Abstract
Vancomycin-resistant Enterococci infections are a significant clinical problem. One proposed solution is to use probiotics, such as lactic acid bacteria, to produce antimicrobial peptides at the site of infection. Enterocin A, a class 2a bacteriocin, exhibits inhibitory activity against E. faecium and E. faecalis, which account for 86% of vancomycin-resistant Enterococci infections. In this study, we aimed to engineer enterocin A mutants with enhanced potency within a lactic acid bacterial production system. Peptide mutants resulting from saturation mutagenesis at sites A24 and T27 were efficiently screened in a 96-well plate assay for inhibition of pathogen growth. Several mutants exhibit increased potency relative to wild-type enterocin A in both liquid- and solid-medium growth assays. In particular, A24P and T27G exhibit enhanced inhibition of multiple strains of E. faecium and E. faecalis, including clinically isolated vancomycin-resistant strains. A24P and T27G enhance killing of E. faecium 8 by 13±3- and 18±4-fold, respectively. The engineered enterocin A/lactic acid bacteria systems offer significant potential to combat antibiotic-resistant infections.
Keywords: antimicrobial peptide, enterocin A, lactic acid bacteria, peptide engineering, saturation mutagenesis
Introduction
Over two million people are infected with antibiotic-resistant bacteria in the United States each year, causing over 23,000 fatalities (CDC, 2013). Antibiotic-resistant bacterial infections undermine many routine medical practices such as chemotherapy, dialysis, transplant, and surgery (CDC, 2013; WHO, 2012). There has been a dramatic increase in antibiotic resistance in the past 50 years, but the development of new classes of antibiotics has not shared the same pace (Amyes, 2000; Laxminarayan et al., 2013). Vancomycin-resistant Enterococci (VRE) are of particular concern to the medical community, as vancomycin is a drug of last resort (CDC, 2013). In 2013, the Centers for Disease Control and Prevention reported 30% of hospital-acquired enterococcal infections were resistant to vancomycin. Of these infections, 86% were attributed to E. faecalis and E. faecium. Both of these species exhibit resistance to multiple antibiotics classes, such as β-lactam, clindamycin, and low levels of aminoglycosides (Arias and Murray, 2012). In addition to antibiotic-resistance and environmental resilience (Hugas et al., 2003; Shepard and Gilmore, 2002), both species of Enterococci have the ability to transfer resistance to other species like Staphylococcus aureus (Arias and Murray, 2012; Huycke et al., 1998; Paulsen et al., 2003). Improved methods to combat VRE are needed.
Antimicrobial peptides (AMPs) are natural products that show promise in antibacterial applications for both the health care and food industries (Drider et al., 2006; Grosu-Tudor et al., 2014; Hu et al., 2014; Kjos et al., 2011a; Snyder and Worobo, 2014; Zhao et al., 2013). AMPs are typically cationic, amphiphilic peptides that serve as defense proteins for the producing organism (Drider et al., 2006; Fimland et al., 2005; Hwang and Vogel, 1998). As AMPs are found in many life forms, there is a wide spectrum of classes that target many different types of cells (Hancock and Diamond, 2000; Peschel and Sahl, 2006). Due to the variety of AMPs that exist, they have the potential to be used for antibacterial treatment (Drider et al., 2006; Hwang and Vogel, 1998; McPhee et al., 2003). Delivery of these AMPs for in vivo applications, however, is not trivial as they can be easily degraded by proteases (Cintas et al., 1997; Giuliani and Rinaldi, 2011; De Kwaadsteniet et al., 2005; Park et al., 2011). One proposed solution is to use lactic acid bacteria (LAB) as a local delivery system to produce the AMPs at the site of infection (Borrero et al., 2014; Geldart et al., 2015). Many LAB are probiotics, including one system used for treatment of Crohn’s disease (Tuohy et al., 2003). Lactococcus lactis, in particular, is used commercially for heterologous production of peptides and has been developed as a delivery system for insulin to the intestinal tract (Morello et al., 2008; Ng and Sarkar, 2011).
Bacteriocins are a specific group of AMPs that are produced by bacteria and, when created by LAB, are typically heat-stable (Drider et al., 2006; Tagg et al., 1976). Bacteriocins have been studied for their potential in antibacterial applications both in the health care and food industries (Drider et al., 2006; Grosu-Tudor et al., 2014; Hu et al., 2014; Kjos et al., 2011a; Snyder and Worobo, 2014; Zhao et al., 2013). Enterocin A (entA) is a class 2a bacteriocin isolated from E. faecium CTC492 and shows inhibition of E. faecalis, Pediococcus, Listeria, and some strains of E. faecium (Aymerich et al., 1996). The potential exists to improve entA’s activity for use in medical applications and food preservation. In general, class 2a bacteriocins are anti-Listeria peptides with up to 80% sequence similarity including β-sheets in the N-terminal portion and amphiphilic α-helixes in the C-terminal portion (Ennahar et al., 2000).
Mutational studies have examined some sequence-function relationships in class 2a bacteriocins. Electrostatic interactions impact binding between cell membranes and AMPs including pediocin PA-1 (Chen et al., 1997) and sakacin P (Kazazic et al., 2002). Reduction in net positive charge of sakacin P decreased activity, but only one in five introductions of a cationic residue (T20K) improved potency (Kazazic et al., 2002). Likewise, only one of three sites of lysine introduction in pediocin PA-1 (S13K) improved efficacy (Song et al., 2014). A disulfide bond in the C-terminal portion of sakacin P and pediocin PA-1 broadens activity and improves activity at an elevated temperature (Fimland et al., 2000). A chimera of the N-terminal portion of enterocin A and the C-terminal portion of pediocin PA-1 was more active than either parent against a strain of Leuconostoc lactis (Tominaga and Hatakeyama, 2007). Shuffling of the N-terminal portion with various class 2a bacteriocins yielded several mutants with varying degrees of improved activity against several bacterial strains. Saturation scanning of pediocin PA-1, albeit with only 35 colonies evaluated per site, indicated mutationally tolerant and intolerant sites, but did not identify improved mutants (Tominaga and Hatakeyama, 2006). Five mutations at each of five sites in pediocin PA-1 revealed a range of tolerant and intolerant mutants but no demonstratively improved mutants (Haugen et al., 2011).
The current study aimed to develop a screen with a sufficient efficiency and throughput to deepen the evaluation of sequence space. Previous efforts to engineer and screen bacterial agents have predominantly relied upon microtiter plate analysis (Geis et al., 1983), agar diffusion tests (Borrero et al., 2011), or colony counting experiments (Deslouches et al., 2014). One example of an innovative way to engineer and test AMPs was used for the peptide bactenecin (Hilpert and Hancock, 2007). Derivative peptides were synthesized on cellulose sheets and tested for activity using a luciferase assay. While the assay enables high-throughput synthesis and analysis of peptide sequences, it does not allow for testing of biological production nor does it yield enough peptide for easily testing multiple pathogens.
