Abstract
Genetic defects of the immune system are referred to as primary immunodeficiencies (PIDs). These immunodeficiencies are clinically and immunologically heterogeneous and, therefore, pose a challenge not only for the clinician but also for the diagnostic immunologist. There are several methodological tools available for evaluation and monitoring of patients with PIDs, and of these tools, flow cytometry has gained prominence, both for phenotyping and functional assays. Flow cytometry allows real-time analysis of cellular composition, cell signaling, and other relevant immunological pathways, providing an accessible tool for rapid diagnostic and prognostic assessment. This minireview provides an overview of the use of flow cytometry in disease-specific diagnosis of PIDs, in addition to other broader applications, which include immune phenotyping and cellular functional measurements.
INTRODUCTION
Primary immunodeficiencies (PIDs) represent more than 300 genetic disorders that affect various components of the immune system (1, 2) and are thus pathologically, clinically, and immunologically heterogeneous. With recent advances in genomics, every year, at least 10 to 15 new genetic defects associated with PIDs are identified (2), making it essential to have nongenetic tests available to analyze the immune system, quantitatively and functionally, to validate new genetic findings and provide additional laboratory data to correlate with the clinical phenotype and genotype. The immunological impact of a monogenic primary immunodeficiency cannot be ascertained by genetic testing alone but requires additional testing, which very often includes flow cytometry. The clinical presentation of a patient is not always directed toward a single genetic defect, and thus, a pregenetic immunological (and other relevant) evaluation is often required to include or eliminate potential genetic candidates. Even when a genetic defect is obvious from the clinical phenotype or family history, basic diagnostic immunological evaluation is almost always performed to ascertain the patient's immune status.
Currently, there are eight broad categories under which PIDs are classified, based on either the immune component affected or the immune/clinical phenotype (2–5). These eight categories are formulated by the International Union of Immunological Societies (IUIS) classification scheme (2, 3, 5) and include PIDs affecting cellular and humoral immunity, combined PIDs with associated or syndromic features, predominantly antibody deficiencies, immune dysregulation PIDs, phagocyte number and function PIDs, innate and intrinsic PIDs, autoinflammatory disorders, and complement disorders. For most of these PIDs, diagnosis at the laboratory level, besides standard biochemical assays and genetic analysis, often involves flow cytometry, in either a disease-specific assessment, or more broadly, in measuring immune phenotype and function.
Flow cytometry represents a methodology that has continuously evolved with the passage of time since its discovery half a century ago (6) with several key technological advances in instrumentation, analysis reagents, and tools in the intervening decades. Flow cytometry has many applications and can be used on virtually on any cellular source—blood, body fluid, tissue, and bone marrow. Flow cytometric assays range from qualitative to quantitative (relative and absolute) and phenotyping to functional, besides being useful for assessing specific protein expression, cell viability, apoptosis and death, cellular interactions and cell enrichment. These characteristics make it an ideal tool for screening, diagnostic and prognostic assays for PIDs. This minireview is divided into four sections, based on the use of flow cytometry in various contexts—disease-specific assessment, functional measurements, cellular phenotyping, and other applications, such as flow-FISH (fluorescence in situ hybridization) for telomere length analysis. This minireview is neither methodological nor comprehensive in scope (there are several other disease-specific, phenotyping, and functional immune-related flow assays that are not covered in this minireview due to space constraints) but rather it provides an overview on the use of flow cytometry in PIDs. The main target audience for this minireview is specialty clinicians who see patients with primary immunodeficiencies fairly routinely and diagnostic laboratory immunologists, who perform and interpret such flow cytometry-based assays. To provide basic information for clinicians who do not typically evaluate PID patients, Table 1 and Table 2 contain broad guidelines on clinical contexts where PID should be considered in the differential diagnosis. All of the tests described within this minireview article are available at at least one or more clinical reference laboratories (academic medical centers and/or commercial) in the United States and Europe. Some of the more esoteric flow tests are less likely to be easily available in developing countries but with dissemination of knowledge and collaboration, this will hopefully be more broadly accessible throughout the globe in the coming decade.
TABLE 1.
An overview of the major clinical phenotypes associated with defects in each of the primary immune system components
| Major clinical phenotypesa associated with defects in: | |||
|---|---|---|---|
| Adaptive immune system (combined PIDs) |
Innate immune system |
||
| B cell defectsb | T cell defectsc | Phagocyte defectsb | Complement defectsd |
| Recurrent bacterial sinopulmonary infections | Failure to thrive (FTT) | Multiple or recurrent soft tissue abscesses, lymphadenitis | Angioedema of face, extremities, and/or GI tract |
| Sepsis (bacterial) | Opportunistic infections (e.g., PJP) | Soft tissue granulomas or infections with catalase-positive organisms or certain fungi (e.g., Aspergillus) | Pyogenic infections |
| Recurrent bacterial sinopulmonary infections or bacterial sepsis with encapsulated organisms | Fungal infections | Improper wound healing | Recurrent or systemic neisserial infections |
| Recurrent or chronic gastroenteritis (e.g., with enteroviruses or Giardia) | Recurrent, severe, or unusual viral infections | Gingivitis and periodontitis, chronic | Atypical hemolytic-uremic syndrome and/or thrombotic microangiopathy |
| Bronchiectasis with no clear cause | Graft-vs-host-type phenotype with elevated liver function tests, chronic GI manifestations (diarrhea) | Ulcerations of the mucosa | |
| Enteroviral meningoencephalitis, usually chronic | Delayed separation of the umbilical cord | ||
PIDs, primary immunodeficiencies: GI, gastrointestinal; PJP, Pneumocystis jirovecii pneumonia.
Found by flow cytometry and genetic testing.
Found by flow cytometry and nongenetic molecular tests to assess thymic function and T cell repertoire diversity and by genetic testing.
Found primarily by serological testing and genetic testing.
TABLE 2.
Evaluation of patients for primary immunodeficienciesa
| Criteria in diagnosis of PIDs | Confounders in diagnosis of PIDs |
|---|---|
| Family history of PID | Large numbers of new genetic defects identified in a relatively short span of time, making it challenging for many specialty practitioners to stay updated |
| Syndromic defect (e.g., ataxia telangiectasia) | Considerable variability in genotype-phenotype correlations |
| Unusual infection (susceptibility to a single microorganism, e.g., Mendelian susceptibility to mycobacterial disease) or recurrent infections | Expanding phenotypic spectrum for known genetic defects |
| Significant autoimmunity (autoimmune cytopenias or multiple organ-specific autoimmunity) | Somatic mosaicism; digenic PIDs (two genetic defects associated with a PID); two-hit hypothesis (underlying monogenic defect that is phenotypically unmasked by infection or other triggering event) |
| Laboratory anomaly (e.g., hypogammaglobulinemia) | Need for functional characterization of new genetic findings; role of epigenetic changes in modulating phenotype, copy number variations (CNV) contributing to phenotype |
| Genetic diagnosis via whole-exome sequencing or whole-genome sequencing | Phenocopies of PIDs (autoantibodies to immunologically relevant proteins that cause clinical conditions phenotypically similar to those caused by known monogenic defects) |
Patients with primary immunodeficiencies (PIDs) may present for evaluation in multiple ways, and the six criteria provided represent some of the possible contexts that should be considered during clinical assessment. There are several confounders that could pose a challenge to the diagnostic evaluation for PIDs, and some of these are presented in this table.
USE OF FLOW CYTOMETRY FOR DISEASE-SPECIFIC DIAGNOSIS OF PIDs
There are several PIDs where it is possible to assess specific protein expression, either as a screening test for phenotype correlation or as a confirmatory test. Four specific examples are provided to discuss the use of specific protein analysis in diagnosis of PIDs: Btk protein expression in X-linked agammaglobulinemia (XLA), CD40 ligand (CD40L) expression in X-linked hyper-IgM syndrome, LRBA (lipopolysaccharide-responsive beige-like anchor) protein expression in LRBA deficiency, and DOCK8 (dedicator of cytokinesis 8) protein expression in DOCK8 deficiency.
(i) X-linked agammaglobulinemia.
XLA is classified under the predominantly antibody deficiencies in the IUIS classification (2) and occurs at a prevalence of ∼1:350,000 to 1:700,000 in males. The immunological characteristics of XLA include profoundly decreased serum immunoglobulins of all isotypes and decreased to absent peripheral B cells (7, 8). Most male patients exhibit symptoms of recurrent bacterial sinopulmonary infections in the first year of life, though there is both a clinical and immunological spectrum observed, depending on the location of the mutation in the BTK gene and effect on protein function (9). Since the Btk protein is expressed intracellularly in B cells, monocytes, and platelets, one or more of these cell subsets can be used for identification of protein. As B cells are significantly reduced or absent in XLA, flow analysis focuses on monocytes and/or platelets (10). The efficiency of the flow cytometry analysis is dependent on the antibody used for intracellular staining, and absent or reduced Btk protein expression is observed in approximately 95% of XLA patients, but 5% may have normal protein expression with abnormal function. Further, the ability to ascertain a mosaic pattern consistent with carrier status in females is also assay and antibody dependent (11). As with other PIDs, XLA also includes the spectrum of classic null mutations and hypomorphic (leaky) defects, and patients with the latter are likely to be diagnosed later in life than the null function patients, due to partially preserved B cell numbers and function (12, 13; R. Pyle, X. Dong, A. Ward, J. B. Hagan, M. van Hee, G. Volcheck, A. Y. Joshi, T. G. Boyce, T. Pozos, L. Hoyt, E. Yousef, S. Bahna, Y. Yilmaz-Demirdag, and R. S. Abraham, unpublished data). It is relevant and useful to perform BTK genetic analysis to identify the specific mutation and correlate it with the phenotype, including Btk protein expression and function for complete genotype-phenotype correlation. Female carriers of XLA are usually asymptomatic unless there is significantly skewed lyonization of the X chromosome (14). BTK genetic analysis is particularly useful in identification of carrier status in female relatives of affected male patients who are of child-bearing age, since the flow cytometric assay is variable in its efficacy in identifying two populations (positive and negative) for protein expression. Therefore, diagnostic evaluation of XLA should include genetic testing once XLA is included in the differential diagnosis, based on clinical phenotype, infection, and family history and/or initial testing of immunoglobulin levels and B cell quantitation, since the Btk flow assay may not be useful in the diagnosis of at least 5% of patients with preserved Btk protein expression. Once genetic testing has been performed and a novel mutation (not previously reported) or variant of uncertain clinical significance (VUS) is identified, then Btk flow cytometry may be valuable in ascertaining whether a genotype-phenotype correlation exists, though several reports indicate that such correlations in XLA are highly variable (15–17).
(ii) X-linked hyper-IgM syndrome (CD40L deficiency).
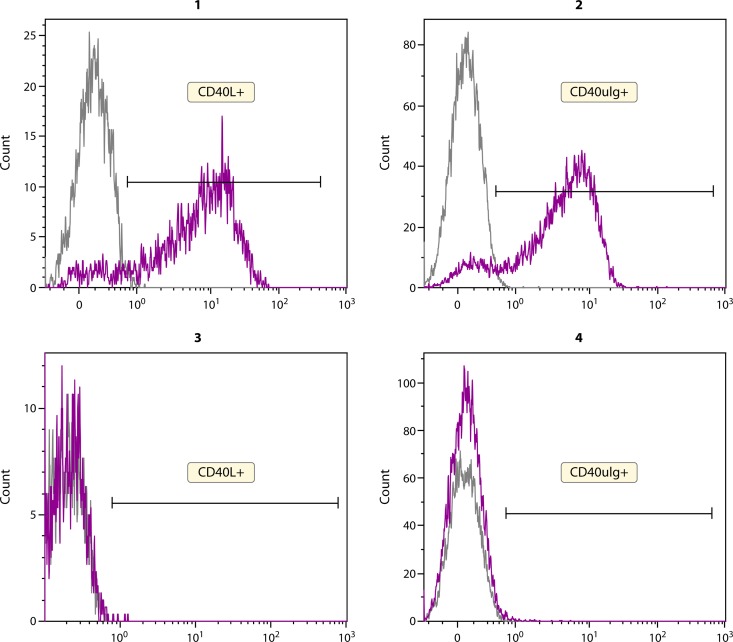
CD40L deficiency represents one of the genetic defects associated with the phenotype of hyper-IgM syndrome and accounts for 70% of such cases (18–23). This disease is caused by mutations in the CD40LG gene, encoding the CD40 ligand protein, a member of the tumor necrosis factor (TNF) family, which is expressed on activated CD4+ T cells. The clinical characteristics of this deficiency include severely impaired production of class-switched immunoglobulins, IgG and IgA with normal or elevated levels of IgM due to impaired class switch recombination (CSR). These patients have both a humoral defect due to the low switched immunoglobulin production and susceptibility to respiratory bacterial infections. However, they also have a cellular defect, due to the importance of CD40L-CD40 interactions for T cell costimulation and activation (24–26), as a result of which, they may present with opportunistic infections, such as Pneumocystis jirovecii, Cryptosporidium, Toxoplasma, etc. Whole blood, which contains T cells, is activated for a few hours to overnight with a mitogen, either phytohemagglutinin (PHA) or phorbol myristate acetate (PMA), following which flow cytometric analysis is performed to examine expression of CD40L (CD154) (Fig. 1). It is appropriate and necessary to include a marker (as a control) for T cell activation, such as CD69 or CD25, when performing the assay. This flow assay can identify ∼80% of CD40L-deficient patients who lack protein expression on the cell surface. However, it will miss ∼20% of patients who have normal protein expression but aberrant function. A valuable addition to the surface flow assessment is the incorporation of functional assessment of the CD40L using a soluble form of the receptor, CD40-muIg (Fig. 1). This flow assay (phenotyping and function) enables detection of all patients with CD40L deficiency without the necessity for additional confirmatory testing though genetic testing has independent value for ascertaining the specific mutation in a given individual. If CD40L protein is absent or if the protein is present but the CD40-muIg functional component is abnormal, indicating a diagnosis of X-linked hyper-IgM syndrome, depending on the age of the patient and clinical severity, hematopoietic cell transplantation is considered for therapeutic intervention (see Treatment of PIDs and the Role of Flow Cytometry in Assessment of Chimerism and Immune Reconstitution, below). Additionally, replacement immunoglobulin therapy and antibiotic prophylaxis are part of standard treatment. If the result of the above flow test is normal, it rules out a diagnosis of X-linked hyper-IgM syndrome (CD40L deficiency), and other PIDs associated with a humoral defect and potential susceptibility to opportunistic infections (impairment of T cell function) should be considered in the differential diagnosis. Since detailing alternate diagnostic possibilities is beyond the scope of this minireview due to space constraints, the reader is encouraged to review the References section for further information.
