]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, Domin M, Grimm O, Hollinshead M, Holmes AJ, Homuth G, Hottenga JJ, Langan C, Lopez LM, Hansell NK, Hwang KS, Kim S, Laje G, Lee PH, Liu X, Loth E, Lourdusamy A, Mattingsdal M, Mohnke S, Maniega SM, Nho K, Nugent AC, O'Brien C, Papmeyer M, Putz B, Ramasamy A, Rasmussen J, Rijpkema M, Risacher SL, Roddey JC, Rose EJ, Ryten M, Shen L, Sprooten E, Strengman E, Teumer A, Trabzuni D, Turner J, van Eijk K, van Erp TG, van Tol MJ, Wittfeld K, Wolf C, Woudstra S, Aleman A, Alhusaini S, Almasy L, Binder EB, Brohawn DG, Cantor RM, Carless MA, Corvin A, Czisch M, Curran JE, Davies G, de Almeida MA, Delanty N, Depondt C, Duggirala R, Dyer TD, Erk S, Fagerness J, Fox PT, Freimer NB, Gill M, Goring HH, Hagler DJ, Hoehn D, Holsboer F, Hoogman M, Hosten N, Jahanshad N, Johnson MP, Kasperaviciute D, Kent JW, Jr, Kochunov P, Lancaster JL, Lawrie SM, Liewald DC, Mandl R, Matarin M, Mattheisen M, Meisenzahl E, Melle I, Moses EK, Muhleisen TW, Nauck M, Nothen MM, Olvera RL, Pandolfo M, Pike GB, Puls R, Reinvang I, Renteria ME, Rietschel M, Roffman JL, Royle NA, Rujescu D, Savitz J, Schnack HG, Schnell K, Seiferth N, Smith C, Steen VM, Valdes Hernandez MC, Van den Heuvel M, van der Wee NJ, Van Haren NE, Veltman JA, Volzke H, Walker R, Westlye LT, Whelan CD, Agartz I, Boomsma DI, Cavalleri GL, Dale AM, Djurovic S, Drevets WC, Hagoort P, Hall J, Heinz A, Jack CR, Jr, Foroud TM, Le Hellard S, Macciardi F, Montgomery GW, Poline JB, Porteous DJ, Sisodiya SM, Starr JM, Sussmann J, Toga AW, Veltman DJ, Walter H, Weiner MW, Alzheimer's Disease Neuro imaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study Group. Bis JC, Ikram MA, Smith AV, Gudnason V, Tzourio C, Vernooij MW, Launer LJ, DeCarli C, Seshadri S, Cohortsfor H, Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Andreassen OA, Apostolova LG, Bastin ME, Blangero J, Brunner HG, Buckner RL, Cichon S, Coppola G, de Zubicaray GI, Deary IJ, Donohoe G, de Geus EJ, Espeseth T, Fernandez G, Glahn DC, Grabe HJ, Hardy J, Hulshoff Pol HE, Jenkinson M, Kahn RS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meyer-Lindenberg A, Morris DW, Muller-Myhsok B, Nichols TE, Ophoff RA, Paus T, Pausova Z, Penninx BW, Potkin SG, Samann PG, Saykin AJ, Schumann G, Smoller JW, Wardlaw JM, Weale ME, Martin NG, Franke B, Wright MJ, Thompson PM Enhancing Neuro Imaging Genetics through Meta Analysis Consortium. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MT, Carroll AJ, Salmeron BJ, Ross TJ, Stein EA. Insula's functional connectivity with ventromedial prefrontal cortex mediates the impact of trait alexithymia on state tobacco craving. Psychopharmacology (Berl) 2013;228:143–155. doi: 10.1007/s00213-013-3018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B, Wright MJ, Martin NG, Agartz I, Alda M, Alhusaini S, Almasy L, Almeida J, Alpert K, Andreasen NC, Andreassen OA, Apostolova LG, Appel K, Armstrong NJ, Aribisala B, Bastin ME, Bauer M, Bearden