In this study, an improved screen was developed for quantifying potency of entA secreted from LAB. Saturation mutagenesis identified several entA/LAB systems with improved activity against E. faecium. In particular, entA mutants A24G, A24P, and T27G showed increased activity. A24P and T27G exhibited improvements against more than one species of Enterococci.
Materials and Methods
Cell Types, Plasmids and Standard Growth Conditions
Supplementary Table S1 lists the bacteria used in this study for production and testing of entA. The production bacteria L. lactis NZ9000 was cultured in GM17, which is M17 media (Oxoid Ltd., Basingstoke, UK) supplemented with 0.5% glucose (Sigma–Aldrich, St. Louis, MO) and 5 μg/mL chloramphenicol (Mediatech, Manassas, VA), at 32°C. Enterococcal strains were grown in brain heart infusion medium (BHI; Becton Dickinson, Franklin Lakes, NJ) at 37°C. For E. faecium 8, BHI media was supplemented with 100 μg/mL ampicillin (Sigma–Aldrich). For plate experiments, the respective media was supplemented with 1.5% wt/vol agar (Sigma–Aldrich).
Assay Development with pBK2idTZ:Bac
The high-throughput assay was designed using pBK2idTZ:Bac (Borrero et al., 2014). AMP production scale was reduced incrementally from 100 mL to 330 μL to ensure AMP activity at each production level including 100 mL, 10 mL, 2 mL and 330 μL (data not shown). Ten microliters of mineral oil (Sigma–Aldrich) was added to the top of 330 μL of growth medium to reduce oxygen transfer. For the mock screen, 62 wells in a 96-well plate were randomly inoculated with L. lactis containing pBK2idTZ:LacZ (producing lacZ as negative control) and 10 wells were inoculated with L. lactis containing pBK2idTZ:Bac (Borrero et al., 2014) (producing entA, hiracin JM79, and enterocin P). Protein production was induced via addition of 50 ng/μL ccF10 pheromone (Mimotopes, Clayton, Australia) when the culture reached an OD600 of 0.4. Production was allowed to continue for 4 h. Cell-free supernatant was obtained by centrifugation of the 96-well plate at 3800g, 4°C for 30 min. The supernatant was then removed and heated at 100°C for 10 min before being stored at −20°C until use. To test for bactericidal activity, 1% v/v supernatant was added to BHI and 106 CFU/mL from an overnight culture of E. faecium. The absorbance at 600 nm was measured at 10, 12, 14, and 16 h on a SpectraMax Plus384 (Molecular Devices, Sunnyvale, CA).
Homology Model
A homology model for entA was prepared from the entA sequence (TTHSGKYYGNGVYCTKNKCTVDWAKATTCIAGMSIGGFLGGAIPGKC) using the I-TASSER server (Roy et al., 2010). The image was rendered using PyMOL.
EntA Construction
The entA gene cassette includes a leader sequence, the bacteriocin, immunity protein, and induction factor (Hancock and Diamond, 2000; Peschel and Sahl, 2006). However, heterologous production of entA can be achieved if the leader sequence is replaced by another signal peptide, in this case usp45 (McPhee et al., 2003; Peschel and Sahl, 2006). This cassette was cloned into a plasmid derivative of pNZ8048, which contains a chloride promoter system (Geldart et al., 2015) (Supplementary Figure S1). Site-specific mutations were made at sites A24 and T27 within the α-helix using NNK degenerate primers from IDT (Supplementary Table S2). The libraries were built using standard molecular cloning techniques (Sambrook et al., 1989). Gel purifications were conducted using Qiagen’s Minelute Kit (Venlo, Netherlands). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA) and digestions were concentrated using Zymo Research’s DNAClean and Concentrator Kit (Irvine, CA). Ligations were transformed into electrocompetent L. lactis NZ9000 using Gene Pulser XCell (Bio-Rad Laboratories, Hercules, CA) and previously described methods (Holo and Nes, 1989).
Production of entA and Mutants with pNZC:entA
L. lactis containing mutant entA plasmids were seeded into a 96-well plate for production and testing. Selecting 90 clones provides a 94% sampling probability for each codon in an NNK library. Saturated overnight cultures were used to inoculate production cultures (at 3% saturation) of 330 μL GM17 (Oxoid) supplemented with 0.1 M NaCl (Sigma–Aldrich) and 5 g/L chloramphenicol and was topped with 10 μL of mineral oil. Protein production was done in triplicate and allowed to continue for 8 h post inoculation. After 8 h, the supernatant was isolated using centrifugation at 3800g, 4°C for 30 min. The supernatant was heated for 10 min at 100°C and then stored at −20°C until tested. AMP was also produced in 2 mL volumes when needed: 1.88 mL GM17 supplemented with 0.1 M NaCl, 3% overnight L. lactis culture and 58 μL mineral oil.
Testing EntA Against E. faecium
The isolated supernatant containing entA was tested in 96 well plates at 10% concentration (90% BHI medium with 100 mg/L ampicillin and pathogen and 10% entA supernatant) against clinical isolate 8 of E. faecium. Growth was monitored every hour for 14 h initially using OD600 on the Synergy H1 152 Multi-Mode Reader (BioTek, Winooski, VT). Derivative entA clones with a comparable (within the parental standard error) or higher activity were isolated for further testing. For every test, new supernatant containing protein was made unless otherwise noted. LacZ, entA, and derivative peptides were then verified by being reproduced at 340 μL scale and tested by monitoring the OD600 of a pathogen culture for 16 h at 3% or 10% v/v concentration. Values presented are the average ± standard error of biological production triplicates evaluated at the average time for lacZ cultures to reach OD600 of 0.8. After the initial screen, each biological production replicate was tested three times in a pathogen test. Statistical significance was calculated in Microsoft Excel, using a one-tailed Student’s t-test with unequal variances.
Stab-on-Agar Tests
BHI was supplemented with 0.8% agar and overnight cultures of 0.5 μL/mL for all E. faecalis strains and 0.75 μL/mL for E. faecium 8. Once the agar solidified, five different colonies of L. lactis expressing A24P, T27G, A24G, entA, or lacZ were spotted onto the plate. The plates were incubated overnight at 37°C. Halo diameter was determined by using ImageJ (Schneider et al., 2012) to calculate the diameter of the inhibition zone minus the size of the colony.