FIG 1.
Assessment of CD40L expression and function for diagnosis of X-linked hyper-IgM syndrome (XL-HIGM). The top left panel (panel 1) shows expression of CD40L (CD154) on activated CD4+ T cells after stimulation with PMA. A control for T cell activation, CD69, is included in the assay (not shown). Functional activity of the ligand is measured by binding to a soluble form of the receptor—CD40muIg (panel 2). The bottom panels show flow cytometric data for a male patient with XL-HIGM with lack of expression of CD40L on activation of CD4+ T cells (panel 3) and therefore, no binding to the soluble receptor (panel 4). The grey peak represents the unstimulated sample. The purple peak represents the specific antibody.
(iii) LRBA deficiency.
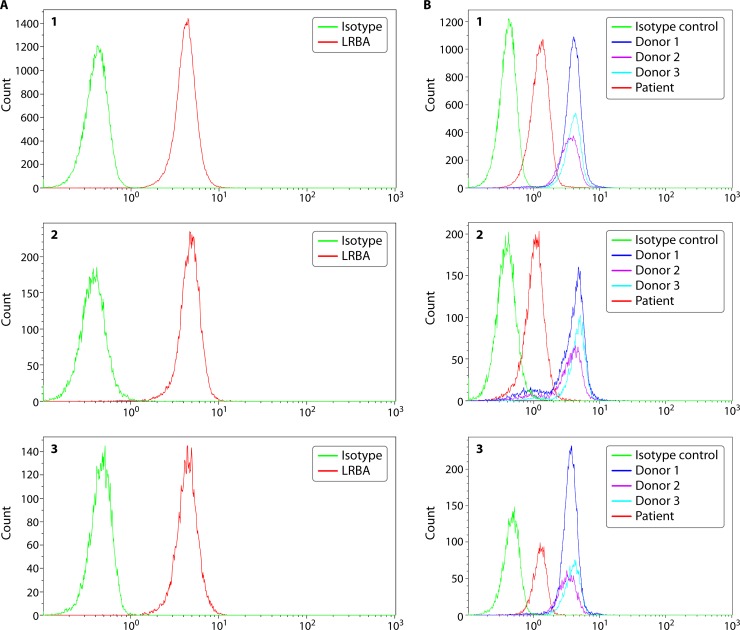
While XLA and CD40L deficiency represent PIDs that have been known and studied for a long time, there are newly described PIDs that constantly challenge the clinical and diagnostic boundaries. LRBA (lipopolysaccharide-responsive beige-like anchor) deficiency was described in 2012 in patients with early onset hypogammaglobulinemia, autoimmunity, and inflammatory bowel disease (27). These patients with germ line LRBA gene mutations affecting LRBA protein expression often have reduced immunoglobulins affecting at least two isotypes and manifest with recurrent infections in addition to severe pulmonary and gastrointestinal complications. However, hypogammaglobulinemia is not a mandatory presenting feature in LRBA-deficient patients (28), though autoimmune complications seem universal. LRBA-deficient patients may also have defective regulatory T cell numbers and function (29), although like many other PIDs, there is a clinical and immunological spectrum observed (30, 31). Assessment of LRBA protein, which is expressed intracellularly in T cells, B cells, monocytes, and NK cells provides a rapid and easy way of determining LRBA deficiency, prior to or while waiting for genetic testing results (which typically takes several weeks). LRBA protein is expressed constitutively in these cell types, and there are modest increments in expression on stimulation of specific cell subsets. In most cases, stimulation may not be necessary to identify absent or reduced protein expression (Fig. 2A). It is useful to quantify mean fluorescence intensity (MFI) as a way to identify reduced protein expression in LRBA-deficient patients (Fig. 2B).
FIG 2.
LRBA protein expression in lymphocytes from a healthy individual and a patient. LRBA protein is expressed intracellularly, and data are shown for T cells (panel 1), B cells (panel 2), and NK cells (panel 3) from a healthy donor (A) and a patient (B). Peripheral blood mononuclear cells (PBMCs) are isolated from blood samples collected in sodium heparin or EDTA and assessed for LRBA expression without stimulation, using an isotype control and a specific primary antibody. Intracellular protein expression is assessed by cell fixation and permeabilization prior to simultaneous staining with cell lineage markers and primary antibody. The protein is visualized using a fluorescently labeled secondary antibody. Both percent-positive lymphocyte subsets (T, B, or NK cells) along with mean fluorescence intensity (MFI) information are captured. LRBA protein is robustly expressed in the majority of lymphocyte subsets without stimulation. In the B panels, data from three healthy donors are represented as donor 1, donor 2, and donor 3. The patient shows a normal proportion of lymphocyte subsets expressing LRBA; however, the mean fluorescence intensity, which correlates with the amount of protein expression, is significantly reduced, which is consistent with LRBA deficiency. The patient had a clinical phenotype of very early onset inflammatory bowel disease, failure to thrive, and multiple autoimmune manifestations. A homozygous 2-bp deletion was identified (c.3958_3986del; p.D1329Yfs*18) in the LRBA gene.
(iv) DOCK8 deficiency.
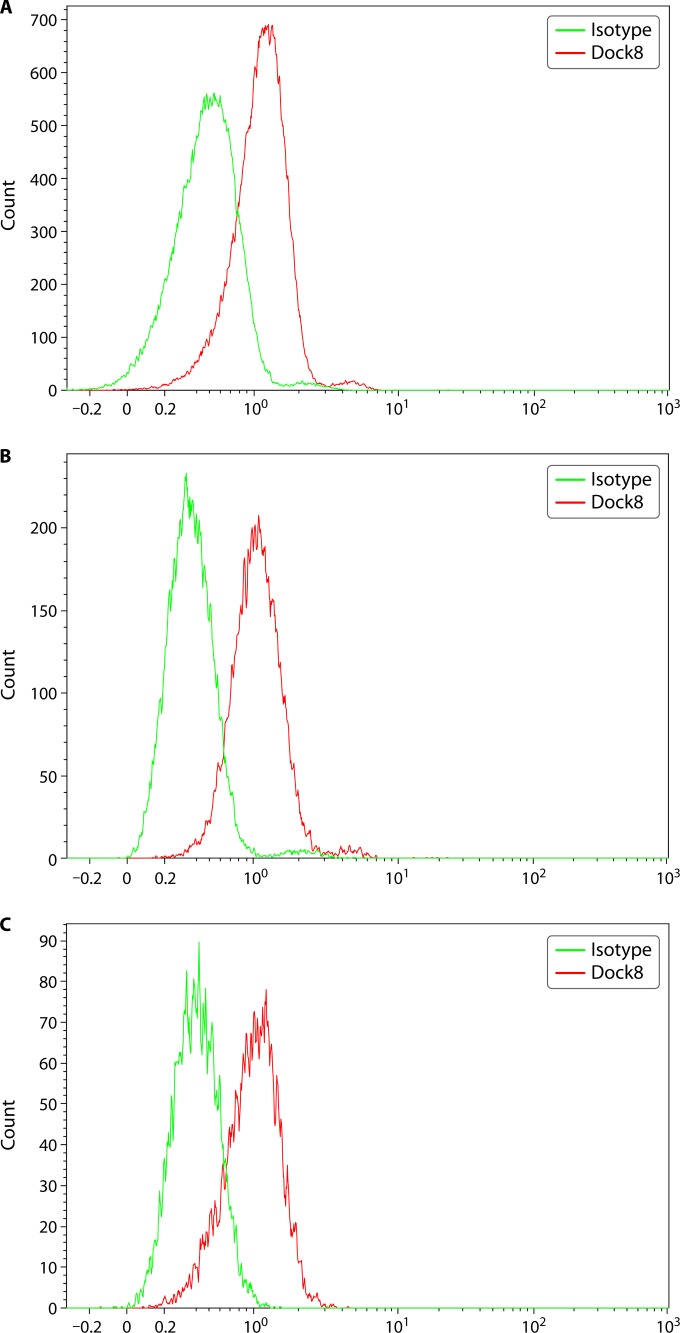
Patients with a combined immunodeficiency phenotype due to mutations in the DOCK8 (dedicator of cytokinesis 8) gene were described 6 years ago (32–35), a PID clinically characterized by recurrent sinopulmonary infections, staphylococcal (Staphylococcus aureus) skin infections, recurrent and severe herpesvirus infections (herpes simplex virus and herpes zoster), molluscum contagiosum, and human papillomavirus (HPV) infections. Immunologically, these patients have significant eosinophilia, elevated serum IgE levels, reduced T and B cells, decreased serum IgM levels, and variably impaired IgG functional antibody responses with reduced in vitro proliferation of activated CD8+ T cells. Additionally, DOCK8-deficient patients have impaired thymic function with decreased TREC (T cell receptor excision circles) (36) consistent with reduced T cell numbers. The susceptibility to herpesvirus and HPV infections could be related to the abnormal NK cell function in these patients (37). Since DOCK8 deficiency shares several clinical and immunological features with severe atopic dermatitis, it is helpful to have a rapid diagnostic test to differentiate the two (38). In fact, the flow cytometric assay for DOCK8 is able to identify all patients with a germ line genetic mutation in DOCK8, making it a reliable and quick diagnostic test, especially since genetic testing is complicated by the large size of the gene (39). It can also be used prognostically to assess protein-specific chimerism postallogeneic hematopoietic cell transplantation (39). DOCK8 protein is expressed intracellularly in a variety of lymphoid and myeloid cell lineages (39) and can be analyzed constitutively (without activation) using a two-step flow cytometry method (Fig. 3) (35, 39). For several of these disease-specific proteins, since there can be variable levels of expression, it is important to include the mean fluorescence intensity data as part of the analysis so that comparisons can be effectively made between the experimental healthy control subjects and patients.
FIG 3.
DOCK8 protein expression in lymphocytes from a healthy individual. DOCK8 protein is expressed intracellularly, and data are shown for T cells (A), B cells (B), and NK cells (C). Peripheral blood mononuclear cells (PBMCs) are isolated from blood samples treated with sodium heparin or EDTA and assessed for DOCK8 expression without stimulation, using an isotype control and a specific primary antibody. Intracellular protein expression is assessed by cell fixation and permeabilization prior to simultaneous staining with cell lineage markers and primary antibody. The protein is visualized using a fluorescently labeled secondary antibody. Both percent-positive lymphocyte subsets (T, B, or NK cells) along with mean fluorescence intensity (MFI) information are captured. DOCK8 protein is robustly expressed in the majority of lymphocyte subsets without stimulation.
While performing disease-specific protein analysis by flow cytometry, it is relevant to note that for some of these proteins, directly conjugated antibodies are not available. While such antibodies improve the efficiency of testing, in some cases with low protein expression, indirectly conjugated antibodies may enhance the sensitivity of detection. Regardless, since comparisons are always made with healthy controls and reference ranges derived thereof, it is relatively straightforward using MFI to identify decreased or absent protein expression.
There are several other PIDs (e.g., IPEX [immune deficiency, polyendocrinopathy, enteropathy, X-linked] syndrome [caused by mutations in the FOXP3 protein], Wiskott-Aldrich syndrome) where protein-specific expression analysis is helpful for diagnosis and monitoring after treatment, and these cannot be described here due to space constraints. It is also important to recognize that the presence of revertant populations resulting in somatic mosaicism (e.g., DOCK8, WASP) can be identified by flow cytometry techniques (40–43).
FUNCTIONAL FLOW CYTOMETRY FOR DISEASE-SPECIFIC PIDs AND IMMUNE COMPETENCE ASSESSMENT
While analysis of specific protein expression as described above is valuable for certain PIDs, in other cases, diagnosis needs to be made on the basis of impairment of functional activity of protein(s). An example of such a PID is chronic granulomatous disease (CGD), which is caused by dysfunctional NADPH oxidase activity in neutrophils. NADPH oxidase is composed of five individual subunits encoded by different genes; therefore, functional assessment of enzyme activity provides a sensitive and specific diagnostic tool. The NADPH oxidase complex is composed of two cell-membrane proteins, gp91phox and p22phox, encoded by CYBB and CYBA genes, respectively. There are three other cytosolic components, p47phox, p67phox, and p40phox encoded by NCF1, NCF2, and NCF4 genes, respectively. Mutations in the CYBB gene (X linked) account for the majority (∼70%) of CGD cases, while autosomal recessive CGD is associated with defect in the other genes. Disease severity is highest with the CYBB (gp91phox) mutations and least with NCF1 (p47phox) defects (44, 45). The majority of infections are caused by five pathogens—Staphylococcus aureus, Nocardia, Burkholderia cepacia, Serratia marcescens, and Aspergillus (44). Though there can be considerable heterogeneity in clinical phenotype, the clinical phenotype includes soft tissue granulomatous disease, cutaneous abscesses, fungal and bacterial infections, autoimmune and inflammatory complications, including colitis (46). Measurement of NADPH oxidase activity is useful not only for diagnosis but also for prognosis (47). For several years, the Nitroblue tetrazolium test (NBT) was used for assessing NADPH oxidase function (48), and it is still used in some clinical and research laboratories. Nearly 2 decades ago, a flow-based assay was validated against the NBT test for measuring the neutrophil respiratory burst and associated NADPH oxidase function (49). This method involves ex vivo activation of neutrophils with a nonspecific stimulant, such as phorbol myristate acetate (PMA) along with the use of dihydrorhodamine 123 (DHR) as the substrate to measure oxidative burst. Nonfluorescent DHR has been shown to be oxidized to fluorescent rhodamine by hydrogen peroxide (50), produced by activation of the NADPH oxidase pathway, and peroxidase produced by neutrophils (myeloperoxidase) and/or eosinophils (eosinophil peroxidase). The ability to perform this analysis with specific evaluation of neutrophils by appropriate side scatter and forward scatter gating on the flow analysis in whole blood (49) provides a relatively quick and robust assay for measuring NADPH oxidase activity. Further, the flow-based DHR assay can also be used to determine the potential genotype of CGD (51), at least for X-linked (gp91phox) CGD, p47phox and p67phox CGD, which accounts for approximately 30% of autosomal recessive CGD (Fig. 4A to D). The pattern of DHR fluorescence associated with the p40phox CGD (52) has also been seen in patients with complete myeloperoxidase deficiency (cMPO) and atypical X-linked CGD in our laboratory (Fig. 4E and F). It should also be noted that cMPO deficiency may show a pattern of DHR fluorescence similar to X-linked CGD (i.e., completely absent DHR [rhodamine] fluorescence on PMA stimulation). Therefore, it is important to bear in mind that cMPO deficiency may give a false-positive (i.e., abnormal) result as illustrated by the example shown in Fig. 4E, which may yield an incorrect diagnosis of CGD that could have major clinical consequences for patients. Therefore, an abnormal DHR result with PMA stimulation should be further evaluated to rule out cMPO deficiency by (i) gating on eosinophils in the DHR flow assay since eosinophil peroxidase can provide a normal DHR result, even when MPO is absent (53) (CGD patients will have abnormal results for DHR flow when gated on eosinophils due to lack of NADPH oxidase activity and production of hydrogen peroxide), or (ii) staining neutrophils for MPO with an anti-MPO antibody, or (iii) performing the NBT test, which is not affected by cMPO deficiency, or (iv) genetic testing (CGD gene panel and MPO gene sequencing), the latter being more expensive. In most cases, it is probably cost-effective and less laborious to stain neutrophils for MPO, which is often routinely performed by many hematopathology laboratories. An abnormal result for MPO staining can be confirmed by genetic testing (it should be kept in mind that most cases of cMPO deficiency are clinically asymptomatic, but a small subset of patients [∼5%], including the example presented here, may develop significant infectious complications, typically with Candida, the control of which appears to require MPO activity in these patients). Most of the patients described in the literature with symptomatic cMPO deficiency have diabetes and Candida infection (54, 55). Therefore, the interpretation of the DHR flow data requires correlation with the clinical phenotype, elimination of false-positive results, and subsequent genetic analysis for confirmation (56). Quantifying the amount of rhodamine fluorescence using mean fluorescence intensity after PMA stimulation is relevant and essential, and it can be correlated to the NADPH oxidase activity.
