Abstract
22q11.2 deletion syndrome (22q11DS) is one of the most common recurrent copy-number variant disorder, caused by a microdeletion in chromosome band 22q11.2 and occurring with a population prevalence of 1 in 2000. Until today there has been no evidence that the size of the deletion has an influence on the clinical phenotype. Most studies report that 22q11DS is associated with mild or borderline intellectual disability. There are a limited number of reports on 22q11DS subjects with moderate or severe intellectual disability.
In this study we describe 63 adult patients with 22q11DS, including 22q11DS patients functioning at a moderate to severe intellectual disabled level. Deletion size was established with an experimental Multiplex ligation-dependent probe amplification (MLPA) mixture (P324) in addition to the commonly used MLPA kit (P250). We compared deletion size with intellectual functioning and presence of psychotic symptoms during life. The use of the experimental MLPA kit gives extra information on deletion size, only when combined with the common MLPA kit. We were able to detect eleven atypical deletions and in two cases the deletion size was shorter than all other “typical ones”. We conclude that the use of the experimental kit P324 gives extra information about the deletion size, but only when used together with the standard P250 kit. We did not found any relation of deletion size with intelligence or presence of psychosis.
Keywords: 22q11 deletion syndrome, cognitive deterioration, intelligence, psychopathology, MLPA, deletion size
Introduction
22q11.2 deletion syndrome (22q11DS) is one of the most common recurrent copy-number variant disorders caused by a microdeletion in chromosome band 22q11.2. Characteristic clinical features include palatal and cardiac anomalies, hypo- or aplasia of the thymus, hypoplasia of the parathyroid, and typical facial features. The physical appearance is known to be highly variable [1–3]. Its prevalence and birth incidence is still under debate. Because of the strong variance in phenotype some patients with a deletion are not immediately identified or diagnosed at birth. Therefore in the literature differences in birth incidence are reported, ranging from 1 in 2000 [4] to 1 in 6000 [5]. Recently Grati et al. [6] reported a birth incidence of 1:992 in a cohort of over 9.500 pregnancies, which indicates that reported prevalence rates are often underestimations of the incidence during pregnancy. As some of the physical abnormalities of 22q11DS are not compatible with life, prevalence rates will be lower, but because of the variability of penetrance of the deletion, some patient with a deletions will remain undiscovered until adulthood. The microdeletion can be diagnosed with e.g. fluorescence in situ hybridization (FISH) [7], multiplex ligation-dependent probe amplification (MLPA) [8] or array comparative genomic hybridization (aCGH) analysis [9].
Approximately half of all subjects with 22q11DS have a normal to borderline intelligence [10–14] and if an intellectual disability is present, this is usually mild [15]. The presence of a profound or severe intellectual disability was thought to be rare. Patients with 22q11DS functioning at a moderate to severe intellectual disability level (full scale intelligence (FSIQ) < 55) have been described less frequently in the literature and concern mostly case reports [16–20] but have recently received more attention [21]. Thus far there has been no evidence that the deletion size is related to the symptoms seen in 22q11DS, nor psychopathology or intelligence [22], but this conclusion is now under debate [23]. Atypical findings in conotruncal heart defects in 22q11DS, are possibly related to atypical deletion sizes [24]. The relation between deletion size and intellectual functioning covering the broader intellectual range has not yet been investigated in 22q11DS. Deletions in the 22q11DS region are described as LCR22-A, LCR22-B, LCR22-C, LCR22-D, LCR22-E, LCR22-F and LCR22-G corresponding to the known Low Copy Repeat (LCR) regions that are involved in the deletion. The deletions described as typical have a length of 3 Mb and range from LCR22-A to LCR22-D [8]. MLPA analysis in 22q11DS is mostly done with the standard MLPA kit (P250, MRC-Holland). In the experimental MLPA P324 kit (MRC-Holland) different probes are used, including 3 Proline Dehydrogenase (PRODH) probes. PRODH is responsible for conversion of proline to glutamate and it has been suggested that it may be of importance in several psychopathological mechanisms in 22q11DS [25–29]. Also, 12 additional T-Box1 (TBX1) probes are included in the P324 kit (the P250 kit has only two TBX1 probes) (supplementary on-line material, table 1). TBX1 is one of the most important candidate genes at 22q11.2 concerning physical abnormalities in 22q11DS [30, 31], but it is also associated with psychiatric features like autism spectrum disorders [32] and in mouse models with social interaction problems [33] and autism [34].