E. faecium Viability Assay
E. faecium 8 was grown overnight and then diluted to 106 CFU/mL in a 5 mL culture containing GM17 supplemented with ampicillin and 10% v/v of supernatant containing either lacZ, entA, A24P, T27G, or entA A24P/T27G. Aliquots were removed every 2 h after the first hour of growth for up to 11 h as well as at 24 h. Ten microliters of tenfold serial dilutions were plated on GM17 medium with ampicillin and allowed to grow for 16 h. Data are presented as average ± standard error of three biological replicates with three technical replicates each.
Results
Development of a High-Throughput Activity Assay for LAB-Generated AMPs
L. lactis have previously been engineered to express three AMPs—entA, hiracin JM79, and enterocin P—under the control of a pathogen-inducible promoter pCF10 (Borrero et al., 2014). Diluted supernatant, at 10% of final culture volume, effectively limits growth of E. faecium 8, a clinical isolate from the University of Minnesota hospital. To enable an efficient high-throughput assay that evaluates pathogen growth inhibition of numerous mutants, we sought to miniaturize and parallelize L. lactis-based production of AMP, processing, and antimicrobial assay. The assay was initially optimized with L. lactis spp. cremoris expressing entA, hiracin JM79, and enterocin P from the pBK2idTZ expression vector inducible by ccF10, an Enterococcal pheromone. In the optimized assay, performed in 96-well plates throughout, (i) L. lactis are cultured in 330 μL of medium plus a mineral oil layer; (ii) AMP production is induced; (iii) cell-free supernatant is acquired by centrifugation and heat killing of L. lactis; (iv) cell-free supernatant is applied to cultures of E. faecium; and (v) E. faecium growth is monitored by optical density. In a mock screen to evaluate system performance, L. lactis harboring the three-AMP plasmid were readily identified versus cells with lacZ control plasmid by dramatically hindering growth of clinical isolate indicator strain, E. faecium 8, at 1% supernatant volume/total volume (v/v) (Fig. 1). Analysis at 10 and 12 h yields no false positives (n = 62 lacZ control samples) and no false negatives (n = 10 AMP samples). At 14 and 16 h, a single false positive or negative (depending on the choice of threshold) was identified.
Figure 1.
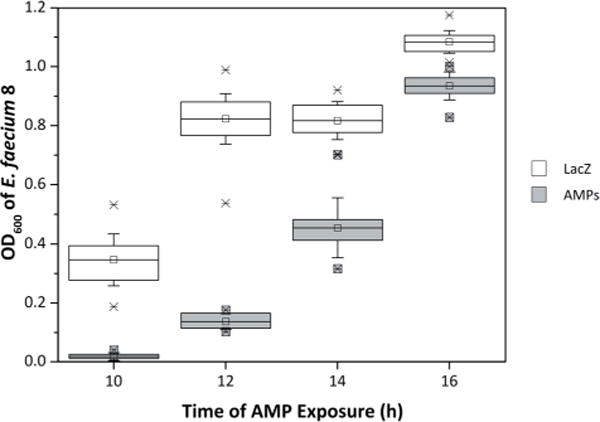
Testing a miniaturized assay using an ideal LAB/AMP system. L. lactis NZ9000 containing plasmid to produce either LacZ (n = 62; white) or a combination of three enterocins (entA, enterocin P, and hiracin JM79; n = 10; gray) were randomly seeded into a 96-well plate. After LAB growth and peptide production, the cell-free supernatant was collected and applied (as 1% v/v) to 106 CFU/mL E. faecium 8 and fresh BHI medium. Pathogen cell density was measured as OD600 at 10, 12, 14, and 16 h. The data are shown in a box and whisker plot where the mean, first quartile, third quartile, standard deviation, and extrema are represented by the centerline, bottom line, top line, whiskers, and X’s, respectively.
Saturation Mutagenesis Analysis of Sites A24 and T27 in EntA
This efficient screen was applied to study sequence/function relationships in entA. The entA gene was cloned with the secretion peptide usp45 and its respective immunity gene into pNZC, a derivative of pNZ8048 containing a chloride inducible promoter (Supplementary Figure S1). The chloride promoter is an environmentally induced promoter that is active within the biological range of chloride concentrations in the intestine (approximately 0.01–0.15 M) (Fordtran and Locklear, 1966; Geldart et al., 2015). Saturation mutagenesis libraries were created at site A24 and, separately, T27 in entA by introducing an NNK codon—which encodes for all 20 natural amino acids—and transforming the resultant plasmids into L. lactis. These two sites were selected because homology modeling predicts that they will be part of the α-helix, which has been implicated in impacting AMP selectivity (Johnsen et al., 2005) (Fig. 2). Moreover, the sites represent two ends of the spectrum for evolutionary conservation. Site 24 is conserved as a small residue (54% G, 22% A, and 14% S, Table I) and has a Shannon entropy (Shannon, 1948) of 1.8. Site 27 is more diverse: 23% T, 21% I, 16% W, and 15% Kwith a Shannon entropy of 2.9. Fifteen sequences from the A24 library and 10 sequences from the T27 library were sequenced with 13 and 9 showing proper construction, respectively. Ninety transformants from each library, which provide an average of 96% probability of sampling for each of the 20 amino acids, were evaluated in triplicate in the assay against E. faecium 8. The E. faecium growth assay used 90% growth medium and 10% supernatant, which is sufficient for parental entA to be modestly potent thereby allowing the ability to observe improved activity. Seven derivatives from the A24X library and 10 derivatives from the T27X library exhibited greater or comparable activity to the parental entA (Fig. 3a). These mutants were sequenced, and their inhibition was tested for reproducibility. Six of seven A24X mutants and nine of ten T24X mutants again inhibited growth (Fig. 3b). Inhibition was generally even stronger in the validation runs. A24P and A24G exhibit especially strong inhibition while A24S and A24T yield intermediate inhibition. T27G, T27N, T27S (TCTor AGT codon but not TCG codon) exhibit strong inhibition while T27E yields intermediate inhibition. Note that TCT and AGT are used in 26% and 23% of L. lactis serine codons whereas TCG is only 5%. Eighteen non-beneficial mutants were also re-tested, which validated reproducibility.
Figure 2.