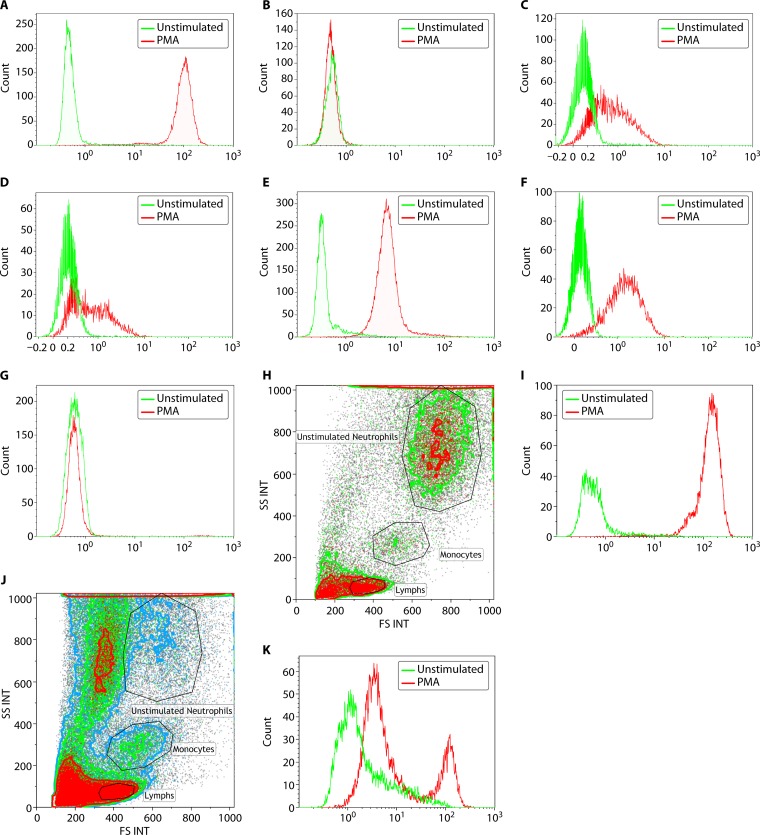
FIG 4.
Assessment of neutrophil oxidative burst using DHR flow cytometry. NADPH oxidase, which produces the respiratory burst in neutrophils, can be assessed following in vitro stimulation with PMA. The oxidation of DHR to fluorescent rhodamine is measured by flow cytometry. The unstimulated fluorescence is represented by the green histogram, while the PMA-stimulated oxidative burst in neutrophils is represented by the red histogram. Both percent-positive neutrophils and mean fluorescence intensity (MFI) are captured for diagnostic interpretation. (A) A normal neutrophil oxidative burst is demonstrated by a complete shift in the stimulated signal. (B) Data on the assessment of neutrophil oxidative burst using DHR flow cytometry in a patient with X-linked CGD. In a patient with a mutation in the CYBB gene (encoding gp91phox protein), there is no evidence of a normal neutrophil oxidative burst, and the unstimulated (green) and stimulated (red) histograms overlap completely. (C and D) Assessment of neutrophil oxidative burst using DHR flow cytometry in a patient with autosomal recessive CGD due to NCF1 and NCF2 mutations, respectively. Patients with autosomal recessive CGD, due to defects in p47phox, have a different pattern of neutrophil oxidative burst, with a proportion of neutrophils negative (stimulated histogram overlapping with unstimulated) and the remaining neutrophils showing significantly reduced fluorescence. Patients with defects in p67phox have a similar pattern of neutrophil oxidative burst to those with NCF1 mutations, with a proportion of neutrophils negative (stimulated histogram overlapping with unstimulated) and the remaining neutrophils showing significantly reduced fluorescence. (E) Assessment of neutrophil oxidative burst using DHR flow cytometry in a patient with complete myeloperoxidase deficiency (cMPO). Patients with cMPO having mutations in the MPO gene may demonstrate a variable pattern of DHR fluorescence, ranging from partially positive to completely absent. In this example, there is complete shift of the neutrophils, indicative of the majority of neutrophils showing DHR fluorescence, but there is an overall significant reduction in the mean fluorescence intensity. This would suggest partially reduced but not completely absent neutrophil oxidative burst in this patient example. (F) Assessment of neutrophil oxidative burst using DHR flow cytometry in a male patient with atypical X-linked CGD. Neutrophil oxidative burst may be preserved in some patients with mutations in the CYBB gene, leading to an atypical clinical phenotype as well as flow cytometric pattern of DHR fluorescence. The pattern is similar to that seen with NCF4 gene mutations (p40phox) and the form of complete MPO deficiency seen in panel E. This is a young male patient with one episode of Burkholderia pneumonia with no other manifestations of CGD who had a missense mutation (c.1061A>G; p.H354R) in the CYBB gene, which has been previously reported to be associated with decreased but not absent NADPH oxidase activity in neutrophils. (G) Assessment of neutrophil oxidative burst using DHR flow cytometry in a female patient with extreme skewing of lyonization, resulting in a phenotype of X-linked CGD. Neutrophil oxidative burst assessment by DHR fluorescence in female carriers of X-linked CGD characteristically shows two populations consistent with the presence of mutant and normal alleles. However, if there is skewing of lyonization, the proportion of neutrophils that are negative for DHR fluorescence can increase. This is an example of an elderly female patient with a history of being a carrier for XL-CGD and who has an affected male offspring demonstrating age-related extreme skewing of lyonization with a DHR flow pattern similar to that seen in a male patient with XL-CGD. (H and I) Side scatter (SSC) and forward scatter (FSC) separation of neutrophils, monocytes, and lymphocytes in whole blood. The DHR flow analysis is performed on neutrophils. (H) A transported sample received within validated stability under optimal conditions with abundant viable neutrophils. INT, integral. (I) DHR fluorescence for PMA-stimulated neutrophils in this sample, indicating a normal and robust result. (J and K) Another transported sample, also received within validated stability, but under suboptimal conditions. (J) There is significant neutrophil cell death observed with reduced viable neutrophils. (K) DHR fluorescence from PMA-stimulated neutrophils of the same sample with high background in unstimulated control (green histogram) and two peaks (red histogram), indicating poor sample quality.
Female carriers for X-linked CGD (XL-CGD) can be identified by the DHR flow cytometry assay, and lyonization (X-chromosome inactivation) patterns can be assessed since there is a relatively high proportion of skewed lyonization observed resulting in symptomatic females (57–59). Further, an age-associated skewing of lyonization in carrier females has been reported (60). In Fig. 4G, complete skewing of lyonization is observed in an elderly carrier female for XL-CGD in the 7th decade of life, consistent with a pattern seen in typical forms for XL-CGD (Fig. 4B). She presented with Burkholderia pneumonia and required a lobectomy and subsequently also developed pyelonephritis. Though the DHR flow assay has very good clinical sensitivity and specificity (keeping in mind the possibility of false-positive results with cMPO deficiency and the necessity of ruling this out via other methods described above), accurate interpretation requires assessment of the sample within 24 to 48 h of blood collection to prevent artifacts due to ex vivo neutrophil activation (the best results are obtained within 24 h, though the analytically validated stability for the assay extends to 48 h, but it is also affected by the quality of the sample, which is dependent on transportation conditions). Examples provided in Fig. 4H and I show a transported sample, which was tested within validated stability (48 h) after blood collection, but the blood was received closer to the 24-h time range. Figure 4H shows the side scatter (SS) and forward scatter (FS) plot for the unstimulated sample. The neutrophils show normal granularity and size, and in Fig. 4I, these neutrophils display normal DHR (rhodamine) fluorescence on PMA stimulation. In contrast, a transported sample (Fig. 4J and K) received within the validated stability of the assay (48 h) but likely subject to temperature fluctuations during transport showed significant neutrophil cell death (Fig. 4J) and two peaks for DHR fluorescence with variable MFI (Fig. 4K), indicative of poor sample quality. Some clinical laboratories control for sample quality by using a cell viability dye in the assay and assessing only viable neutrophils. This is helpful in the interpretation of the flow data in transported samples.
Patients with other neutrophil defects, such as Rac2 deficiency, show normal neutrophil oxidative burst and DHR fluorescence after PMA stimulation but abnormal respiratory burst and superoxide production when stimulated with fMLP (N-formylmethionyl-leucyl-phenylalanine) (61–63). However, Rac2 deficiency is far less common than CGD, which has an estimated incidence of 1:200,000 (64), and therefore, in standard clinical laboratory practice, PMA-stimulated assessment of neutrophil respiratory burst is more relevant than the fMLP-based version, which should be used in clinically ambiguous cases or if there is other evidence to support evaluation for a possible Rac2 deficiency.
Besides disease-specific functional assessments, flow cytometry can also be used for measurement of other cellular immune functions, such as lymphocyte proliferation, cytokine production, and NK cell cytotoxicity (65), and modification of cell signaling proteins, such as phosphorylation (66–71). Phospho-flow can be used in the diagnostic evaluation of PIDs associated with defects in the STAT (signal transducer and activator of transcription factor) molecules, e.g., loss-of-function (LOF) STAT1 mutations seen in patients with susceptibility to intracellular bacterial and viral infections (72, 73). Similarly, gain-of-function (GOF) mutations seen in the same gene, STAT1, associated with autosomal dominant chronic mucocutaneous candidiasis (CMC) and defective Th17 immunity (74–78) can also be assessed by changes in phosphorylation by flow cytometry. It is important to recognize that for accurate diagnostic use of phospho-flow assays, only the appropriate cell subset after careful selection of the stimulus (typically cytokines) is evaluated. Though LOF mutations in STAT3, as seen in autosomal dominant hyper-IgE syndrome (79, 80), and GOF mutations in STAT3, associated with autoimmunity, lymphoproliferation, and infection (81, 82) have been reported, assessment of STAT3 phosphorylation in either context is of questionable and limited clinical utility.
This is not a comprehensive listing of all the possible functional immune applications of flow cytometry but rather examples that highlight the versatility and value of this method beyond assessment of protein expression.
CELLULAR IMMUNOPHENOTYPING IN PIDs
There is significant cellular heterogeneity in both the lymphoid and myeloid compartments of the hematopoietic system, especially in the subsets circulating in blood. Phenotypic analysis and quantitation of peripheral blood lymphocyte subsets have been shown to be either diagnostically and/or prognostically useful in several PIDs. One representative example is B cell subset phenotyping in the classification and prognostic assessment of patients with common variable immunodeficiency (CVID). Among adults, CVID is probably the most commonly represented PID with an incidence of 1:25,000 to 1:50,000 depending on the population studied (83–87). CVID is a term that encompasses an extremely heterogeneous clinical, immunological, and genetic group of diseases, but some of the common characteristics include a B cell deficiency associated with primary hypogammaglobulinemia and impaired ability to mount functional antibody responses to vaccines, in addition to certain key clinical manifestations, including susceptibility to sinopulmonary infections, granulomatous disease, lymphoproliferation, and autoimmunity (in subsets of patients) (88–90). Attempts have been made over the past decade or longer to classify CVID patients based on differences in peripheral B cell subsets, and some of these have provided clinical correlations, which may have prognostic value in subgroups of patients (91–98). B cell subset analysis, in particular evaluation of memory B cells and switched memory B cells, has been incorporated into the newer diagnostic criteria as an accessory criterion (88), as the majority of CVID patients have impaired peripheral B cell differentiation and reduced class-switched memory B cells. Most clinical laboratories that offer B cell subset immunophenotyping include assessment of various memory B cell subsets, including marginal zone B cells (CD19+ CD27+ IgM+ IgD+), class-switched memory B cells (C19+ CD27+ IgM− IgD−), transitional B cells (CD19+ CD27− CD24hi CD38hi IgMhi CD10+), plasmablasts (CD19+ CD20− IgM− CD38int CD27+), and CD21− and CD21+ B cells (includes CD21dim B cells).
In addition to B cell subset evaluation in CVID, quantitative defects (both increase and decrease in specific cell populations) in other lymphocyte subsets have also been reported in subsets of CVID patients, including T cells (99–104) and NK cells (105). CVID patients with severe defects in naive T cell differentiation are now often classified as a separate entity, late-onset combined immunodeficiency (LOCID), as they likely represent another genetically distinct subset of CVID (106). In such patients, assessment of T cell subsets, e.g., naive and memory T cells, is relevant.
