Abstract
The scientific literature teems with clique-centric clustering strategies. In this paper we analyze one such method, the paraclique algorithm. Paraclique has found practical utility in a variety of application domains, and has been successfully employed to reduce the effects of noise. Nevertheless, its formal analysis and worst-case guarantees have remained elusive. We address this issue by deriving a series of lower bounds on paraclique densities.
Keywords: clique, paraclique, graph density, clustering
1. Introduction
Clique-centric methods have long played an important role in data science and engineering. Classic techniques include algorithms for 𝒩𝒫-hard problems such as maximal clique [1] and maximum clique [2]. The availability of high-throughput data has prompted interest in noise-abatement relaxations, most notably k-clique communities [3] (more recently also called clique percolation) and paraclique [4]. These algorithms have been used for biological data clustering, and been found superior to traditional methods [5]. Although similar in objective, k-clique communities is hampered in practice by its bottom up approach relying on an exhaustive enumeration of maximal cliques. Paraclique, in contrast, applies top down design principles and employs maximum clique, for which there are highly efficient and reasonably scalable algorithms [6], plus viable alternatives based on duality and parameterized complexity [7].
Paraclique can be formulated in a variety of ways. The general idea is to expand a maximum clique by augmenting it with non-clique vertices adjacent to most, but not all, members of the clique. The motivation for deriving dense subgraphs in this fashion is based on the fact that so-called “missing” edges, while relevant, are often lost due to noise, improper thresholding, weak experimental design, and numerous other causes. A classic example of this phenomenon can be found in the use of DNA microarrays for transcriptomic data analysis. In this setting, vertices represent genes, edges signify co-expression, and paracliques denote molecular response networks differentially (in)activated by stimulus [4]. Depending on a variety of factors, most but not all network elements may be highly intercorrelated at any particular time.
Previous paraclique studies have focused mainly on practical results. Representative examples include [8, 9, 10]. Instead, our primary goal in this paper is to investigate paraclique’s theoretical basis. In so doing, we seek to derive bounds on its worst-case behavior, applying density as the classic clustering metric (we compute a subgraph’s density in the traditional way, as the number of edges present divided by the maximum number possible). In the original paraclique formulation, the total number of missing edges was left unchecked. Density could, therefore, in principle be driven to zero. By limiting paraclique size to at most twice the maximum clique size, however, and by requiring that a new non-clique vertex be adjacent to all but one vertex in the growing paraclique, it is known that density is maintained at no less than 50% [4]. Here, we greatly expand upon such density results.
In the next section, we formalize definitions, describe relevant background, and establish several helpful preliminary results. In Sections 3 and 4, we derive bounds on general and special cases, respectively. In a final section we draw conclusions and discuss directions for future research.
2. Preliminaries
Let G denote a finite, simple, undirected graph. A clique is a subgraph of G in which every pair of vertices is connected by an edge. A paraclique, P, is constructed by first finding a clique C of maximum size, then glomming onto non-clique vertices in a controlled fashion. An integer glom term, g, is used to accomplish this. In the original algorithmic formulation [4], a non-clique vertex was chosen if and only if it was adjacent to g or more vertices in P. The number of required adjacencies does not scale with the size of P using this approach, however, so we generally invert this comparison. Thus, we glom onto a non-clique vertex if and only if it is adjacent to all but at most g vertices in P. In applications, g is usually some small value. In any case we insist that 0 < g < k, where k denotes the number of vertices in C.
Pseudocode for the paraclique procedure is displayed in Algorithm 1. A sample paraclique construction is illustrated in Figure 1. For the reader’s convenience, definitions employed in the sequel are summarized in Table 1.
Algorithm 1.