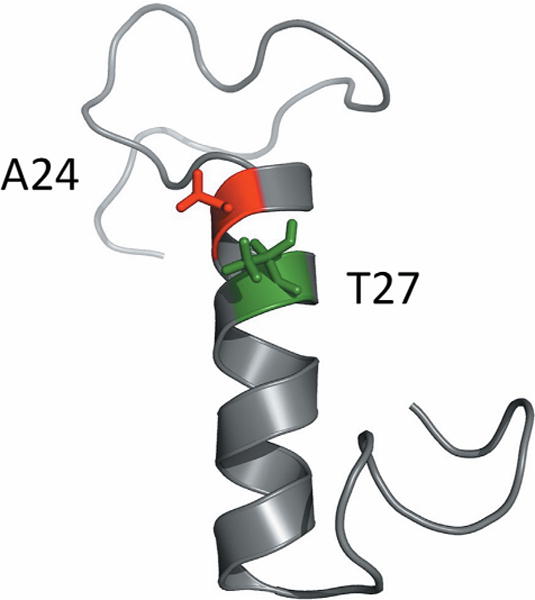
Homology model of entA structure. A homology model for entA was prepared from the entA sequence using the I-TASSER server. The image was rendered using PyMOL. Sites A24 and T27 are labeled.
Table I.
Amino acid frequencies at sites 24 and 27 in entA homologs.
| Amino Acid | Site 24 | Site 27 |
|---|---|---|
| A | 22% | 6% |
| C | 0% | 0% |
| D | 0% | 0% |
| E | 0% | 2% |
| F | 0% | 5% |
| G | 54% | 0% |
| H | 0% | 0% |
| I | 0% | 21% |
| K | 0% | 15% |
| L | 0% | 0% |
| M | 0% | 0% |
| N | 9% | 2% |
| P | 1% | 0% |
| Q | 1% | 1% |
| R | 0% | 3% |
| S | 14% | 5% |
| T | 0% | 23% |
| V | 0% | 0% |
| W | 0% | 16% |
| Y | 0% | 0% |
Natural homologs of entA (n = 138) were identified in the Pfam database. Amino acid frequencies at sites 24 and 27 were calculated. The impact of identical sequences was dampened by counting the square root of their occurrence.
Figure 3.
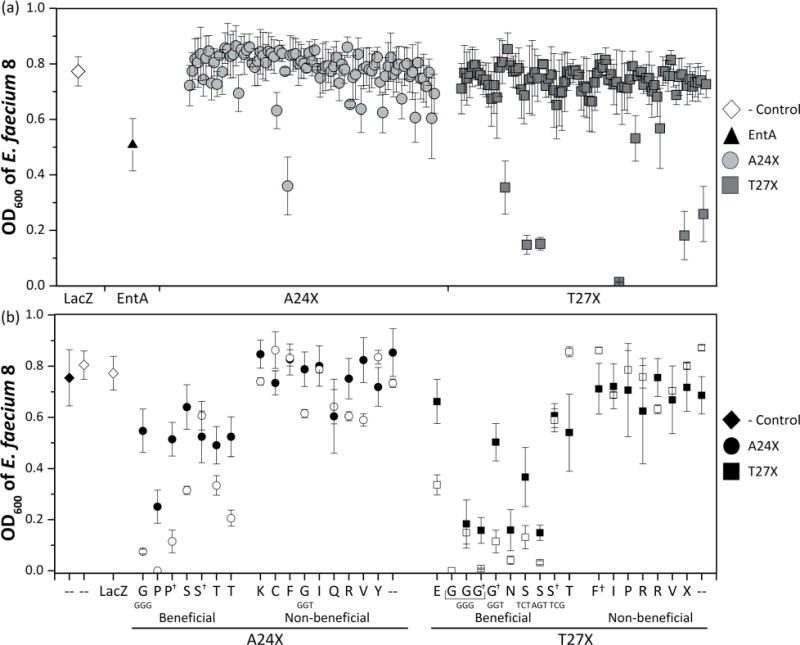
Screening of A24X and T27X libraries. (a) A24X and T27X mutant libraries were constructed and 90 colonies were seeded into 96-well plates. Biological triplicates of AMP were produced and applied (as 10% v/v) to 106 CFU/mL E. faecium 8. Pathogen cell density (OD600) after 9 h of AMP exposure is plotted as average ± standard error. Seventeen derivative AMPs were selected for additional testing based on their activity compared to the parental enterocin A. (b) The 17 functional clones were retested to identify false positives, and eighteen random ineffective AMPs were retested to identify false negatives. Average and standard error (n = 3) are shown for both the initial (solid) and the verification (empty) assays. The mutant amino acid and codon (if different—otherwise Supplementary Table S3) are indicated. † denotes a mutation in the amino acid sequence in either the secretion peptide or immunity gene for entA.
Four beneficial mutants exhibited mutations in the immunity gene in addition to site 24 or 27. In two cases, the corresponding entA mutant (A24S or T27G) was also observed with a wild-type immunity gene allowing direct evaluation of the immunity gene mutation, however no impact was observed. The immunity mechanism is not fully understood (Kjos et al., 2011a), but an immunity gene mutation could perhaps hinder L. lactis health or entA production. The growth of the L. lactis was monitored for significant variations, but none were observed. A24P+ exhibited a mutation in the secretion peptide causing reduced, but functional, pathogen inhibition.
To identify the most potent A24X and T27X mutants for further evaluation and development, the stringency of the assay was increased by reducing the supernatant concentration to 3% v/v in the pathogen culture. Three mutants maintained strong inhibition relative to parental activity: A24G (P = 0.0035), A24P (P = 0.0097), and T27G (P = 0.059) (Fig. 4).
Figure 4.
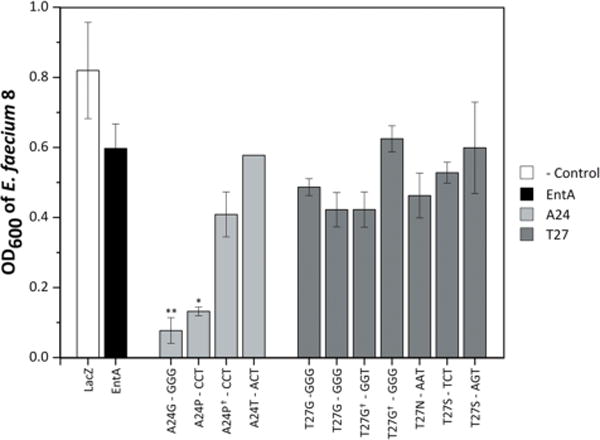
Supernatant activity at 3%. For increased stringency, the eleven AMP derivatives maintaining pathogen density at or below OD600 = 0.2 with 10% supernatant (Fig. 3) were tested at 3% v/v against E. faecium 8. The supernatant production and processing were conducted as previously described (n = 3 with standard error bars for all supernatant production except A24T for which n =2 therefore no error bars were added). Cell densities were monitored every hour for 16 h; the data shown is at the average time it takes lacZ to reach an OD600 of 0.8 to allow for comparison across multiple days. A one-tailed Student’s t-test compared mutants to entA. *indicates P≤0.05, and **indicates P≤0.01.