T cell subsets, like B cell subsets, can be effectively quantitated in peripheral blood by flow cytometry, and a typical analytical profile includes naive (CD45RA+ CD62L+ CCR7+) and memory (CD45RO+) T cell subsets in both the CD4+ and CD8+ T cell compartments (107, 108). Memory T cells can be further subdivided into central (CD45RO+ CD62L+ CD27+) and effector (CD45RO+ CD62L− CD27−) cells based on location and functional specialization (109–112). There are memory CD4+ and CD8+ T cells that can reexpress CD45RA after antigenic stimulation; these cells are referred to as EMRA T cells (CD4+/CD8+ CD45RA+ CD27−), and they demonstrate robust effector T cell activity (113, 114). In addition to the classic naive and memory T cell subsets, activated T cells expressing major histocompatibility complex (MHC) class II molecules (HLA DR) (115, 116) are often included in a T cell subset profile, as these may be expanded in PIDs during infection and/or inflammation or posthematopoietic cell transplant in the context of graft-versus-host disease (GVHD) (117).
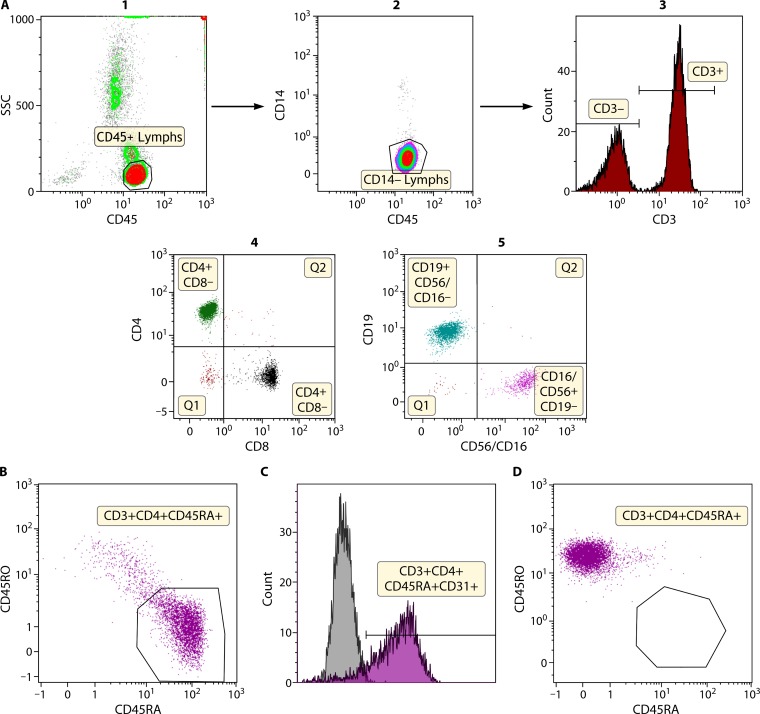
The distribution of naive and memory T cells is particularly relevant for the diagnosis of severe combined immunodeficiency (SCID) and its variants, Omenn syndrome and leaky SCID, in young infants especially with the introduction of newborn screening for SCID (NBS SCID) (118, 119). Flow cytometric lymphocyte subset quantitation (Fig. 5A) and analysis of naive and memory T cell subset (Fig. 5B) distribution is part of the first level of confirmatory testing performed on infants identified as having T cell lymphopenia as part of the newborn screen. Additionally, flow analysis for recent thymic emigrants (Fig. 5C) can be performed to evaluate and corroborate thymic function, since CD31 expression on naive CD45RA+ CD4+ T cells is associated with nascent T cells (120, 121). This is particularly useful in the identification of SCID infants with Omenn syndrome or leaky SCID who may have oligoclonal T cell expansion and a skewed distribution of activated and memory T cells but no naive T cells due to absent thymic output (Fig. 5D) (122–124).
FIG 5.
Immunophenotyping of lymphocyte subsets. (A) Lymphocyte subset quantitation. T, B, and NK cells can be quantitated by flow cytometry. The top panels (panels 1 to 3) show identification of lymphocytes using CD45 and side scatter (SSC). The use of CD14 allows discrimination between monocytes that may inadvertently be included in the lymphocyte population. CD45+ lymphocytes are further subdivided into CD3− lymphocytes and CD3+ T cells. The CD3+ T cells are further analyzed for CD4+ and CD8+ T cells (panel 4), and the CD3− lymphocytes are further assessed for B and NK cells (panel 5). Absolute quantitation (number of cells per microliter) can be performed with the use of fluorescent beads. (B) Identification of naive and memory T cells by flow cytometry. CD3+ T cells can be further divided based on specific cell markers (typically CD45RA and CD45RO though additional markers can be used) into naive and memory subsets. A healthy newborn infant shows predominantly naive CD45RA+ T cells in the CD4+ T cell compartment with only a few memory CD45RO+ T cells. (C) Identification of naive recent thymic emigrants by flow cytometry. CD4+ T cells are newly derived from thymic output and have typically not undergone antigen-induced cell expansion in the periphery and express CD31 (recent thymic emigrant marker). A large proportion of the naive CD45RA+ CD4+ T cells in a healthy newborn infant express CD31. (D) Identification of naive and memory T cells by flow cytometry in a patient with Omenn syndrome. In infants with leaky SCID or Omenn syndrome, there are usually no naive CD45RA+ CD4+ T cells, and the majority of T cells that are present are oligoclonally expanded and express the memory marker, CD45RO. They also have no recent thymic emigrants due to absent thymic output. This patient shows that more than 99% of CD4+ T cells present in blood express CD45RO with no CD45RA+ expression. Further, this patient demonstrated severely skewed T cell receptor repertoire diversity and absent thymic function, in addition to the phenotypic features associated with Omenn syndrome. This female patient had a RAG1 gene mutation associated with a hypomorphic form of SCID.
Immunophenotyping can be performed by flow cytometry on virtually any cell subset in blood, besides those described above, including regulatory T cells (29, 125–129), for example, in diagnosis of IPEX (immune deficiency, polyendocrinopathy, enteropathy, X-linked) syndrome, and dendritic cells (130–134) in the diagnosis of GATA2 deficiency. There are broader applications of the phenotyping assays beyond diagnostic purposes described in this minireview, and an example is provided below of the use of several of these flow assays in therapeutic monitoring.
flow-FISH LEUKOCYTE TELOMERE LENGTH MEASUREMENTS AS A DIAGNOSTIC TEST FOR TELOMERE BIOLOGY DISORDERS
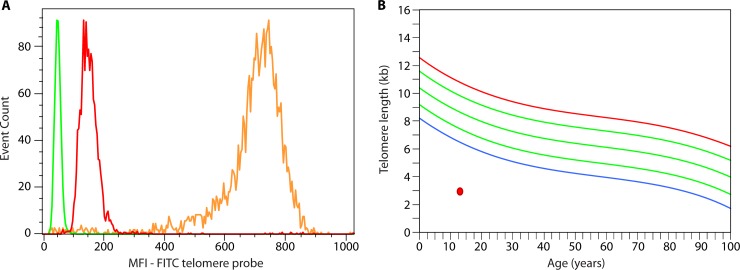
Telomeres are located at the ends of linear chromosomes and are composed of arrays of specific repetitive nucleic acid sequence (TTAGGG) and associated multiprotein complexes (shelterin). Their functions are essential to the maintenance of cellular genomic stability and to continued proliferative capacity. Telomere biology disorders are inherited or de novo systemic conditions that affect the functions of telomeres and display complex etiology with multiple modes of genetic inheritance and variable penetrance. These factors, compounded by the fact that clinical presentation can be first apparent in distinct organ systems, can make their diagnosis challenging. The most common manifestation of telomere biology disorders is bone marrow failure; however, primary immunodeficiency is part of the clinical spectrum for these disorders and was first described about a decade ago (135). Deficient telomerase was then implicated in causing lymphopenia and hypogammaglobulinemia in the context of dyskeratosis congenita (DC) caused by a mutation in the telomerase long noncoding RNA component TERC. Immune deficiency was more recently systematically characterized in the context of inherited bone marrow failure syndromes, including DC (136), and has also been described as a primary presentation which can be severe (137, 138). Further, a recent classification of primary immunodeficiencies summarizes the main genetic etiologies and immunological features of the classical telomere biology disorder syndrome dyskeratosis congenita along with other PIDs (3). Irrespective of the genetic etiology (when known), leukocyte telomere length deficit has been a consistent laboratory finding in telomere biology disorders. This can be assessed by flow-FISH, a quantitative assay that can identify individuals with telomere biology gene mutations from noncarrier direct relatives (139). flow-FISH combines quantitative fluorescence in situ hybridization to a fluorescently labeled telomere repeat-specific peptide nucleic acid (PNA) probe to quantify telomere repeats and together with limited immunophenotyping can differentiate between different blood cell subsets under conditions that maintain cellular integrity so the cells can be analyzed by flow cytometry. DNA content is assessed simultaneously to control for cell ploidy so that only cells that have 2N DNA content are included in the data analysis (i.e., are not actively dividing and contain a known number of chromosome ends or telomeres). The median telomere fluorescence of the cell population of interest is used to calculate the mean telomere length of this cell population (Fig. 6A). These leukocyte telomere length data are then represented in relation to the fairly wide range of normal telomere length distribution and expected decline for age seen in healthy individuals (Fig. 6B). The clinical diagnostic sensitivity and specificity criteria for this assay as applied to the diagnosis of DC have been established (140) and allow for this assay to be applied to the differential diagnosis approach of inherited bone marrow failure syndromes. In this approach, the cell subsets analyzed are representative of different hematopoietic or immune cell compartments as well as several differentiation states (as analyzed according to their cytometry physical properties and limited immunophenotype); very low telomere length (i.e., below the first centile of distribution in healthy individuals) in most or all cell populations support a systemic telomere biology disorder diagnosis.
FIG 6.
Assessment of lymphocyte telomere length in a patient with dyskeratosis congenita. (A) Leukocyte telomere length assessment is performed by measuring the median fluorescence intensity (MFI) of distinct cell subpopulations while controlling for DNA content (not shown) and controlling for the intrinsic fluorescent properties of the cell type analyzed (green [performed in a separate tube and overlaid on the graph]). The MFI of the gated lymphocyte cell population is shown on the graph (red), along with the internal positive hybridization control cells (fixed cow thymocytes [orange]). FITC, fluorescein isothiocyanate. (B) An example of a young patient with dyskeratosis congenita showing in the selected lymphocyte cell population example very short lymphocyte telomere length (red circle) compared to the reference curve summarizing data from healthy individuals – or below the first percentile of distribution for age (blue curve); green curves represent the 10th, 50th, and 90th percentiles, and the red curve represents the 99th percentile of distribution.
TREATMENT OF PIDs AND THE ROLE OF FLOW CYTOMETRY IN ASSESSMENT OF CHIMERISM AND IMMUNE RECONSTITUTION
The genetic etiology of PIDs permits correction of the immune abnormalities by hematopoietic cell transplantation (HCT) for the more-severe diseases, which cannot be managed adequately in the long-term by relatively passive therapy (referring to the amount of intervention required and relative risk of complications), such as immunoglobulin replacement and antibiotics (141–143). For some PIDs, gene therapy is emerging as a successful alternative (144–150). To measure efficacy of the therapeutic intervention, it is important to assess specific protein recovery and chimerism (e.g., recovery of NADPH oxidase function in CGD; CD40L expression and function in X-linked hyper-IgM syndrome) in the appropriate cell subsets, which can be performed by the same flow assays used for diagnostic purposes. The difference lies in the interpretation of the results based on clinical context and serial assessment, based on time since initiation of treatment. Similarly, recovery of cell subsets (phenotypic quantitation) and cell-specific immune functions, in both innate and adaptive compartments (immune reconstitution) (151–153) can be measured by the flow cytometric methods described herein. Posttreatment evaluation of immune recovery and competence is crucial in assessing outcomes posttherapy and the necessity for additional intervention and/or management and long-term prognosis.
INTERPRETATION OF FLOW CYTOMETRY DATA
All clinical laboratories that perform flow testing have typically conducted appropriate analytical and clinical validation studies as per regulatory guidelines before introducing these tests for routine patient diagnosis and monitoring. Typically, analytical validation of flow assays includes preanalytical, analytical, and postanalytical variables that are assessed for its impact on performance and interpretation of results (154–157). While validation includes common variables that may affect interpretation of results, including but not limited to, stability of sample, not all possible conditions, such as types of infections, severity of infections, therapeutic drugs or combinations of drugs, and other immunological conditions, especially those encountered in clinical practice in patients with immunodeficiencies can be exhaustively analyzed before performing clinical testing. Most clinical laboratories establish reference intervals or values based on healthy individuals without underlying immunological conditions so as to provide a frame of reference for interpretation of patient data. For example, very short-term and low-dosage use of steroids does not appear to affect global lymphocyte functional assays; however, long-term or high-dose steroid use can interfere with such lymphocyte proliferation assays. Similarly, the use of biological immunomodulatory agents (including monoclonal antibodies) may affect specific immunophenotyping or functional assays, and these are assessed or interpreted accordingly, based on the information available at the time of testing. For many therapeutic agents that have immunosuppressive or immunomodulatory properties, the mechanism of action is known, and therefore, if the information is available, a customized patient-specific interpretation may be provided. Similarly, in the setting of acute infection, a recommendation is often provided to repeat testing after recovery to measure changes in the functional or phenotypic immune response. Another example is T cell lymphopenia, which may be observed in premature and/or low-birth-weight infants resulting in false-positive tests on the newborn screen for SCID (NBS SCID) (119). For most of these infants, the T cell lymphopenia resolves with maturation of the immune system; however, it is possible for SCID to be present in a premature infant, and therefore, follow-up for a defined period of observation is typically recommended until a diagnosis can be included or excluded. For complex flow cytometry testing as described in this minireview, the most useful interpretations are provided when there is a dialogue between the clinician and the laboratory clinical immunologist, which includes information on clinical history, other comorbidities, family history, medications that may confound interpretation of results, potential interferences, false-positive results, and biological factors, like circadian rhythms and exercise (158, 159). These sorts of advanced and personalized reports are most often provided by larger academic reference laboratories, though additional guidance in interpretation could potentially be obtained by a clinician from any laboratory performing the testing. As discussed in the section on functional testing by flow cytometry for chronic granulomatous disease, an ambiguous or uninterpretable result due to poor sample quality usually results in the request for a fresh specimen. However, large reference laboratories have well-developed methods in place for transport (special boxes to maintain temperature conditions, which avoid extreme fluctuations), receipt, and handling of patient samples that are submitted for testing within the country, and in some cases, even internationally, and it is fairly routine (except in severe weather or other unforeseen conditions) to obtain samples within the validated stability requirements. In contexts where samples are received outside analytically validated conditions or demonstrate suboptimal viability or questionable results, testing is either typically not performed or a fresh sample is requested that is submitted under appropriate conditions of packaging, transport, and time. There are many elements that may affect performance and interpretation of flow cytometry testing, especially for immunological studies, and other than those alluded to above, are outside the scope and intent of this minireview.