The Paraclique Algorithm
| input : graph G, glom term g |
| output: paraclique P, a subgraph of G |
| C ← maximum clique of G |
| V ← vertex set of C |
| while V̅ contains a vertex v adjacent to all but at most g vertices |
| in V do |
| | V ← V ∪ {v} |
| end |
| P ← subgraph induced by V |
| return P |
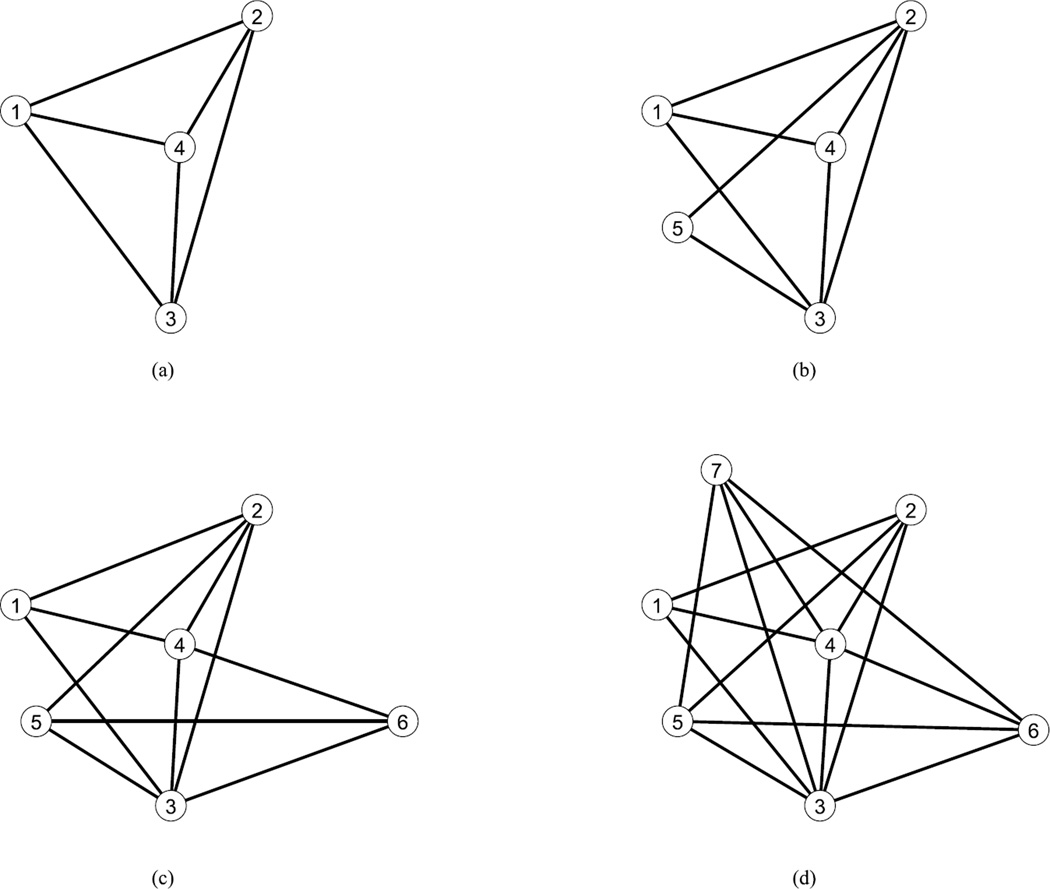
Figure 1.
An example paraclique grown with glom term g = 2 from an initial maximum clique of size four {1,2,3,4} (a) and successively adding vertices 5 (b), 6 (c) and 7 (d).
Table 1.
Definitions used in this paper
| term | meaning |
|---|---|
| G | a finite, simple, undirected graph |
| C | a maximum clique in G |
| k | the number of vertices in C |
| P | a paraclique as produced by Algorithm 1 |
| p | the number of vertices in P |
| g | the glom term used in Algorithm 1 |
| d(P) | the density of P |
We start by establishing lower and upper bounds on maximum paraclique size.
Lemma 1
A paraclique may contain as many as (g + 1)k vertices.
Proof
To construct a paraclique P that satisfies this bound, we begin with g+1 disjoint cliques of size k, denoting them C0, C1, …, Cg, and labeling the vertices of each Ci as υik, υik+1, …, υ(i+1)k−1. To this we add edges connecting vertices υr and υs provided they are in different cliques and r ≢ s modulo k. The maximum clique size has not changed, since any set of k+1 vertices will contain at least two whose indices are in the same equivalance class modulo k (and are thus non-adjacent). Given a graph containing this structure, the paraclique algorithm may return P because υi is adjacent to all but at most g lower-indexed vertices for any 0 < i ≤ (g + 1)k − 1.