Growth Inhibition of E. faecium and E. faecalis Strains
The three lead mutants, A24G, A24P, and T27G, were evaluated in spot-on-agar tests in which AMP-producing L. lactis is cultured in a spot on an agar plate harboring E. faecium. Potent LAB/AMP systems are capable of inhibiting pathogen growth in a zone surrounding the L. lactis spot (Fig. 5a). All three entA mutants exhibited superior growth inhibition of E. faecium 8 relative to wild-type entA (Fig. 5b). The spot-on-agar assays were expanded to five E. faecalis strains (Fig. 5b). T27G showed improved growth inhibition, relative to wild-type entA, against all five E. faecalis strains (P < 0.03), while A24P showed improved activity against three of the five E. faecalis strains (OG1RF, V583, and CH116) and comparable activity on Com1 and JH2-2. A24G showed reduced activity, yet still inhibited growth relative to lacZ control, against all strains of E. faecalis (Fig. 5b).
Figure 5.
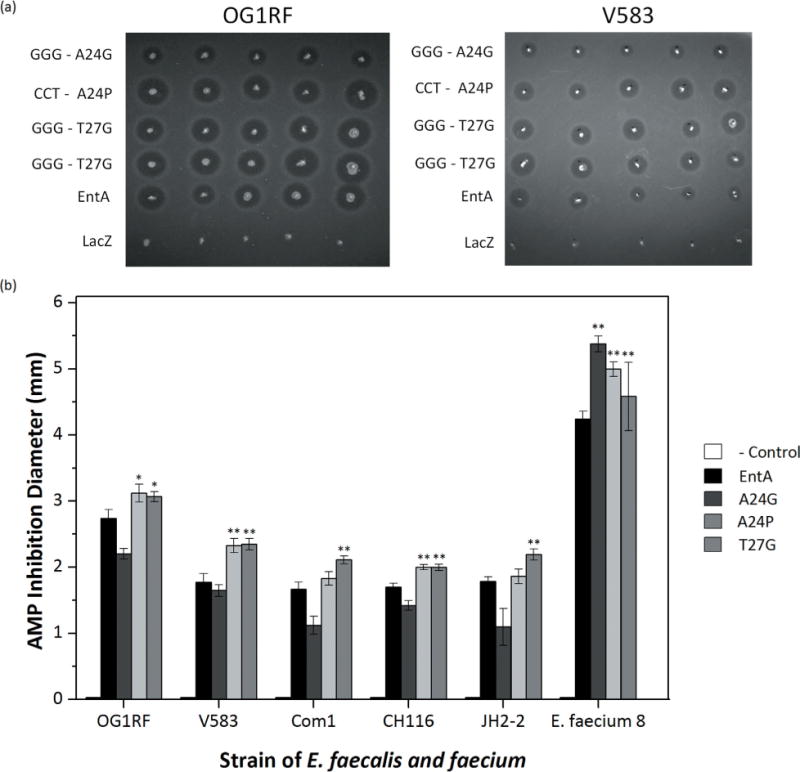
EntA and derivatives tested against multiple pathogens. (a) A24G, A24P, and T27G constructs were tested against multiple species of E. faecalis using a stab-on agar test. BHI with 0.8% agar and 0.5 μL/mL (E. faecalis) or 0.75 μL/mL (E. faecium) were allowed to solidify. Five colonies of L. lactis containing each construct were stabbed on to agar containing pathogen and allowed to grow at 37°C overnight. (b) The average halo diameter was measured using ImageJ. A one-tailed Student’s t-test compared mutants to entA. *indicates P≤0.05, and **indicates P≤0.01.
A24P and T27G were also evaluated in liquid pathogen inhibition assays against several strains of E. faecalis and E. faecium. The mutants exhibited improved efficacy, relative to wild-type entA, against two of three strains of E. faecalis (Com1 and OG1RF whereas comparable inhibition was observed for V583) and two additional clinically isolated strains of E. faecium (E. faecium 6 and 7) (Fig. 6).
Figure 6.
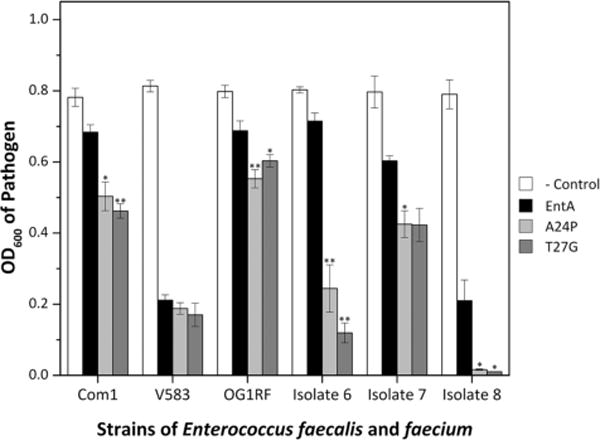
EntA and derivatives tested against multiple pathogens in liquid culture. The A24G, A24P, and T27G constructs were evaluated in liquid phase pathogen inhibition assays as in Figure 3 but against six strains of Enterococci. The average and standard error (n = 3) of pathogen cell density is shown at the time for which lacZ control reached OD600 = 0.8. A one-tailed Student’s t-test compared mutants to entA. *indicates P≤0.05, and **indicates P≤0.01.
Pathogen Viability
The reduction in pathogen cell density several hours post-exposure to entA and its mutants could result from cell death, reduction in growth rate, or a combination thereof. To evaluate these possibilities, viable cell density was measured dynamically starting at the time of initial exposure to 10% v/v entA-containing supernatant. The viable E. faecium density decreases to 5.4% ± 0.2% of initial upon exposure to entA (Fig. 7). A24P and T27G exhibit greater potency with a reduction of viable E. faecium density to 0.4% ± 0.1% and 0.31% ± 0.06% of initial, respectively (Fig. 7a). After 7 h, entA and the two mutants exhibit cell growth rates comparable to lacZ control. At 24 h, inhibition is not evident, which is likely due to pathogen resistance as previously demonstrated for class 2a bacteriocins (Geldart et al., 2015; Kjos et al., 2011a,b).
Figure 7.