THE FUTURE OF FLOW CYTOMETRY IN IMMUNE ANALYSIS: MASS CYTOMETRY
The limitation of analyzing multiple cellular markers by flow cytometry due to the lack of a large repertoire of fluorochromes has been overcome by a new technology, mass cytometry, which epitomizes advances in multiparametric flow with use of rare earth lanthanide-labeled antibodies as detecting reagents (160–162). Mass cytometry enables analysis of up to 45 different parameters simultaneously; however, the throughput is lower (∼500 to 1,000 cells/s) than a conventional flow cytometer (∼50,000 cells/s). Another caveat in its implementation for routine diagnostic evaluation in PIDs is the relatively large cell acquisition requirement, which can be difficult to achieve, especially with pediatric samples. Though mass cytometry has not yet gained access into routine diagnostic flow cytometry in the clinical immunology laboratory, the next decade may very well see such advances translated from the research setting into the diagnostic arena.
SUMMARY AND CONCLUSIONS
Flow cytometry has proven to be a technique that is integral to the repertoire of the clinical laboratory immunologist, particularly with regard to diagnosis and follow-up of patients with PIDs. The rapid advances in this field with identification of more than 300 genetic defects pose a considerable challenge to ensuring an accurate diagnosis and appropriate genotype-phenotype correlations. Though genetic analyses can facilitate the discernment of a molecular diagnosis, there is without a doubt a need to immunologically characterize the defect and link the molecular etiology to abnormalities in function and immunophenotype, besides clinical phenotype. Flow cytometry can provide a cheaper and faster approach to ascertain potential genetic defects, for example, in severe combined immunodeficiency (SCID), lymphocyte subset quantitation (T, B, and NK cell subsets) can subcategorize specific genetic defects, based on which lymphocyte subsets are numerically affected (T−B−NK− SCID, T−B−NK+ SCID, T−B+NK− SCID, and T−B+NK+ SCID). Also, some of the disease-specific examples provided in this minireview showcase how flow can be used to identify a genetic defect (e.g., LRBA and DOCK8 deficiency), which can subsequently be confirmed by actual gene sequencing methods. In other situations, where the genetic diagnosis is not apparent, based either on clinical or immunological phenotype or initial genetic testing, whole-exome sequencing or larger targeted gene panels could be assessed; however, in all cases, immunological characterization of such new or uncertain defects would involve flow cytometry assessment to different extents, in addition to other testing methodologies, depending on the specific context. Regardless, relevant laboratory data have to be correlated with the clinical phenotype. Therefore, in summary, multiparametric flow cytometry, as described in this minireview, provides the necessary methodological platform whereby to achieve this in a manner suitable for rapid implementation in patient care and management.
REFERENCES
- 1.Bonilla FA, Khan DA, Ballas ZK, Chinen J, Frank MM, Hsu JT, Keller M, Kobrynski LJ, Komarow HD, Mazer B, Nelson RP Jr, Orange JS, Routes JM, Shearer WT, Sorensen RU, Verbsky JW, Bernstein DI, Blessing-Moore J, Lang D, Nicklas RA, Oppenheimer J, Portnoy JM, Randolph CR, Schuller D, Spector SL, Tilles S, Wallace D, Khan D. 2015. Practice parameter for the diagnosis and management of primary immunodeficiency. J Allergy Clin Immunol 136:1186–1205.e1-78. doi: 10.1016/j.jaci.2015.04.049. [DOI] [PubMed] [Google Scholar]
- 2.Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, Cunningham-Rundles C, Etzioni A, Holland SM, Klein C, Nonoyama S, Ochs HD, Oksenhendler E, Puck JM, Sullivan KE, Tang ML, Franco JL, Gaspar HB. 2015. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol 35:696–726. doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, Cunningham-Rundles C, Etzioni A, Franco JL, Gaspar HB, Holland SM, Klein C, Nonoyama S, Ochs HD, Oksenhendler E, Picard C, Puck JM, Sullivan K, Tang ML. 2014. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for primary immunodeficiency. Front Immunol 5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bousfiha A, Jeddane L, Al-Herz W, Ailal F, Casanova JL, Chatila T, Conley ME, Cunningham-Rundles C, Etzioni A, Franco JL, Gaspar HB, Holland SM, Klein C, Nonoyama S, Ochs HD, Oksenhendler E, Picard C, Puck JM, Sullivan KE, Tang ML. 2015. The 2015 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol 35:727–738. doi: 10.1007/s10875-015-0198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bousfiha AA, Jeddane L, Ailal F, Al Herz W, Conley ME, Cunningham-Rundles C, Etzioni A, Fischer A, Franco JL, Geha RS, Hammarstrom L, Nonoyama S, Ochs HD, Roifman CM, Seger R, Tang ML, Puck JM, Chapel H, Notarangelo LD, Casanova JL. 2013. A phenotypic approach for IUIS PID classification and diagnosis: guidelines for clinicians at the bedside. J Clin Immunol 33:1078–1087. doi: 10.1007/s10875-013-9901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson JP, Roederer M. 2015. Flow cytometry strikes gold. Science 350:739–740. doi: 10.1126/science.aad6770. [DOI] [PubMed] [Google Scholar]
- 7.Conley ME, Cooper MD. 1998. Genetic basis of abnormal B cell development. Curr Opin Immunol 10:399–406. doi: 10.1016/S0952-7915(98)80112-X. [DOI] [PubMed] [Google Scholar]
- 8.Ochs HD, Smith CI. 1996. X-linked agammaglobulinemia. A clinical and molecular analysis. Medicine 75:287–299. [DOI] [PubMed] [Google Scholar]
- 9.Lee PP, Chen TX, Jiang LP, Chan KW, Yang W, Lee BW, Chiang WC, Chen XY, Fok SF, Lee TL, Ho MH, Yang XQ, Lau YL. 2010. Clinical characteristics and genotype-phenotype correlation in 62 patients with X-linked agammaglobulinemia. J Clin Immunol 30:121–131. doi: 10.1007/s10875-009-9341-5. [DOI] [PubMed] [Google Scholar]
- 10.Kanegane H, Futatani T, Wang Y, Nomura K, Shinozaki K, Matsukura H, Kubota T, Tsukada S, Miyawaki T. 2001. Clinical and mutational characteristics of X-linked agammaglobulinemia and its carrier identified by flow cytometric assessment combined with genetic analysis. J Allergy Clin Immunol 108:1012–1020. [DOI] [PubMed] [Google Scholar]
- 11.Futatani T, Miyawaki T, Tsukada S, Hashimoto S, Kunikata T, Arai S, Kurimoto M, Niida Y, Matsuoka H, Sakiyama Y, Iwata T, Tsuchiya S, Tatsuzawa O, Yoshizaki K, Kishimoto T. 1998. Deficient expression of Bruton's tyrosine kinase in monocytes from X-linked agammaglobulinemia as evaluated by a flow cytometric analysis and its clinical application to carrier detection. Blood 91:595–602. [PubMed] [Google Scholar]
- 12.Conley ME, Farmer DM, Dobbs AK, Howard V, Aiba Y, Shurtleff SA, Kurosaki T. 2008. A minimally hypomorphic mutation in Btk resulting in reduced B cell numbers but no clinical disease. Clin Exp Immunol 152:39–44. doi: 10.1111/j.1365-2249.2008.03593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nonoyama S, Tsukada S, Yamadori T, Miyawaki T, Jin YZ, Watanabe C, Morio T, Yata J, Ochs HD. 1998. Functional analysis of peripheral blood B cells in patients with X-linked agammaglobulinemia. J Immunol 161:3925–3929. [PubMed] [Google Scholar]
- 14.Takada H, Kanegane H, Nomura A, Yamamoto K, Ihara K, Takahashi Y, Tsukada S, Miyawaki T, Hara T. 2004. Female agammaglobulinemia due to the Bruton tyrosine kinase deficiency caused by extremely skewed X-chromosome inactivation. Blood 103:185–187. doi: 10.1182/blood-2003-06-1964. [DOI] [PubMed] [Google Scholar]
- 15.Broides A, Yang W, Conley ME. 2006. Genotype/phenotype correlations in X-linked agammaglobulinemia. Clin Immunol 118:195–200. doi: 10.1016/j.clim.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Granados E, Perez de Diego R, Ferreira Cerdan A, Fontan Casariego G, Garcia Rodriguez MC. 2005. A genotype-phenotype correlation study in a group of 54 patients with X-linked agammaglobulinemia. J Allergy Clin Immunol 116:690–697. [DOI] [PubMed] [Google Scholar]
- 17.Teimourian S, Nasseri S, Pouladi N, Yeganeh M, Aghamohammadi A. 2008. Genotype-phenotype correlation in Bruton's tyrosine kinase deficiency. J Pediatr Hematol Oncol 30:679–683. doi: 10.1097/MPH.0b013e318180bb45. [DOI] [PubMed] [Google Scholar]
- 18.Davies EG, Thrasher AJ. 2010. Update on the hyper immunoglobulin M syndromes. Br J Haematol 149:167–180. doi: 10.1111/j.1365-2141.2010.08077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durandy A, Peron S, Fischer A. 2006. Hyper-IgM syndromes. Curr Opin Rheumatol 18:369–376. doi: 10.1097/01.bor.0000231905.12172.b5. [DOI] [PubMed] [Google Scholar]
- 20.Gulino AV, Notarangelo LD. 2003. Hyper IgM syndromes. Curr Opin Rheumatol 15:422–429. doi: 10.1097/00002281-200307000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Hennig C, Happle C, Hansen G. 2011. “A bad wound may heal, but a bad name can kill”–lessons learned from “hyper-IgM syndrome.” J Allergy Clin Immunol 128:1380–1382. doi: 10.1016/j.jaci.2011.07.041 (Reply, 128: 1382–1383. doi:. [DOI] [PubMed] [Google Scholar]
- 22.Lee WI, Torgerson TR, Schumacher MJ, Yel L, Zhu Q, Ochs HD. 2005. Molecular analysis of a large cohort of patients with the hyper immunoglobulin M (IgM) syndrome. Blood 105:1881–1890. doi: 10.1182/blood-2003-12-4420. [DOI] [PubMed] [Google Scholar]
- 23.van Zelm MC, Bartol SJ, Driessen GJ, Mascart F, Reisli I, Franco JL, Wolska-Kusnierz B, Kanegane H, Boon L, van Dongen JJ, van der Burg M. 2014. Human CD19 and CD40L deficiencies impair antibody selection and differentially affect somatic hypermutation. J Allergy Clin Immunol 134:135–144. doi: 10.1016/j.jaci.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Cabral-Marques O, Schimke LF, Pereira PV, Falcai A, de Oliveira JB, Hackett MJ, Errante PR, Weber CW, Ferreira JF, Kuntze G, Rosario-Filho NA, Ochs HD, Torgerson TR, Carvalho BT, Condino-Neto A. 2012. Expanding the clinical and genetic spectrum of human CD40L deficiency: the occurrence of paracoccidioidomycosis and other unusual infections in Brazilian patients. J Clin Immunol 32:212–220. doi: 10.1007/s10875-011-9623-6. [DOI] [PubMed] [Google Scholar]
- 25.Jain A, Atkinson TP, Lipsky PE, Slater JE, Nelson DL, Strober W. 1999. Defects of T-cell effector function and post-thymic maturation in X-linked hyper-IgM syndrome. J Clin Invest 103:1151–1158. doi: 10.1172/JCI5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsui-Sekinaka K, Imai K, Sato H, Tomizawa D, Kajiwara M, Nagasawa M, Morio T, Nonoyama S. 2015. Clinical features and hematopoietic stem cell transplantations for CD40 ligand deficiency in Japan. J Allergy Clin Immunol 136:1018–1024. doi: 10.1016/j.jaci.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Herrera G, Tampella G, Pan-Hammarstrom Q, Herholz P, Trujillo-Vargas CM, Phadwal K, Simon AK, Moutschen M, Etzioni A, Mory A, Srugo I, Melamed D, Hultenby K, Liu C, Baronio M, Vitali M, Philippet P, Dideberg V, Aghamohammadi A, Rezaei N, Enright V, Du L, Salzer U, Eibel H, Pfeifer D, Veelken H, Stauss H, Lougaris V, Plebani A, Gertz EM, Schaffer AA, Hammarstrom L, Grimbacher B. 2012. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am J Hum Genet 90:986–1001. doi: 10.1016/j.ajhg.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns SO, Zenner HL, Plagnol V, Curtis J, Mok K, Eisenhut M, Kumararatne D, Doffinger R, Thrasher AJ, Nejentsev S. 2012. LRBA gene deletion in a patient presenting with autoimmunity without hypogammaglobulinemia. J Allergy Clin Immunol 130:1428–1432. doi: 10.1016/j.jaci.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charbonnier LM, Janssen E, Chou J, Ohsumi TK, Keles S, Hsu JT, Massaad MJ, Garcia-Lloret M, Hanna-Wakim R, Dbaibo G, Alangari AA, Alsultan A, Al-Zahrani D, Geha RS, Chatila TA. 2015. Regulatory T-cell deficiency and immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like disorder caused by loss-of-function mutations in LRBA. J Allergy Clin Immunol 135:217–227. doi: 10.1016/j.jaci.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo B, Zhang K, Lu W, Zheng L, Zhang Q, Kanellopoulou C, Zhang Y, Liu Z, Fritz JM, Marsh R, Husami A, Kissell D, Nortman S, Chaturvedi V, Haines H, Young LR, Mo J, Filipovich AH, Bleesing JJ, Mustillo P, Stephens M, Rueda CM, Chougnet CA, Hoebe K, McElwee J, Hughes JD, Karakoc-Aydiner E, Matthews HF, Price S, Su HC, Rao VK, Lenardo MJ, Jordan MB. 2015. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science 349:436–440. doi: 10.1126/science.aaa1663. [DOI] [PubMed] [Google Scholar]
- 31.Revel-Vilk S, Fischer U, Keller B, Nabhani S, Gamez-Diaz L, Rensing-Ehl A, Gombert M, Honscheid A, Saleh H, Shaag A, Borkhardt A, Grimbacher B, Warnatz K, Elpeleg O, Stepensky P. 2015. Autoimmune lymphoproliferative syndrome-like disease in patients with LRBA mutation. Clin Immunol 159:84–92. doi: 10.1016/j.clim.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Aydin SE, Kilic SS, Aytekin C, Kumar A, Porras O, Kainulainen L, Kostyuchenko L, Genel F, Kutukculer N, Karaca N, Gonzalez-Granado L, Abbott J, Al-Zahrani D, Rezaei N, Baz Z, Thiel J, Ehl S, Marodi L, Orange JS, Sawalle-Belohradsky J, Keles S, Holland SM, Sanal O, Ayvaz DC, Tezcan I, Al-Mousa H, Alsum Z, Hawwari A, Metin A, Matthes-Martin S, Honig M, Schulz A, Picard C, Barlogis V, Gennery A, Ifversen M, van Montfrans J, Kuijpers T, Bredius R, Duckers G, Al-Herz W, Pai SY, Geha R, Notheis G, Schwarze CP, Tavil B, Azik F, Bienemann K, Grimbacher B, Heinz V, et al. 2015. DOCK8 deficiency: clinical and immunological phenotype and treatment options - a review of 136 patients. J Clin Immunol 35:189–198. doi: 10.1007/s10875-014-0126-0. [DOI] [PubMed] [Google Scholar]