Lemma 2
A paraclique cannot contain more than 2gk vertices.
Proof
Let P denote a paraclique of size p > k. By construction, the number of edges in P is at least
Since P has no clique of size k +1, we know by Turán’s Theorem [11] that it contains at most edges. Combining the two edge counts produces
Because p > k, we conclude that p ≤ 2gk.
3. General Case
Let us suppose that C has been isolated, and that g has been chosen. By comparing g and p, we will now prove that as P grows its density approaches 1.0. On the other hand, by comparing g and k, we will also prove that no matter how P changes its density never falls below 0.5.
Theorem 3
A paraclique’s density is at least .
Proof
As we have previously shown, the number of edges in P is at least
Combining that with Lemma 2 ensures
Theorem 4
A paraclique’s density is at least .
Proof
P is missing at most g(p−k) edges. From this it follows that d(P) is bounded below by
Using basic calculus plus the fact that p ≥ k, we see that this function takes its minimum on the interval [k, 2gk] at . Because for all k ≥ 2, d(P) is minimized when p is either 2k − 1 or 2k. We know from Lemma 1 that both values are possible, and in either case we find that d(P) is at least
Sometimes Theorem 3 provides the better guarantee. This happens, for example, when k = 6, g = 2 and p = 20. At other times, say when k = 5, g = 4 and p = 10, Theorem 4 provides the tighter result. In any event, the following overall lower bound on density is obtained from Theorem 4 coupled with the fact that 1 ≤ g < k.
Corollary 5
A paraclique’s density always exceeds 1/2.
4. Special Case
The glom term setting g = 1 is frequently used in practice. Paraclique structure is considerably more scrutable in this special case. See Figure 2. In what follows, we say that P is nontrivial if it does not equal C.
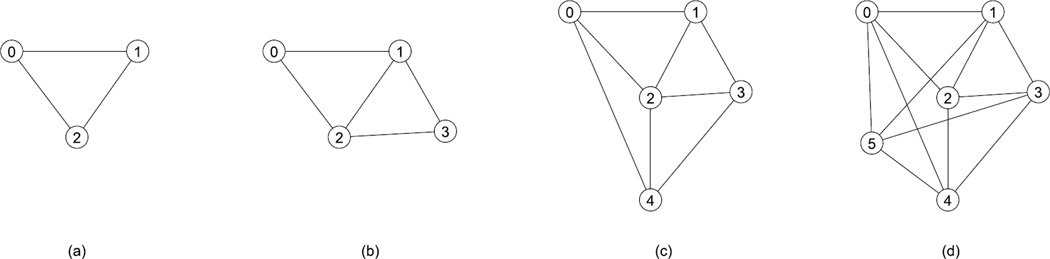
Figure 2.
An example of a paraclique with g=1, starting from maximum clique {0,1,2} (a), and successively adding vertices 3 (b), 4 (c), 5 (d).
Theorem 6
When g = 1, a nontrivial paraclique’s density lies between and , inclusive.
Proof
Suppose P is such a paraclique. From Lemma 2 and the nontriviality of P we know . P must be missing exactly (p − k) edges, else G would have a clique of size k + 1. The number of edges in P is thus , and so
Given p, this is just a linear function of k with a positive slope. It’s minimum therefore occurs when , ensuring
and its maximum occurs when k = p − 1, ensuring
Theorem 7
When g = 1, a nontrivial paraclique’s density lies between and , inclusive.
Proof
From Lemma 2 and the nontriviality of P we know k + 1 ≤ p ≤ 2k. Again we note that P must be missing exactly (p − k) edges, and so . As in the proof of Theorem 4, we find from basic calculus that this function is minimized at , which occurs at both p = 2k − 1 and p = 2k. It is maximized at when p = k + 1.