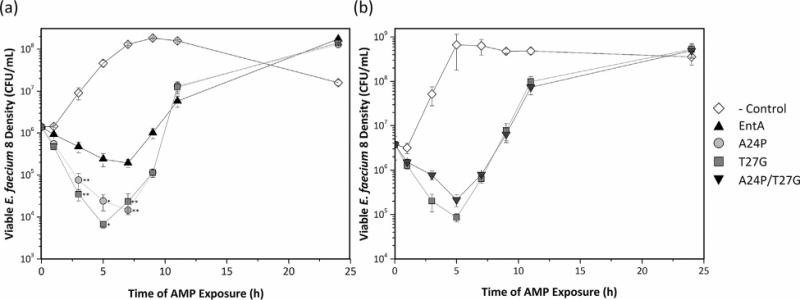
E. faecium viability assays. LAB/AMP supernatant was produced at 2 mL scale and applied (at 10% v/v) to 5 mL E. faecium 8 (initially at 0.6% density from an overnight culture). Cultures were incubated at 37°C for 24 h. Viable cell density was measured by dilution plating on GM17 medium at 0, 1, 3, 5, 7, 9, 11, and 24 h. (a) A24P and T27G constructs are compared to those of the parental entA and the control protein, lacZ. (b) The double mutant containing both mutations was compared to T27G and lacZ.
A24P/T27G Double Mutant
A double mutant containing both mutations A24P and T27G was built and tested to see if the mutational benefits were additive. When monitoring OD600, entA A24P/T27G exhibited similar behavior to both T27G and A24P (data not shown). E. faecium viability measurements were performed to compare the double mutant to lacZ and T27G. In these tests entA A24P/T27G did not perform as well as T27G, with 2.2 ± 0.6-fold less killing (Fig. 7b). After 5 h, A24P/T27G and T27G grew similarly to the lacZ control.
Discussion
VRE pose a significant threat to the modern healthcare system (CDC, 2013); as such, innovative methods for discovery of new or improved antimicrobial systems is imperative. AMPs are one proposed solution to treating antibiotic-resistant infections. LAB have been posed as a potential production host (Bermúdez-Humarán et al., 2011; Martín et al., 2007; Morello et al., 2008) and delivery vehicle (Borrero et al., 2014; Geldart et al., 2015; Ng and Sarkar, 2011). Enhanced potency of LAB/AMP systems would be advantageous. Previous studies have predominantly reported production of AMPs in cultures greater than 10 mL (Aymerich et al., 1996; Van Den Berghe et al., 2006; Borrero et al., 2011; Casaus et al., 1997; Rehaiem et al., 2011), which are inefficient for combinatorial library-based approaches. Miniaturization of the assay drives efficient parallelization, which accommodates a more extensive search of derivative AMPs, production strains, and different types of pathogens. Use of a liquid assay allows for dynamic growth evaluation using simple OD600 measurements. The developed assay performed well in the mock screen with the triple-AMP system (no false positives or negatives at 10 and 12 h; a single false positive/negative at 14 and 16 h). When used for discovery of entA mutants with enhanced inhibition of E. faecium 8, initial hits of substantial inhibition were validated in a follow-up assay while mutants with slight inhibition yielded modest variance (Fig. 3b).
The parallelized assay empowered site saturation mutagenesis (probability of sampling each amino acid is 94%, on average, and 92%, minimum when accounting for library construction quality) at sites 24 and 27. Despite focusing on only two sites in this initial study, mutants of substantial improvement were identified. A24P and T27G kill E. faecium 8 to a 13 ± 3-fold and 18 ± 4-fold greater extent than parental entA, respectively, (Fig. 7) and yield significant potency even at 3% v/v (Fig. 4). Moreover, the mutants exhibit enhanced activity against numerous strains of E. faecium and E. faecalis in both liquid and spot-on-agar assays (Figures 5 and 6). The lone exceptionwas E. faecalis V583, against which the mutants exhibited comparable activity relative to entA. This could be due to a number of reasons including, but not limited to: downregulation of the mannose phosphotransferase system for sugar transport; strain-specific variations within the mannose phosphotransferase sequence; or use of a wider variety of sugar transporters (Kjos et al., 2011a,b). Importantly, the mutant plasmids were also rebuilt to verify that the indicated entA mutations were responsible for the improved performance rather than extraneous mutations or LAB strain modifications; inhibitory performance was maintained with the rebuilt plasmids introduced into fresh L. lactis. Thus, saturation mutagenesis at even two sites is sufficient to identify improvements in antimicrobial activity. Future studies can explore additional sites and mutant combinations, empowered by the efficient screening assay. It remains unclear whether the peptides show increased activity due to increased production, increased specific activity, or a combination thereof.
The utility of an efficient screen to assess diverse mutants in a combinatorial manner is noteworthy since the most effective mutants were unlikely to be identified via rational approaches. Although sites 24 and 27 have different diversities in natural homologs, both sites offer significant evolvability relative to wild-type as four amino acids at each site provided superior inhibition relative to wild-type. It is intriguing that, despite the presumed existence—based on homology modeling—of A24 and T27 within the core of the entA alpha helix (Fig. 2), the three most effective mutations (A24G, T27G, and A24P) introduce amino acids that are less frequently observed in α helices (Blaber et al., 1993; Pace and Scholtz, 1998). Structural studies will be needed to evaluate the impacts on this particular helix. All four improved mutations from A24 (P, G, T, and S) introduce a side chain with increased hydrophilicity relative to wild-type. Similarly, three of the four improved mutations from T27 (N, S, and E) increase hydrophilicity, while the best mutation, T27G, provides comparable hydrophilicity.
One of the top mutants, A24G introduces a mutation that is dominant among entA homologs (57% G at site 24 within 138 natural entA homologs (Table 1)). While this result is consistent with the concept of consensus design (Steipe et al., 1994), the other top mutations conflicted with this approach (Fig. 8). At site 24, proline is observed only once in 138 natural entA homologs. Similarly, glycine is not seen at site 27 in any of the homologous peptides. Yet site T28 is glycine 18% of the time, suggesting local tolerance for conformationally flexible glycine. In short, a consensus design approach would have yielded false negative designs (A24P and T27G)—mutants predicted to be non-functional that instead were advantageous. Moreover, the thorough search of sequence space enables identification of false positive consensus designs—mutants that are frequent in natural homologs but not advantageous in inhibiting E. faecium: T27I (21% naturally but non-inhibitory (Fig. 3) as well as T27W and T27K, which are present 16% and 15% naturally but were not identified as inhibitory mutants (each with a 92% likelihood of being evaluated in the 90 clones tested). Thus, amino acid frequency in natural homologs is insufficient for identifying beneficial mutations; combinatorial library screening provides an efficient means to evaluate sequence-function relationships and identify functional AMPs.
Figure 8.