- 33.Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, Chen A, Kim HS, Lloret MG, Schulze I, Ehl S, Thiel J, Pfeifer D, Veelken H, Niehues T, Siepermann K, Weinspach S, Reisli I, Keles S, Genel F, Kutukculer N, Camcioglu Y, Somer A, Karakoc-Aydiner E, Barlan I, Gennery A, Metin A, Degerliyurt A, Pietrogrande MC, Yeganeh M, Baz Z, Al-Tamemi S, Klein C, Puck JM, Holland SM, McCabe ER, Grimbacher B, Chatila TA. 2009. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol 124:1289–1302.e4. doi: 10.1016/j.jaci.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su HC. 2010. Dedicator of cytokinesis 8 (DOCK8) deficiency. Curr Opin Allergy Clin Immunol 10:515–520. doi: 10.1097/ACI.0b013e32833fd718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, Matthews HF, Davis J, Turner ML, Uzel G, Holland SM, Su HC. 2009. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med 361:2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dasouki M, Okonkwo KC, Ray A, Folmsbeel CK, Gozales D, Keles S, Puck JM, Chatila T. 2011. Deficient T Cell Receptor Excision Circles (TRECs) in autosomal recessive hyper IgE syndrome caused by DOCK8 mutation: implications for pathogenesis and potential detection by newborn screening. Clin Immunol 141:128–132. doi: 10.1016/j.clim.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizesko MC, Banerjee PP, Monaco-Shawver L, Mace EM, Bernal WE, Sawalle-Belohradsky J, Belohradsky BH, Heinz V, Freeman AF, Sullivan KE, Holland SM, Torgerson TR, Al-Herz W, Chou J, Hanson IC, Albert MH, Geha RS, Renner ED, Orange JS. 2013. Defective actin accumulation impairs human natural killer cell function in patients with dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol 131:840–848. doi: 10.1016/j.jaci.2012.12.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janssen E, Tsitsikov E, Al-Herz W, Lefranc G, Megarbane A, Dasouki M, Bonilla FA, Chatila T, Schneider L, Geha RS. 2014. Flow cytometry biomarkers distinguish DOCK8 deficiency from severe atopic dermatitis. Clin Immunol 150:220–224. doi: 10.1016/j.clim.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pai SY, de Boer H, Massaad MJ, Chatila TA, Keles S, Jabara HH, Janssen E, Lehmann LE, Hanna-Wakim R, Dbaibo G, McDonald DR, Al-Herz W, Geha RS. 2014. Flow cytometry diagnosis of dedicator of cytokinesis 8 (DOCK8) deficiency. J Allergy Clin Immunol 134:221–223. doi: 10.1016/j.jaci.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis BR, Yan Q, Bui JH, Felix K, Moratto D, Muul LM, Prokopishyn NL, Blaese RM, Candotti F. 2010. Somatic mosaicism in the Wiskott-Aldrich syndrome: molecular and functional characterization of genotypic revertants. Clin Immunol 135:72–83. doi: 10.1016/j.clim.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Jing H, Zhang Q, Zhang Y, Hill BJ, Dove CG, Gelfand EW, Atkinson TP, Uzel G, Matthews HF, Mustillo PJ, Lewis DB, Kavadas FD, Hanson IC, Kumar AR, Geha RS, Douek DC, Holland SM, Freeman AF, Su HC. 2014. Somatic reversion in dedicator of cytokinesis 8 immunodeficiency modulates disease phenotype. J Allergy Clin Immunol 133:1667–1675. doi: 10.1016/j.jaci.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai S, Minegishi M, Ohashi Y, Sasahara Y, Kumaki S, Konno T, Miki H, Derry J, Nonoyama S, Miyawaki T, Horibe K, Tachibana N, Kudoh E, Yoshimura Y, Izumikawa Y, Sako M, Tsuchiya S. 2002. Flow cytometric determination of intracytoplasmic Wiskott-Aldrich syndrome protein in peripheral blood lymphocyte subpopulations. J Immunol Methods 260:195–205. doi: 10.1016/S0022-1759(01)00549-X. [DOI] [PubMed] [Google Scholar]
- 43.Nakajima M, Yamada M, Yamaguchi K, Sakiyama Y, Oda A, Nelson DL, Yawaka Y, Ariga T. 2009. Possible application of flow cytometry for evaluation of the structure and functional status of WASP in peripheral blood mononuclear cells. Eur J Haematol 82:223–230. doi: 10.1111/j.1600-0609.2008.01180.x. [DOI] [PubMed] [Google Scholar]
- 44.Kang EM, Marciano BE, DeRavin S, Zarember KA, Holland SM, Malech HL. 2011. Chronic granulomatous disease: overview and hematopoietic stem cell transplantation. J Allergy Clin Immunol 127:1319–1326. doi: 10.1016/j.jaci.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenzweig SD, Holland SM. 2004. Phagocyte immunodeficiencies and their infections. J Allergy Clin Immunol 113:620–626. doi: 10.1016/j.jaci.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Martire B, Rondelli R, Soresina A, Pignata C, Broccoletti T, Finocchi A, Rossi P, Gattorno M, Rabusin M, Azzari C, Dellepiane RM, Pietrogrande MC, Trizzino A, Di Bartolomeo P, Martino S, Carpino L, Cossu F, Locatelli F, Maccario R, Pierani P, Putti MC, Stabile A, Notarangelo LD, Ugazio AG, Plebani A, De Mattia D. 2008. Clinical features, long-term follow-up and outcome of a large cohort of patients with chronic granulomatous disease: an Italian multicenter study. Clin Immunol 126:155–164. doi: 10.1016/j.clim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Kuhns DB, Alvord WG, Heller T, Feld JJ, Pike KM, Marciano BE, Uzel G, DeRavin SS, Priel DA, Soule BP, Zarember KA, Malech HL, Holland SM, Gallin JI. 2010. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med 363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gentle TA, Thompson RA. 1990. Neutrophil function tests in clinical immunology, p 51 In Gooi HC, Chapel H (ed), Clinical immunology: a practical approach. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 49.Richardson MP, Ayliffe MJ, Helbert M, Davies EG. 1998. A simple flow cytometry assay using dihydrorhodamine for the measurement of the neutrophil respiratory burst in whole blood: comparison with the quantitative nitroblue tetrazolium test. J Immunol Methods 219:187–193. doi: 10.1016/S0022-1759(98)00136-7. [DOI] [PubMed] [Google Scholar]
- 50.Vowells SJ, Sekhsaria S, Malech HL, Shalit M, Fleisher TA. 1995. Flow cytometric analysis of the granulocyte respiratory burst: a comparison study of fluorescent probes. J Immunol Methods 178:89–97. doi: 10.1016/0022-1759(94)00247-T. [DOI] [PubMed] [Google Scholar]
- 51.Vowells SJ, Fleisher TA, Sekhsaria S, Alling DW, Maguire TE, Malech HL. 1996. Genotype-dependent variability in flow cytometric evaluation of reduced nicotinamide adenine dinucleotide phosphate oxidase function in patients with chronic granulomatous disease. J Pediatr 128:104–107. doi: 10.1016/S0022-3476(96)70437-7. [DOI] [PubMed] [Google Scholar]
- 52.Matute JD, Arias AA, Wright NA, Wrobel I, Waterhouse CC, Li XJ, Marchal CC, Stull ND, Lewis DB, Steele M, Kellner JD, Yu W, Meroueh SO, Nauseef WM, Dinauer MC. 2009. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood 114:3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauch L, Lun A, O'Gorman MR, Harris JS, Schulze I, Zychlinsky A, Fuchs T, Oelschlagel U, Brenner S, Kutter D, Rosen-Wolff A, Roesler J. 2007. Chronic granulomatous disease (CGD) and complete myeloperoxidase deficiency both yield strongly reduced dihydrorhodamine 123 test signals but can be easily discerned in routine testing for CGD. Clin Chem 53:890–896. doi: 10.1373/clinchem.2006.083444. [DOI] [PubMed] [Google Scholar]
- 54.Cech P, Stalder H, Widmann JJ, Rohner A, Miescher PA. 1979. Leukocyte myeloperoxidase deficiency and diabetes mellitus associated with Candida albicans liver abscess. Am J Med 66:149–153. doi: 10.1016/0002-9343(79)90507-2. [DOI] [PubMed] [Google Scholar]
- 55.Nauseef WM. 1988. Myeloperoxidase deficiency. Hematol Oncol Clin North Am 2:135–158. [PubMed] [Google Scholar]
- 56.Abraham RS. 2011. Relevance of laboratory testing for the diagnosis of primary immunodeficiencies: a review of case-based examples of selected immunodeficiencies. Clin Mol Allergy 9:6. doi: 10.1186/1476-7961-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis EM, Singla M, Sergeant S, Koty PP, McPhail LC. 2008. X-linked chronic granulomatous disease secondary to skewed X chromosome inactivation in a female with a novel CYBB mutation and late presentation. Clin Immunol 129:372–380. doi: 10.1016/j.clim.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lun A, Roesler J, Renz H. 2002. Unusual late onset of X-linked chronic granulomatous disease in an adult woman after unsuspicious childhood. Clin Chem 48:780–781. [PubMed] [Google Scholar]
- 59.Roesler J. 2009. Carriers of X-linked chronic granulomatous disease at risk. Clin Immunol 130:233.doi: 10.1016/j.clim.2008.09.013 (Reply, 130: 234.) [DOI] [PubMed] [Google Scholar]
- 60.Rosen-Wolff A, Soldan W, Heyne K, Bickhardt J, Gahr M, Roesler J. 2001. Increased susceptibility of a carrier of X-linked chronic granulomatous disease (CGD) to Aspergillus fumigatus infection associated with age-related skewing of lyonization. Ann Hematol 80:113–115. doi: 10.1007/s002770000230. [DOI] [PubMed] [Google Scholar]
- 61.Ambruso DR, Knall C, Abell AN, Panepinto J, Kurkchubasche A, Thurman G, Gonzalez-Aller C, Hiester A, deBoer M, Harbeck RJ, Oyer R, Johnson GL, Roos D. 2000. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci U S A 97:4654–4659. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurkchubasche AG, Panepinto JA, Tracy TF Jr, Thurman GW, Ambruso DR. 2001. Clinical features of a human Rac2 mutation: a complex neutrophil dysfunction disease. J Pediatr 139:141–147. doi: 10.1067/mpd.2001.114718. [DOI] [PubMed] [Google Scholar]
- 63.Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, Jia B, Zheng Y, Ambruso DR, Lowe JB, Atkinson SJ, Dinauer MC, Boxer L. 2000. Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood 96:1646–1654. [PubMed] [Google Scholar]
- 64.Winkelstein JA, Marino MC, Johnston RB Jr, Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, Buckley RH, Foster CB, Chanock SJ, Dickler H. 2000. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine 79:155–169. [DOI] [PubMed] [Google Scholar]
- 65.Abraham RS. 2016. Lymphocyte activation. In Detrick B, Hamilton RG, Folds JL (ed), Manual of molecular and clinical laboratory immunology, 8th ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 66.Krutzik PO, Irish JM, Nolan GP, Perez OD. 2004. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol 110:206–221. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 67.Krutzik PO, Nolan GP. 2003. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A 55:61–70. [DOI] [PubMed] [Google Scholar]
- 68.Lafarge S, Hamzeh-Cognasse H, Chavarin P, Genin C, Garraud O, Cognasse F. 2007. A flow cytometry technique to study intracellular signals NF-kappaB and STAT3 in peripheral blood mononuclear cells. BMC Mol Biol 8:64. doi: 10.1186/1471-2199-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montag DT, Lotze MT. 2006. Rapid flow cytometric measurement of cytokine-induced phosphorylation pathways [CIPP] in human peripheral blood leukocytes. Clin Immunol 121:215–226. doi: 10.1016/j.clim.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 70.Montag DT, Lotze MT. 2006. Successful simultaneous measurement of cell membrane and cytokine induced phosphorylation pathways [CIPP] in human peripheral blood mononuclear cells. J Immunol Methods 313:48–60. doi: 10.1016/j.jim.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 71.Perez OD, Nolan GP. 2006. Phospho-proteomic immune analysis by flow cytometry: from mechanism to translational medicine at the single-cell level. Immunol Rev 210:208–228. doi: 10.1111/j.0105-2896.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 72.Chapgier A, Kong XF, Boisson-Dupuis S, Jouanguy E, Averbuch D, Feinberg J, Zhang SY, Bustamante J, Vogt G, Lejeune J, Mayola E, de Beaucoudrey L, Abel L, Engelhard D, Casanova JL. 2009. A partial form of recessive STAT1 deficiency in humans. J Clin Invest 119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kong XF, Ciancanelli M, Al-Hajjar S, Alsina L, Zumwalt T, Bustamante J, Feinberg J, Audry M, Prando C, Bryant V, Kreins A, Bogunovic D, Halwani R, Zhang XX, Abel L, Chaussabel D, Al-Muhsen S, Casanova JL, Boisson-Dupuis S. 2010. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood 116:5895–5906. doi: 10.1182/blood-2010-04-280586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frans G, Moens L, Schaballie H, Van Eyck L, Borgers H, Wuyts M, Dillaerts D, Vermeulen E, Dooley J, Grimbacher B, Cant A, Declerck D, Peumans M, Renard M, De Boeck K, Hoffman I, Francois I, Liston A, Claessens F, Bossuyt X, Meyts I. 2014. Gain-of-function mutations in signal transducer and activator of transcription 1 (STAT1): chronic mucocutaneous candidiasis accompanied by enamel defects and delayed dental shedding. J Allergy Clin Immunol 134:1209–1213. doi: 10.1016/j.jaci.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulacsy V, Chernyshova L, Chernyshov V, Bondarenko A, Grimaldo RM, Blancas-Galicia L, Beas IM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel PR, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bue M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, et al. 2011. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med 208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mizoguchi Y, Tsumura M, Okada S, Hirata O, Minegishi S, Imai K, Hyakuna N, Muramatsu H, Kojima S, Ozaki Y, Imai T, Takeda S, Okazaki T, Ito T, Yasunaga S, Takihara Y, Bryant VL, Kong XF, Cypowyj S, Boisson-Dupuis S, Puel A, Casanova JL, Morio T, Kobayashi M. 2014. Simple diagnosis of STAT1 gain-of-function alleles in patients with chronic mucocutaneous candidiasis. J Leukoc Biol 95:667–676. doi: 10.1189/jlb.0513250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, Kullberg BJ, van der Meer JW, Lilic D, Veltman JA, Netea MG. 2011. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med 365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 78.Yamazaki Y, Yamada M, Kawai T, Morio T, Onodera M, Ueki M, Watanabe N, Takada H, Takezaki S, Chida N, Kobayashi I, Ariga T. 2014. Two novel gain-of-function mutations of STAT1 responsible for chronic mucocutaneous candidiasis disease: impaired production of IL-17A and IL-22, and the presence of anti-IL-17F autoantibody. J Immunol 193:4880–4887. doi: 10.4049/jimmunol.1401467. [DOI] [PubMed] [Google Scholar]