Theorem 6 tends to provide a better lower bound when p is at the lower end of its range relative to k, while Theorem 7 tends to produce a better upper bound when p is at the upper end of its range. In any event, the following overall lower bound on density is obtained from Theorem 7 coupled with the fact that C must contain at least one edge.
Corollary 8
When g = 1, a paraclique’s density is always at least 2/3.
5. Conclusions and Directions for Further Research
We have derived density bounds for the paraclique algorithm, a noise-resilient clique-centric technique designed for dense subgraph extraction. To the best of our knowledge, other than the elementary result from [4], these are the first formal density limits for what have come to be popularly known as network community methods. This gives paraclique another potential practical endorsement, in addition to those due to biological enrichment as discussed in [5].
Although we remain primarily concerned with lower bounds, we were able to prove asymptotically tight lower and upper bounds for the special case g = 1. Proving better bounds for the general case remains an elusive open problem. Our lower bounds are not tight for arbitrary g > 1; our only upper bounds are weak because they are inherited from the special case. If formal analysis proves too difficult, and it may well might, then an alternate approach could center on empirical testing. Both real and synthetic data might be employed to estimate average densities and their expected deviations from our worst-case guarantees.
Acknowledgements
This research has been supported by the National Institutes of Health under grants P20MD000516 and R01AA018776.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ronald D. Hagan, Email: rhagan@tennessee.edu.
Michael A. Langston, Email: langston@tennessee.edu.
Kai Wang, Email: kwang11@tennessee.edu.
References
- 1.Bron C, Kerbosch J. Algorithm 457: Finding all cliques of an undirected graph. Commun. ACM. 1973;16:575–577. [Google Scholar]
- 2.Bomze IM, Budinich M, Pardalos PM, Pelillo M. Handbook of Combinatorial Optimization. Kluwer Academic Publishers; 1999. The maximum clique problem; pp. 1–74. [Google Scholar]
- 3.Palla G, Dernyi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 4.Chesler EJ, Langston MA. Combinatorial genetic regulatory network analysis tools for high throughput transcriptomic data. In: Eskin E, Ideker T, Raphael B, Workman C, editors. Systems Biology and Regulatory Genomics, volume 4023 of Lecture Notes in Computer Science. Berlin Heidelberg: Springer; 2006. pp. 150–165. [Google Scholar]
- 5.Jay J, Eblen J, Zhang Y, Benson M, Perkins A, Saxton A, Voy B, Chesler E, Langston M. A systematic comparison of genome-scale clustering algorithms. BMC Bioinformatics. 2012;13:S7. doi: 10.1186/1471-2105-13-S10-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomita E, Akutsu T, Matsunaga T. Efficient algorithms for finding maximum and maximal cliques: Effective tools for bioinformatics. Biomedical Engineering, Trends in Electronics, Communications and Software. 2011;32 [Google Scholar]
- 7.Abu-Khzam FN, Langston MA, Shanbhag P, Symons CT. Scalable parallel algorithms for fpt problems. Algorithmica. 2006;45:269–284. [Google Scholar]
- 8.Eblen JD, Gerling IC, Saxton AM, Wu J, Snoddy JR, Langston MA. Graph Algorithms for Integrated Biological Analysis. with Applications to Type 1 Diabetes Data. 2009:207–222. [Google Scholar]
- 9.Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, York TP, Williams RW, Miles MF. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: Functional and mechanistic implications. PLoS ONE. 2012;7:e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha T, Swanson DJ, Larouche M, Glenn HR, Weeden D, Zhang P, Hamre K, Langston MA, Phillips CA, Song M, Ouyang Z, Chesler EJ, Duvvurru S, Yordanova R, Cui Y, Campbell K, Ricker G, Phillips CR, Homayouni R, Goldowitz DA. Cbgrits: Cerebellar gene regulation in time and space. Developmental Biology. 2015;397:18–30. doi: 10.1016/j.ydbio.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Turán P. Eine extremalaufgabe aus der graphentheorie. Mat. Fiz. Lapok. 1941;48:61. [Google Scholar]