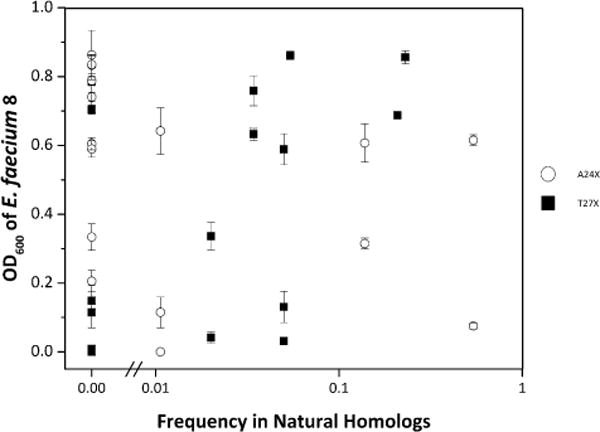
Inefficiency of consensus design. The E. faecium density after exposure to 10% v/v supernatant (from Fig. 3b) is plotted versus the frequency of the mutant amino acid in natural homologs.
The thorough evaluation of sequence space is also beneficial for evaluating codon-dependent effects. For one of the top amino acid mutations, T27G, both of the possible NNK-encoded codons (GGG and GGT) were identified as improved relative to parental entA. Conversely, for A24P, the CCT codon mutant was identified but CCG was not. This can potentially be explained by the rare use of the CCG codon in L. lactis (only 5% of P encoded by CCG vs. 20% by CCT). Alternatively, it is possible that the CCG codon mutant was not present in the 90 clones analyzed (8% likelihood of omission). For A24G, the GGG mutant was identified as a significant improvement relative to parental entA whereas the GGT mutant was ineffective. The failure of the GGT mutant cannot be explained by rare codon usage as GGT is used to encode for 39% of glycines in L. lactis. While further experimentation would be needed to elucidate the alternative regulatory mechanisms responsible for these results, it is important to note the value of thorough analysis of genetic sequence space in addition to amino acid sequence space.
Conclusions
VRE pose a significant threat to the modern healthcare system both through the increased risk of an antibiotic-resistant infection and the possibility of horizontal gene transfer (Huycke et al., 1998; Palmer et al., 2010). Historically, optimizing compounds from other organisms’ defense mechanisms has led to the development of effective antibiotics. In a similar fashion, we have worked to optimize entA efficacy against multiple strains of E. faecalis and E. faecium. We developed a high-throughput assay to analyze the antimicrobial activity of an LAB/AMP system and applied it to saturation mutagenesis studies at A24 and T27 of entA. Through this system we identified two mutants—A24P and T27G—that kill pathogenic Enterococcus strains to a greater extent than native entA.
Supplementary Material
Acknowledgments
We thank Dr. Patricia Ferrieri and Dr. Gary Dunny at the University of Minnesota Medical School for supplying the Entercoccal strains. This work was supported by a National Institutes of Health Biotechnology Training Grant (GM008347). This work was partially supported by a grant from the National Institutes of Health (GM111358) and by a grant from the National Science Foundation (CBET-1412283).
Contract grant sponsor: National Institutes of Health Biotechnology Training
Contract grant number: GM008347
Contract grant sponsor: National Institutes of Health
Contract grant number: GM111358
Contract grant sponsor: National Science Foundation
Contract grant number: CBET-1412283
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Amyes S. The rise in bacterial resistance. Br Med J. 2000;320:199–200. doi: 10.1136/bmj.320.7229.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Murray BE. The rise of the Enterococcus: Beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich T, Holo H, Håvarstein LS, Hugas M, Garriga M, Nes IF. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol. 1996;62:1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-Humarán LG, Kharrat P, Chatel J-M, Langella P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb Cell Fact. 2011;10:S4. doi: 10.1186/1475-2859-10-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaber M, Zhang X-J, Matthews BW. Structural basis of amino acid α-helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- Borrero J, Chen Y, Dunny GM, Kaznessis YN. Modified lactic acid bacteria detect and inhibit multiresistant enterococci. ACS Synth Biol. 2014;4:299–306. doi: 10.1021/sb500090b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrero J, Jiménez JJ, Gútiez L, Herranz C, Cintas LM, Hernández PE. Protein expression vector and secretion signal peptide optimization to drive the production, secretion, and functional expression of the bacteriocin enterocin A in lactic acid bacteria. J Biotechnol. 2011;156:76–86. doi: 10.1016/j.jbiotec.2011.07.038. [DOI] [PubMed] [Google Scholar]
- Casaus P, Nilsen T, Cintas LM, Nes IF, Herndndez PE, Holo H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology. 1997;143:2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- CDC. Antibiotic resistance threats in the United States, 2013. Washington, DC: CDC; 2013. [Google Scholar]
- Chen Y, Ludescher RD, Montville TJ. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl Environ Microbiol. 1997;63:4770–4777. doi: 10.1128/aem.63.12.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintas LM, Casaus P, Håvarstein LS. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol. 1997;63:4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kwaadsteniet M, Todorov SD, Knoetze H, Dicks LMT. Characterization of a 3944 Da bacteriocin, produced by Enterococcus mundtii ST15, with activity against Gram-positive and Gram-negative bacteria. Int J Food Microbiol. 2005;105:433–444. doi: 10.1016/j.ijfoodmicro.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL, Montelaro RC. Engineered cationic antimicrobial peptides (eCAPs) to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother. 2014;59:1329–1333. doi: 10.1128/AAC.03937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev. 2006;70:564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennahar S, Sashihara T, Sonomoto K, Ishizaki A. Class IIa bacteriocins: Biosynthesis, structure and activity. FEMS Microbiol Rev. 2000;24:85–106. doi: 10.1111/j.1574-6976.2000.tb00534.x. [DOI] [PubMed] [Google Scholar]
- Fimland G, Johnsen L, Axelsson L, May B, Nes IF, Eijsink VGH. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J Bacteriol. 2000;182:2643–2648. doi: 10.1128/jb.182.9.2643-2648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimland G, Johnsen L, Dalhus B, Nissen-Meyer J. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: Biosynthesis, structure, and mode of action. J Pept Sci. 2005;11:688–696. doi: 10.1002/psc.699. [DOI] [PubMed] [Google Scholar]
- Fordtran JS, Locklear TW. Ionic constituents and osmolality of gastric and small-intestinal fluids after eating. Am J Dig Dis. 1966;11:503–521. doi: 10.1007/BF02233563. [DOI] [PubMed] [Google Scholar]