- 79.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schaffer AA, Puck JM, Grimbacher B. 2007. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 80.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, Metin A, Karasuyama H. 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 81.Haapaniemi EM, Kaustio M, Rajala HL, van Adrichem AJ, Kainulainen L, Glumoff V, Doffinger R, Kuusanmaki H, Heiskanen-Kosma T, Trotta L, Chiang S, Kulmala P, Eldfors S, Katainen R, Siitonen S, Karjalainen-Lindsberg ML, Kovanen PE, Otonkoski T, Porkka K, Heiskanen K, Hanninen A, Bryceson YT, Uusitalo-Seppala R, Saarela J, Seppanen M, Mustjoki S, Kere J. 2015. Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood 125:639–648. doi: 10.1182/blood-2014-04-570101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, Lyons JJ, Engelhardt KR, Zhang Y, Topcagic N, Roberson ED, Matthews H, Verbsky JW, Dasu T, Vargas-Hernandez A, Varghese N, McClain KL, Karam LB, Nahmod K, Makedonas G, Mace EM, Sorte HS, Perminow G, Rao VK, O'Connell MP, Price S, Su HC, Butrick M, McElwee J, Hughes JD, Willet J, Swan D, Xu Y, Santibanez-Koref M, Slowik V, Dinwiddie DL, Ciaccio CE, Saunders CJ, Septer S, Kingsmore SF, White AJ, Cant AJ, Hambleton S, Cooper MA. 2015. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood 125:591–599. doi: 10.1182/blood-2014-09-602763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cunningham-Rundles C, Maglione PJ. 2012. Common variable immunodeficiency. J Allergy Clin Immunol 129:1425–1426. doi: 10.1016/j.jaci.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 84.Ochs HD. 2014. Common variable immunodeficiency (CVID): new genetic insight and unanswered questions. Clin Exp Immunol 178(Suppl 1):5–6. doi: 10.1111/cei.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park MA, Li JT, Hagan JB, Maddox DE, Abraham RS. 2008. Common variable immunodeficiency: a new look at an old disease. Lancet 372:489–502. doi: 10.1016/S0140-6736(08)61199-X. [DOI] [PubMed] [Google Scholar]
- 86.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. 2012. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood 119:1650–1657. doi: 10.1182/blood-2011-09-377945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yong PF, Thaventhiran JE, Grimbacher B. 2011. “A rose is a rose is a rose,” but CVID is Not CVID common variable immune deficiency (CVID), what do we know in 2011? Adv Immunol 111:47–107. doi: 10.1016/B978-0-12-385991-4.00002-7. [DOI] [PubMed] [Google Scholar]
- 88.Ameratunga R, Brewerton M, Slade C, Jordan A, Gillis D, Steele R, Koopmans W, Woon ST. 2014. Comparison of diagnostic criteria for common variable immunodeficiency disorder. Front Immunol 5:415. doi: 10.3389/fimmu.2014.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ameratunga R, Woon ST, Gillis D, Koopmans W, Steele R. 2013. New diagnostic criteria for common variable immune deficiency (CVID), which may assist with decisions to treat with intravenous or subcutaneous immunoglobulin. Clin Exp Immunol 174:203–211. doi: 10.1111/cei.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chapel H, Lucas M, Lee M, Bjorkander J, Webster D, Grimbacher B, Fieschi C, Thon V, Abedi MR, Hammarstrom L. 2008. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood 112:277–286. doi: 10.1182/blood-2007-11-124545. [DOI] [PubMed] [Google Scholar]
- 91.Al Kindi M, Mundy J, Sullivan T, Smith W, Kette F, Smith A, Heddle R, Hissaria P. 2012. Utility of peripheral blood B cell subsets analysis in common variable immunodeficiency. Clin Exp Immunol 167:275–281. doi: 10.1111/j.1365-2249.2011.04507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferry BL, Jones J, Bateman EA, Woodham N, Warnatz K, Schlesier M, Misbah SA, Peter HH, Chapel HM. 2005. Measurement of peripheral B cell subpopulations in common variable immunodeficiency (CVID) using a whole blood method. Clin Exp Immunol 140:532–539. doi: 10.1111/j.1365-2249.2005.02793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kalina T, Stuchly J, Janda A, Hrusak O, Ruzickova S, Sediva A, Litzman J, Vlkova M. 2009. Profiling of polychromatic flow cytometry data on B-cells reveals patients' clusters in common variable immunodeficiency. Cytometry A 75:902–909. doi: 10.1002/cyto.a.20801. [DOI] [PubMed] [Google Scholar]
- 94.Kutukculer N, Gulez N, Karaca NE, Aksu G, Berdeli A. 2012. Three different classifications, B lymphocyte subpopulations, TNFRSF13B (TACI), TNFRSF13C (BAFF-R), TNFSF13 (APRIL) gene mutations, CTLA-4 and ICOS gene polymorphisms in Turkish patients with common variable immunodeficiency. J Clin Immunol 32:1165–1179. doi: 10.1007/s10875-012-9717-9. [DOI] [PubMed] [Google Scholar]
- 95.Piqueras B, Lavenu-Bombled C, Galicier L, Bergeron-van der Cruyssen F, Mouthon L, Chevret S, Debre P, Schmitt C, Oksenhendler E. 2003. Common variable immunodeficiency patient classification based on impaired B cell memory differentiation correlates with clinical aspects. J Clin Immunol 23:385–400. doi: 10.1023/A:1025373601374. [DOI] [PubMed] [Google Scholar]
- 96.Rosel AL, Scheibenbogen C, Schliesser U, Sollwedel A, Hoffmeister B, Hanitsch L, von Bernuth H, Kruger R, Warnatz K, Volk HD, Thomas S. 2015. Classification of common variable immunodeficiencies using flow cytometry and a memory B-cell functionality assay. J Allergy Clin Immunol 135:198–208. doi: 10.1016/j.jaci.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 97.Warnatz K, Denz A, Drager R, Braun M, Groth C, Wolff-Vorbeck G, Eibel H, Schlesier M, Peter HH. 2002. Severe deficiency of switched memory B cells (CD27+IgM−IgD−) in subgroups of patients with common variable immunodeficiency: a new approach to classify a heterogeneous disease. Blood 99:1544–1551. doi: 10.1182/blood.V99.5.1544. [DOI] [PubMed] [Google Scholar]
- 98.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, Vlkova M, Hernandez M, Detkova D, Bos PR, Poerksen G, von Bernuth H, Baumann U, Goldacker S, Gutenberger S, Schlesier M, Bergeron-van der Cruyssen F, Le Garff M, Debre P, Jacobs R, Jones J, Bateman E, Litzman J, van Hagen PM, Plebani A, Schmidt RE, Thon V, Quinti I, Espanol T, Webster AD, Chapel H, Vihinen M, Oksenhendler E, Peter HH, Warnatz K. 2008. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood 111:77–85. doi: 10.1182/blood-2007-06-091744. [DOI] [PubMed] [Google Scholar]
- 99.Carbone J, Sarmiento E, Micheloud D, Rodriguez-Molina J, Fernandez-Cruz E. 2006. Elevated levels of activated CD4 T cells in common variable immunodeficiency: association with clinical findings. Allergol Immunopathol 34:131–135. doi: 10.1157/13091037. [DOI] [PubMed] [Google Scholar]
- 100.Fevang B, Yndestad A, Sandberg WJ, Holm AM, Muller F, Aukrust P, Froland SS. 2007. Low numbers of regulatory T cells in common variable immunodeficiency: association with chronic inflammation in vivo. Clin Exp Immunol 147:521–525. doi: 10.1111/j.1365-2249.2006.03314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fulcher DA, Avery DT, Fewings NL, Berglund LJ, Wong S, Riminton DS, Adelstein S, Tangye SG. 2009. Invariant natural killer (iNK) T cell deficiency in patients with common variable immunodeficiency. Clin Exp Immunol 157:365–369. doi: 10.1111/j.1365-2249.2009.03973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Giovannetti A, Pierdominici M, Mazzetta F, Marziali M, Renzi C, Mileo AM, De Felice M, Mora B, Esposito A, Carello R, Pizzuti A, Paggi MG, Paganelli R, Malorni W, Aiuti F. 2007. Unravelling the complexity of T cell abnormalities in common variable immunodeficiency. J Immunol 178:3932–3943. doi: 10.4049/jimmunol.178.6.3932. [DOI] [PubMed] [Google Scholar]
- 103.Kamae C, Nakagawa N, Sato H, Honma K, Mitsuiki N, Ohara O, Kanegane H, Pasic S, Pan-Hammarstrom Q, van Zelm MC, Morio T, Imai K, Nonoyama S. 2013. Common variable immunodeficiency classification by quantifying T-cell receptor and immunoglobulin kappa-deleting recombination excision circles. J Allergy Clin Immunol 131:1437–1440. doi: 10.1016/j.jaci.2012.10.059. [DOI] [PubMed] [Google Scholar]
- 104.Viallard JF, Blanco P, Andre M, Etienne G, Liferman F, Neau D, Vidal E, Moreau JF, Pellegrin JL. 2006. CD8+HLA-DR+ T lymphocytes are increased in common variable immunodeficiency patients with impaired memory B-cell differentiation. Clin Immunol 119:51–58. doi: 10.1016/j.clim.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 105.Aspalter RM, Sewell WA, Dolman K, Farrant J, Webster AD. 2000. Deficiency in circulating natural killer (NK) cell subsets in common variable immunodeficiency and X-linked agammaglobulinaemia. Clin Exp Immunol 121:506–514. doi: 10.1046/j.1365-2249.2000.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malphettes M, Gerard L, Carmagnat M, Mouillot G, Vince N, Boutboul D, Berezne A, Nove-Josserand R, Lemoing V, Tetu L, Viallard JF, Bonnotte B, Pavic M, Haroche J, Larroche C, Brouet JC, Fermand JP, Rabian C, Fieschi C, Oksenhendler E. 2009. Late-onset combined immune deficiency: a subset of common variable immunodeficiency with severe T cell defect. Clin Infect Dis 49:1329–1338. doi: 10.1086/606059. [DOI] [PubMed] [Google Scholar]
- 107.De Rosa SC, Herzenberg LA, Roederer M. 2001. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med 7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 108.Jameson SC, Masopust D. 2009. Diversity in T cell memory: an embarrassment of riches. Immunity 31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RA. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med 186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lefrancois L, Marzo AL. 2006. The descent of memory T-cell subsets. Nat Rev Immunol 6:618–623. doi: 10.1038/nri1866. [DOI] [PubMed] [Google Scholar]
- 111.Pepper M, Jenkins MK. 2011. Origins of CD4+ effector and central memory T cells. Nat Immunol 12:467–471. doi: 10.1038/nrm3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sallusto F, Geginat J, Lanzavecchia A. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol 22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 113.Di Mitri D, Azevedo RI, Henson SM, Libri V, Riddell NE, Macaulay R, Kipling D, Soares MV, Battistini L, Akbar AN. 2011. Reversible senescence in human CD4+CD45RA+CD27- memory T cells. J Immunol 187:2093–2100. doi: 10.4049/jimmunol.1100978. [DOI] [PubMed] [Google Scholar]
- 114.Takata H, Naruto T, Takiguchi M. 2012. Functional heterogeneity of human effector CD8+ T cells. Blood 119:1390–1398. doi: 10.1182/blood-2011-03-343251. [DOI] [PubMed] [Google Scholar]
- 115.Holling TM, van der Stoep N, Quinten E, van den Elsen PJ. 2002. Activated human T cells accomplish MHC class II expression through T cell-specific occupation of class II transactivator promoter III. J Immunol 168:763–770. doi: 10.4049/jimmunol.168.2.763. [DOI] [PubMed] [Google Scholar]
- 116.Wang R, Green DR. 2012. Metabolic checkpoints in activated T cells. Nat Immunol 13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 117.Stanzani M, Martins SL, Saliba RM, St John LS, Bryan S, Couriel D, McMannis J, Champlin RE, Molldrem JJ, Komanduri KV. 2004. CD25 expression on donor CD4+ or CD8+ T cells is associated with an increased risk for graft-versus-host disease after HLA-identical stem cell transplantation in humans. Blood 103:1140–1146. [DOI] [PubMed] [Google Scholar]
- 118.Brown L, Xu-Bayford J, Allwood Z, Slatter M, Cant A, Davies EG, Veys P, Gennery AR, Gaspar HB. 2011. Neonatal diagnosis of severe combined immunodeficiency leads to significantly improved survival outcome: the case for newborn screening. Blood 117:3243–3246. doi: 10.1182/blood-2010-08-300384. [DOI] [PubMed] [Google Scholar]