CE, Bergmann O, Binder EB, Blangero J, Bockholt HJ, Boen E, Bois C, Boomsma DI, Booth T, Bowman IJ, Bralten J, Brouwer RM, Brunner HG, Brohawn DG, Buckner RL, Buitelaar J, Bulayeva K, Bustillo JR, Calhoun VD, Cannon DM, Cantor RM, Carless MA, Caseras X, Cavalleri GL, Chakravarty MM, Chang KD, Ching CR, Christoforou A, Cichon S, Clark VP, Conrod P, Coppola G, Crespo-Facorro B, Curran JE, Czisch M, Deary IJ, de Geus EJ, den Braber A, Delvecchio G, Depondt C, de Haan L, de Zubicaray GI, Dima D, Dimitrova R, Djurovic S, Dong H, Donohoe G, Duggirala R, Dyer TD, Ehrlich S, Ekman CJ, Elvsashagen T, Emsell L, Erk S, Espeseth T, Fagerness J, Fears S, Fedko I, Fernandez G, Fisher SE, Foroud T, Fox PT, Francks C, Frangou S, Frey EM, Frodl T, Frouin V, Garavan H, Giddaluru S, Glahn DC, Godlewska B, Goldstein RZ, Gollub RL, Grabe HJ, Grimm O, Gruber O, Guadalupe T, Gur RE, Gur RC, Goring HH, Hagenaars S, Hajek T, Hall GB, Hall J, Hardy J, Hartman CA, Hass J, Hatton SN, Haukvik UK, Hegenscheid K, Heinz A, Hickie IB, Ho BC, Hoehn D, Hoekstra PJ, Hollinshead M, Holmes AJ, Homuth G, Hoogman M, Hong LE, Hosten N, Hottenga JJ, Hulshoff Pol HE, Hwang KS, Jack CR, Jr, Jenkinson M, Johnston C, Jonsson EG, Kahn RS, Kasperaviciute D, Kelly S, Kim S, Kochunov P, Koenders L, Kramer B, Kwok JB, Lagopoulos J, Laje G, Landen M, Landman BA, Lauriello J, Lawrie SM, Lee PH, Le Hellard S, Lemaitre H, Leonardo CD, Li CS, Liberg B, Liewald DC, Liu X, Lopez LM, Loth E, Lourdusamy A, Luciano M, Macciardi F, Machielsen MW, Macqueen GM, Malt UF, Mandl R, Manoach DS, Martinot JL, Matarin M, Mather KA, Mattheisen M, Mattingsdal M, Meyer-Lindenberg A, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meisenzahl E, Melle I, Milaneschi Y, Mohnke S, Montgomery GW, Morris DW, Moses EK, Mueller BA, Munoz Maniega S, Muhleisen TW, Muller-Myhsok B, Mwangi B, Nauck M, Nho K, Nichols TE, Nilsson LG, Nugent AC, Nyberg L, Olvera RL, Oosterlaan J, Ophoff RA, Pandolfo M, Papalampropoulou-Tsiridou M, Papmeyer M, Paus T, Pausova Z, Pearlson GD, Penninx BW, Peterson CP, Pfennig A, Phillips M, Pike GB, Poline JB, Potkin SG, Putz B, Ramasamy A, Rasmussen J, Rietschel M, Rijpkema M, Risacher SL, Roffman JL, Roiz-Santianez R, Romanczuk-Seiferth N, Rose EJ, Royle NA, Rujescu D, Ryten M, Sachdev PS, Salami A, Satterthwaite TD, Savitz J, Saykin AJ, Scanlon C, Schmaal L, Schnack HG, Schork AJ, Schulz SC, Schur R, Seidman L, Shen L, Shoemaker JM, Simmons A, Sisodiya SM, Smith C, Smoller JW, Soares JC, Sponheim SR, Sprooten E, Starr JM, Steen VM, Strakowski S, Strike L, Sussmann J, Samann PG, Teumer A, Toga AW, Tordesillas-Gutierrez D, Trabzuni D, Trost S, Turner J, Van den Heuvel M, van der Wee NJ, van Eijk K, van Erp TG, van Haren NE, van't Ent D, van Tol MJ, Valdes Hernandez MC, Veltman DJ, Versace A, Volzke H, Walker R, Walter H, Wang L, Wardlaw JM, Weale ME, Weiner MW, Wen W, Westlye LT, Whalley HC, Whelan CD, White T, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Zilles D, Zwiers MP, Thalamuthu A, Schofield PR, Freimer NB, Lawrence NS, Drevets W Alzheimer's Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, Komiyama T, Harano M, Sekine Y, Inada T, Ozaki N, Iyo M, Iwata N, Yamada M, Sora I, Chen CK, Liu HC, Ujike H, Lin SK. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008a;65:345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008b;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay J, Maleki N, Potter J, Elman I, Rudrauf D, Knudsen J, Wallin D, Pendse G, McDonald L, Griffin M, Anderson J, Nutile L, Renshaw P, Weiss R, Becerra L, Borsook D. Alterations in brain structure and functional connectivity in prescription opioid-dependent patients. Brain. 2010;133:2098–2114. doi: 10.1093/brain/awq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, Agartz I, Westlye LT, Haukvik UK, Dale AM, Melle I, Hartberg CB, Gruber O, Kraemer B, Zilles D, Donohoe G, Kelly S, McDonald C, Morris DW, Cannon DM, Corvin A, Machielsen MW, Koenders L, de Haan L, Veltman DJ, Satterthwaite TD, Wolf DH, Gur RC, Gur RE, Potkin SG, Mathalon DH, Mueller BA, Preda A, Macciardi F, Ehrlich S, Walton E, Hass J, Calhoun VD, Bockholt HJ, Sponheim SR, Shoemaker JM, van Haren NE, Pol HE, Ophoff RA, Kahn RS, Roiz-Santianez R, Crespo-Facorro B, Wang L, Alpert KI, Jonsson EG, Dimitrova R, Bois C, Whalley HC, McIntosh AM, Lawrie SM, Hashimoto R, Thompson PM, Turner JA. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.118. http://dx.doi.org/10.1038/mp.2015.63. [DOI] [PMC free article] [PubMed]
- van Soelen IL, Brouwer RM, Peper JS, van Leeuwen M, Koenis MM, van Beijsterveldt TC, Swagerman SC, Kahn RS, Hulshoff Pol HE, Boomsma DI. Brain SCALE: brain structure and cognition: an adolescent longitudinal twin study into the genetic etiology of individual differences. Twin Res Hum Genet. 2012;15:453–467. doi: 10.1017/thg.2012.4. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Zeng M. ANAPC1 and SLCO3A1 are associated with nicotine dependence: meta-analysis of genome-wide association studies. Drug Alcohol Depend. 2012a;124:325–332. doi: 10.1016/j.drugalcdep.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Wang X, Li B, Zhou X, Liao Y, Tang J, Liu T, Hu D, Hao W. Changes in brain gray matter in abstinent heroin addicts. Drug Alcohol Depend. 2012b;126:304–308. doi: 10.1016/j.drugalcdep.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel M, Solowij N, Respondek C, Whittle S, Fornito A, Pantelis C, Lubman DI. Regional brain abnormalities associated with long-term heavy cannabis use. Arch Gen Psychiatry. 2008;65:694–701. doi: 10.1001/archpsyc.65.6.694. [DOI] [PubMed] [Google Scholar]
- Zhang X, Salmeron BJ, Ross TJ, Geng X, Yang Y, Stein EA. Factors underlying prefrontal and insula structural alterations in smokers. Neuroimage. 2011;54:42–48. doi: 10.1016/j.neuroimage.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res. 2013;37:730–739. doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]