We explored whether the research MLPA kit could give additional information about the deletion size compared with the standardized MLPA kit. In this study also 22q11DS subjects functioning at an intellectual level lower than 55 were included, covering a broader range of this syndrome. The aim was to explore a potential relation of deletion size and intelligence, and furthermore a potential relationships between deletion size and presence of psychosis during life, as a relation between psychosis and intellectual decline has recently been described [21].
Method
The study was approved by the Medical Ethics Committee of the University of Maastricht, The Netherlands and the Academic Medical Centre, Amsterdam, The Netherlands and is part of larger research project in 22q11DS patients.
Study population
Participants were recruited through the Dutch 22q11DS family network, a specialized psychiatric 22q11DS outpatient clinic, and through several learning disability centres in The Netherlands. Patients with a confirmed deletion at chromosome 22q11.2 (established with MLPA P250 kit) were included. Fifty-four out of sixty-three (86%) had the typical deletion of 3 Mb length from CLTCL1 to LZTR1 (type A–D deletion). Atypical deletions were: six deletions from CTLC1 to DGCR8 (type A–B deletion); two deletions from CTLC1 to PCQAP (type A–C deletion); one deletion ranging from CLTCL1 to HIC2 (A–D deletion including HIC2) and one A–D deletion with a concomitant distal duplication (from SMARCB1 to SNRPD3, F–G duplication). Those aged under 18 years and over the age of 65 years were excluded. A number of participants gave written consent, and where participants were not able to do so, consent was obtained from carers. We included sixty-four patients with 22q11DS.
FSIQ measurements
Full Scale Intelligence Quotient (FSIQ) scores were obtained in the 22q11DS group using a shortened version of the Wechsler Adult Intelligence Scale (WAIS) version III [35]. Patients unable to perform a WAIS, were investigated using a Vineland-screener. The outcome of this test was converted to a FSIQ rating as described earlier [36].
Psychosis outcome measures
All patients were assessed for life-time presence of a psychotic disorder, based on information obtained from medical records and/or the present score on the Mini PAS-ADD, (Psychiatric Assessment Schedules for Adults with Developmental Disabilities) [37] or MINI (Mini-International Neuropsychiatric Interview) [38].
Molecular cytogenetics
Multiplex ligation-dependent probe amplification (MLPA) was performed according to the manufacturer’s specifications (MRC Holland). To determine the size of the deletion we used the P324 22q11 MLPA mix containing primers specific for the CECR1, BCL2L13, BID, PEX26, CGT3P, PRODH (3 probes), GSC2, TBX1 (12 probes), GNB1L, COMT (2 probes), ARVCF, DGCR8, FLJ42953, RIMBP3C, VPREB1, RAB36, BCR (2 probes), MIF, SNRPD3, and SEZ6L genes (supplementary on-line material, table 1).
Statistical analyses
Coffalyser software (MLPA-Holland) was used to analyse and give an interpretation of the raw MLPA data [39].
The remaining data were statistical analyzed with Stata version 12.1 [40]. Age and IQ are analysed with T-test analysis and displayed in means with 95% confidence interval. The relation between deletion type with IQ was done in a simple regression model and the relation between deletion type with presence of psychosis during life was investigated with Pearson Chi-square analysis.
Results
Participants
Sixty-three participants [34 women (54 %), 29 men (46 %)] with confirmed 22q11DS participated in this study. The mean age of participants was 35 years (CI 32–38, SD= 11, range 19–59 years). Mean IQ was 51 (CI 44–58, SD=25). Thirty-eight out of 63 patients (60%) had a history of psychotic symptoms. Regression analysis of deletion type with IQ revealed no significant difference (p = 0.799). Pearson chi-square analysis of the relation of deletion type with psychotic problems during life revealed no significant differences (p =0.371).