- 119.Kwan A, Abraham RS, Currier R, Brower A, Andruszewski K, Abbott JK, Baker M, Ballow M, Bartoshesky LE, Bonilla FA, Brokopp C, Brooks E, Caggana M, Celestin J, Church JA, Comeau AM, Connelly JA, Cowan MJ, Cunningham-Rundles C, Dasu T, Dave N, De La Morena MT, Duffner U, Fong CT, Forbes L, Freedenberg D, Gelfand EW, Hale JE, Hanson IC, Hay BN, Hu D, Infante A, Johnson D, Kapoor N, Kay DM, Kohn DB, Lee R, Lehman H, Lin Z, Lorey F, Abdel-Mageed A, Manning A, McGhee S, Moore TB, Naides SJ, Notarangelo LD, Orange JS, Pai SY, Porteus M, Rodriguez R, et al. 2014. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA 312:729–738. doi: 10.1001/jama.2014.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kimmig S, Przybylski GK, Schmidt CA, Laurisch K, Mowes B, Radbruch A, Thiel A. 2002. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J Exp Med 195:789–794. doi: 10.1084/jem.20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kohler S, Thiel A. 2009. Life after the thymus: CD31+ and CD31- human naive CD4+ T-cell subsets. Blood 113:769–774. doi: 10.1182/blood-2008-02-139154. [DOI] [PubMed] [Google Scholar]
- 122.Honig M, Schwarz K. 2006. Omenn syndrome: a lack of tolerance on the background of deficient lymphocyte development and maturation. Curr Opin Rheumatol 18:383–388. doi: 10.1097/01.bor.0000231907.50290.6f. [DOI] [PubMed] [Google Scholar]
- 123.Santagata S, Villa A, Sobacchi C, Cortes P, Vezzoni P. 2000. The genetic and biochemical basis of Omenn syndrome. Immunol Rev 178:64–74. doi: 10.1034/j.1600-065X.2000.17818.x. [DOI] [PubMed] [Google Scholar]
- 124.Villa A, Notarangelo LD, Roifman CM. 2008. Omenn syndrome: inflammation in leaky severe combined immunodeficiency. J Allergy Clin Immunol 122:1082–1086. doi: 10.1016/j.jaci.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 125.Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, Dagna-Bricarelli F, Sartirana C, Matthes-Martin S, Lawitschka A, Azzari C, Ziegler SF, Levings MK, Roncarolo MG. 2006. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest 116:1713–1722. doi: 10.1172/JCI25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu W, Putnam AL, Zhou X-Y, Szot GL, Lee MR, Zhu S, Gottlieb PA, Kapranov P, Gingeras TR, Fazekas de St Groth B, Clayberger C, Soper DM, Ziegler SF, Bluestone JA. 2006. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med 203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Otsubo K, Kanegane H, Kamachi Y, Kobayashi I, Tsuge I, Imaizumi M, Sasahara Y, Hayakawa A, Nozu K, Iijima K, Ito S, Horikawa R, Nagai Y, Takatsu K, Mori H, Ochs HD, Miyawaki T. 2011. Identification of FOXP3-negative regulatory T-like (CD4+CD25+CD127low) cells in patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Clin Immunol 141:111–120. doi: 10.1016/j.clim.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 128.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. 2010. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol 10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 129.Torgerson TR, Ochs HD. 2007. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked: forkhead box protein 3 mutations and lack of regulatory T cells. J Allergy Clin Immunol 120:744–750. doi: 10.1016/j.jaci.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 130.Dickinson RE, Griffin H, Bigley V, Reynard LN, Hussain R, Haniffa M, Lakey JH, Rahman T, Wang XN, McGovern N, Pagan S, Cookson S, McDonald D, Chua I, Wallis J, Cant A, Wright M, Keavney B, Chinnery PF, Loughlin J, Hambleton S, Santibanez-Koref M, Collin M. 2011. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118:2656–2658. doi: 10.1182/blood-2011-06-360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Harman AN, Bye CR, Nasr N, Sandgren KJ, Kim M, Mercier SK, Botting RA, Lewin SR, Cunningham AL, Cameron PU. 2013. Identification of lineage relationships and novel markers of blood and skin human dendritic cells. J Immunol 190:66–79. doi: 10.4049/jimmunol.1200779. [DOI] [PubMed] [Google Scholar]
- 132.Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, Frucht DM, Vinh DC, Auth RD, Freeman AF, Olivier KN, Uzel G, Zerbe CS, Spalding C, Pittaluga S, Raffeld M, Kuhns DB, Ding L, Paulson ML, Marciano BE, Gea-Banacloche JC, Orange JS, Cuellar-Rodriguez J, Hickstein DD, Holland SM. 2011. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, Shortman K, Wu L, Harrison LC. 2011. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J Immunol 186:6207–6217. doi: 10.4049/jimmunol.1002632. [DOI] [PubMed] [Google Scholar]
- 134.Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR, Arthur DC, Gu W, Gould CM, Brewer CC, Cowen EW, Freeman AF, Olivier KN, Uzel G, Zelazny AM, Daub JR, Spalding CD, Claypool RJ, Giri NK, Alter BP, Mace EM, Orange JS, Cuellar-Rodriguez J, Hickstein DD, Holland SM. 2014. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood 123:809–821. doi: 10.1182/blood-2013-07-515528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Knudson M, Kulkarni S, Ballas ZK, Bessler M, Goldman F. 2005. Association of immune abnormalities with telomere shortening in autosomal-dominant dyskeratosis congenita. Blood 105:682–688. doi: 10.1182/blood-2004-04-1673. [DOI] [PubMed] [Google Scholar]
- 136.Giri N, Alter BP, Penrose K, Falk RT, Pan Y, Savage SA, Williams M, Kemp TJ, Pinto LA. 2015. Immune status of patients with inherited bone marrow failure syndromes. Am J Hematol 90:702–708. doi: 10.1002/ajh.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ballew BJ, Joseph V, De S, Sarek G, Vannier JB, Stracker T, Schrader KA, Small TN, O'Reilly R, Manschreck C, Harlan Fleischut MM, Zhang L, Sullivan J, Stratton K, Yeager M, Jacobs K, Giri N, Alter BP, Boland J, Burdett L, Offit K, Boulton SJ, Savage SA, Petrini JH. 2013. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson syndrome. PLoS Genet 9:e1003695. doi: 10.1371/journal.pgen.1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walter JE, Armanios M, Shah U, Friedmann AM, Spitzer T, Sharatz SM, Hagen C. 2015. Case 41-2015. A 14-year-old boy with immune and liver abnormalities. N Engl J Med 373:2664–2676. doi: 10.1056/NEJMcpc1408595. [DOI] [PubMed] [Google Scholar]
- 139.Aubert G, Baerlocher GM, Vulto I, Poon SS, Lansdorp PM. 2012. Collapse of telomere homeostasis in hematopoietic cells caused by heterozygous mutations in telomerase genes. PLoS Genet 8:e1002696. doi: 10.1371/journal.pgen.1002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, Peters JA, Giri N, Lansdorp PM. 2007. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood 110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Buckley RH. 2011. Transplantation of hematopoietic stem cells in human severe combined immunodeficiency: longterm outcomes. Immunol Res 49:25–43. doi: 10.1007/s12026-010-8191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Griffith LM, Cowan MJ, Notarangelo LD, Kohn DB, Puck JM, Pai SY, Ballard B, Bauer SC, Bleesing JJ, Boyle M, Brower A, Buckley RH, van der Burg M, Burroughs LM, Candotti F, Cant AJ, Chatila T, Cunningham-Rundles C, Dinauer MC, Dvorak CC, Filipovich AH, Fleisher TA, Gaspar HB, Gungor T, Haddad E, Hovermale E, Huang F, Hurley A, Hurley M, Iyengar S, Kang EM, Logan BR, Long-Boyle JR, Malech HL, McGhee SA, Modell F, Modell V, Ochs HD, O'Reilly RJ, Parkman R, Rawlings DJ, Routes JM, Shearer WT, Small TN, Smith H, Sullivan KE, Szabolcs P, Thrasher A, Torgerson TR, Veys P, et al. 2014. Primary Immune Deficiency Treatment Consortium (PIDTC) report. J Allergy Clin Immunol 133:335–347. doi: 10.1016/j.jaci.2013.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Griffith LM, Cowan MJ, Notarangelo LD, Puck JM, Buckley RH, Candotti F, Conley ME, Fleisher TA, Gaspar HB, Kohn DB, Ochs HD, O'Reilly RJ, Rizzo JD, Roifman CM, Small TN, Shearer WT. 2009. Improving cellular therapy for primary immune deficiency diseases: recognition, diagnosis, and management. J Allergy Clin Immunol 124:1152–1160. doi: 10.1016/j.jaci.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aiuti A, Roncarolo MG. 2009. Ten years of gene therapy for primary immune deficiencies. Hematology Am Soc Hematol Educ Program 2009:682–689. doi: 10.1182/asheducation-2009.1.682. [DOI] [PubMed] [Google Scholar]
- 145.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 2010. 20 years of gene therapy for SCID. Nat Immunol 11:457–460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- 146.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 2011. Gene therapy for primary adaptive immune deficiencies. J Allergy Clin Immunol 127:1356–1359. doi: 10.1016/j.jaci.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 147.Hacein-Bey-Abina S, Pai SY, Gaspar HB, Armant M, Berry CC, Blanche S, Bleesing J, Blondeau J, de Boer H, Buckland KF, Caccavelli L, Cros G, De Oliveira S, Fernandez KS, Guo D, Harris CE, Hopkins G, Lehmann LE, Lim A, London WB, van der Loo JC, Malani N, Male F, Malik P, Marinovic MA, McNicol AM, Moshous D, Neven B, Oleastro M, Picard C, Ritz J, Rivat C, Schambach A, Shaw KL, Sherman EA, Silberstein LE, Six E, Touzot F, Tsytsykova A, Xu-Bayford J, Baum C, Bushman FD, Fischer A, Kohn DB, Filipovich AH, Notarangelo LD, Cavazzana M, Williams DA, Thrasher AJ. 2014. A modified gamma-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med 371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ott de Bruin LM, Volpi S, Musunuru K. 2015. Novel genome-editing tools to model and correct primary immunodeficiencies. Front Immunol 6:250. doi: 10.3389/fimmu.2015.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pessach IM, Notarangelo LD. 2011. Gene therapy for primary immunodeficiencies: looking ahead, toward gene correction. J Allergy Clin Immunol 127:1344–1350. doi: 10.1016/j.jaci.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pessach IM, Ordovas-Montanes J, Zhang SY, Casanova JL, Giliani S, Gennery AR, Al-Herz W, Manos PD, Schlaeger TM, Park IH, Rucci F, Agarwal S, Mostoslavsky G, Daley GQ, Notarangelo LD. 2011. Induced pluripotent stem cells: a novel frontier in the study of human primary immunodeficiencies. J Allergy Clin Immunol 127:1400–1407. doi: 10.1016/j.jaci.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bosch M, Khan FM, Storek J. 2012. Immune reconstitution after hematopoietic cell transplantation. Curr Opin Hematol 19:324–335. doi: 10.1097/MOH.0b013e328353bc7d. [DOI] [PubMed] [Google Scholar]
- 152.Gambineri E, Ciullini Mannurita S, Robertson H, Vignoli M, Haugk B, Lionetti P, Hambleton S, Barge D, Gennery AR, Slatter M, Nademi Z, Flood TJ, Jackson A, Abinun M, Cant AJ. 2015. Gut immune reconstitution in immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome after hematopoietic stem cell transplantation. J Allergy Clin Immunol 135:260–262. doi: 10.1016/j.jaci.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seggewiss R, Einsele H. 2010. Immune reconstitution after allogeneic transplantation and expanding options for immunomodulation: an update. Blood 115:3861–3868. doi: 10.1182/blood-2009-12-234096. [DOI] [PubMed] [Google Scholar]
- 154.Cunliffe J, Derbyshire N, Keeler S, Coldwell R. 2009. An approach to the validation of flow cytometry methods. Pharm Res 26:2551–2557. doi: 10.1007/s11095-009-9972-5. [DOI] [PubMed] [Google Scholar]
- 155.Davis BH, Dasgupta A, Kussick S, Han JY, Estrellado A. 2013. Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS - part II - preanalytical issues. Cytometry B Clin Cytom 84:286–290. doi: 10.1002/cyto.b.21105. [DOI] [PubMed] [Google Scholar]
- 156.Owens MA, Vall HG, Hurley AA, Wormsley SB. 2000. Validation and quality control of immunophenotyping in clinical flow cytometry. J Immunol Methods 243:33–50. doi: 10.1016/S0022-1759(00)00226-X. [DOI] [PubMed] [Google Scholar]
- 157.Wood B, Jevremovic D, Bene MC, Yan M, Jacobs P, Litwin V. 2013. Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS - part V - assay performance criteria. Cytometry B Clin Cytom 84:315–323. doi: 10.1002/cyto.b.21108. [DOI] [PubMed] [Google Scholar]
- 158.Pedersen BK, Hoffman-Goetz L. 2000. Exercise and the immune system: regulation, integration, and adaptation. Physiol Rev 80:1055–1081. [DOI] [PubMed] [Google Scholar]
- 159.Suzuki S, Toyabe S, Moroda T, Tada T, Tsukahara A, Iiai T, Minagawa M, Maruyama S, Hatakeyama K, Endoh K, Abo T. 1997. Circadian rhythm of leucocytes and lymphocyte subsets and its possible correlation with the function of the autonomic nervous system. Clin Exp Immunol 110:500–508. doi: 10.1046/j.1365-2249.1997.4411460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bjornson ZB, Nolan GP, Fantl WJ. 2013. Single-cell mass cytometry for analysis of immune system functional states. Curr Opin Immunol 25:484–494. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mei HE, Leipold MD, Schulz AR, Chester C, Maecker HT. 2015. Barcoding of live human peripheral blood mononuclear cells for multiplexed mass cytometry. J Immunol 194:2022–2031. doi: 10.4049/jimmunol.1402661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. 2010. Highly multiparametric analysis by mass cytometry. J Immunol Methods 361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]