- Geis A, Singh J, Teuber M. Potential of lactic streptococci to produce bacteriocin. Appl Environ Microbiol. 1983;45:205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldart K, Borrero J, Kaznessis YN. A chloride-inducible expression vector for delivery of antimicrobial peptides against antibiotic-resistant Enterococcus faecium. Appl Environ Microbiol. 2015;81:3889–3897. doi: 10.1128/AEM.00227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani A, Rinaldi AC. Beyond natural antimicrobial peptides: Multimeric peptides and other peptidomimetic approaches. Cell Mol Life Sci. 2011;68:2255–2266. doi: 10.1007/s00018-011-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosu-Tudor S-S, Stancu M-M, Pelinescu D, Zamfir M. Characterization of some bacteriocins produced by lactic acid bacteria isolated from fermented foods. World J Microbiol Biotechnol. 2014;30:2459–2469. doi: 10.1007/s11274-014-1671-7. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- Haugen HS, Fimland G, Nissen-Meyer J. Mutational analysis of residues in the helical region of the class IIa bacteriocin pediocin PA-1. Appl Environ Microbiol. 2011;77:1966–1972. doi: 10.1128/AEM.02488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilpert K, Hancock REW. Use of luminescent bacteria for rapid screening and characterization of short cationic antimicrobial peptides synthesized on cellulose using peptide array technology. Nat Protoc. 2007;2:1652–1660. doi: 10.1038/nprot.2007.203. [DOI] [PubMed] [Google Scholar]
- Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Mao R, Zhang Y, Teng D, Wang X, Xi D, Huang J, Wang J. Biotechnical paving of recombinant enterocin A as the candidate of anti-Listeria agent. BMC Microbiol. 2014;14:220. doi: 10.1186/s12866-014-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugas M, Garriga M, Aymerich MT. Functionalty of enterococci in meat products. Int J Food Microbiol. 2003;88:223–233. doi: 10.1016/s0168-1605(03)00184-3. [DOI] [PubMed] [Google Scholar]
- Huycke MM, Sahm DF, Gilmore MS. Multiple-drug resistant enterococci: The nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PM, Vogel HJ. Structure-function relationships of antimicrobial peptides. Biochem Cell Biol. 1998;76:235–246. doi: 10.1139/bcb-76-2-3-235. [DOI] [PubMed] [Google Scholar]
- Johnsen L, Fimland G, Nissen-Meyer J. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem. 2005;280:9243–9250. doi: 10.1074/jbc.M412712200. [DOI] [PubMed] [Google Scholar]
- Kazazic M, Nissen-Meyer J, Fimland G. Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology. 2002;148:2019–2027. doi: 10.1099/00221287-148-7-2019. [DOI] [PubMed] [Google Scholar]
- Kjos M, Borrero J, Opsata M, Birri DJ, Holo H, Cintas LM, Snipen L, Hernández PE, Nes IF, Diep DB. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology. 2011a;157:3256–3267. doi: 10.1099/mic.0.052571-0. [DOI] [PubMed] [Google Scholar]
- Kjos M, Nes IF, Diep DB. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol. 2011b;77:3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ. Antibiotic resistance — the need for global solutions. Lancet Infect Dis Comm. 2013;3099:1–42. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Martín M, Gutiérrez J, Criado R, Herranz C, Cintas LM, Hernández PE. Cloning, production and expression of the bacteriocin enterocin A produced by Enterococcus faecium PLBC21 in Lactococcus lactis. Appl Microbiol Biotechnol. 2007;76:667–675. doi: 10.1007/s00253-007-1044-3. [DOI] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, Hancock REW. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- Morello E, Bermúdez-Humarán LG, Llull D, Solé V, Miraglio N, Langella P, Poquet I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol. 2008;14:48–58. doi: 10.1159/000106082. [DOI] [PubMed] [Google Scholar]
- Ng DTW, Sarkar CA. Nisin-inducible secretion of a biologically active single-chain insulin analog by Lactococcus lactis NZ9000. Biotechnol Bioeng. 2011;108:1987–1996. doi: 10.1002/bit.23130. [DOI] [PubMed] [Google Scholar]
- Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol. 2010;13:632–639. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SC, Park Y, Hahm KS. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int J Mol Sci. 2011;12:5971–5992. doi: 10.3390/ijms12095971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Peschel A, Sahl H-G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- Rehaiem A, Guerra NP, Belgacem Z, Ben Bernárdez PF, Castro LP, Manai M. Enhancement of enterocin A production by Enterococcus faecium MMRA and determination of its stability to temperature and pH. Biochem Eng J. 2011;56:94–106. [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Edward F, Maniatis T, Laboratory CSH. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:623–656. [Google Scholar]
- Shepard BD, Gilmore MS. Antibiotic-resistant enterococci: The mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 2002;4:215–224. doi: 10.1016/s1286-4579(01)01530-1. [DOI] [PubMed] [Google Scholar]
- Snyder AB, Worobo RW. Chemical and genetic characterization of bacteriocins: Antimicrobial peptides for food safety. J Sci Food Agric. 2014;94:28–44. doi: 10.1002/jsfa.6293. [DOI] [PubMed] [Google Scholar]
- Song DF, Li X, Zhang YH, Zhu MY, Gu Q. Mutational analysis of positively charged residues in the N-terminal region of the class IIa bacteriocin pediocin PA-1. Lett Appl Microbiol. 2014;58:356–361. doi: 10.1111/lam.12197. [DOI] [PubMed] [Google Scholar]
- Steipe B, Schiller B, Pluckthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J Mol Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- Tagg JR, Dajani AS, Wannamaker LW. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Hatakeyama Y. Determination of essential and variable residues in pediocin PA-1 by NNK scanning. Appl Environ Microbiol. 2006;72:1141–1147. doi: 10.1128/AEM.72.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Hatakeyama Y. Development of innovative pediocin PA-1 by DNA shuffling among class IIa bacteriocins. Appl Environ Microbiol. 2007;73:5292–5299. doi: 10.1128/AEM.00558-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuohy KM, Probert HM, Smejkal CW, Gibson GR. Using probiotics and prebiotics to improve gut health. Drug Discov Today. 2003;8:692–700. doi: 10.1016/s1359-6446(03)02746-6. [DOI] [PubMed] [Google Scholar]
- Van Den Berghe E, De Winter T, De Vuyst L. Enterocin A production by Enterococcus faecium FAIR-E 406 is characterised by a temperature- and pH-dependent switch-off mechanism when growth is limited due to nutrient depletion. Int J Food Microbiol. 2006;107:159–170. doi: 10.1016/j.ijfoodmicro.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Martinez Lindsay., editor. WHO. The evolving threat of antimicrobial resistance: Options for action. 1st. Geneva, Switzerland: World Health Organization; 2012. [Google Scholar]
- Zhao J, Zhao C, Liang G, Zhang M, Zheng J. Engineering antimicrobial peptides with improved antimicrobial and hemolytic activities. J Chem Inf Model. 2013;53:3280–3296. doi: 10.1021/ci400477e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