Molecular cytogenetics
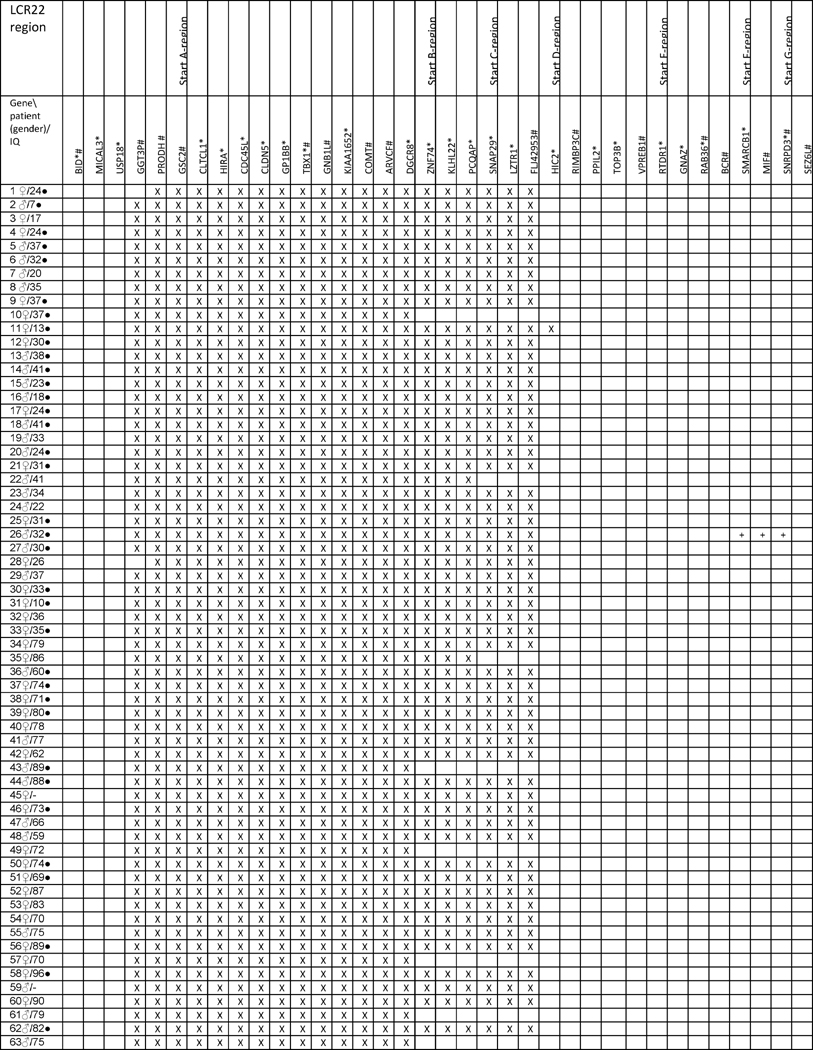
MLPA analysis with the P324 kit showed that there were 52 typical deletions (82.5 %) (from GGT3P to FLJ42953) and one typical deletion with a concomitant a duplication (Table 1). Two patients had a deletion extending from PRODH to FLJ42953. Eight patients (12.6%) had a deletion from GGT3P to ARVCF.
Table 1.
Combined MLPA results with two Salsa kits (P250 and P324)
Blanc: two copies; X: One copy deleted (hemizygous deletion); +: duplication (three copies present)
genes covered by the P250 kit;
genes covered by the P324 kit;
suffered from psychotic problems during life
Discussion
In this study we included 63 patients with 22q11DS and determined the deletion size with the experimental P324 MLPA kit. This reveals extra information about the deletion endpoints, especially in the A–B region, but fails to detect subtle differences in the B–C–D region. The original A–D deletion is described as a deletion from CLTCL1 to LZTR1, but with the P324 kit, probes proximal to the A-region (GSC2, PRODH and GGT3P) and one probe distal in the D-region (FLJ42953) were deleted in the majority of 22q11DS patients. This suggests that the description of the typical A–D deletion can be extended, including at the proximal end GGT3P, PRODH, GSC2, and at the distal end FLJ42953 and that some of the deletions described until now as typical, could be shorter, atypical. Two patients” with deletions characterised as “typical deletions” with P250, turned out to be shorter after analysis with P324 (from PRODH to FLJ42953) compared with the remaining 52.
12 out of 63 patients (19%) had a different deletion size compared to the typical deletion of 3 Mb length. A percentage that can be considered normal [23], especial taken into account that the use of an alternative MLPA kit delivered 2 extra patients (3%) with atypical deletion. The atypical deletions found in this cohort (n=11,) had three different sizes, two extending from PRODH to FLJ42953, eight from GGT3P to ARVCF and one duplication (combined with the typical A–D deletion). With the P324 kit there was no differentiation of the GGT3P-ARVCF deletion, while with the P250 kit six of them were determined as A–B deletion and two as A–C deletion. Analysis with the P324 kit did detect a typical A–D deletion, which was detected as A–D + HIC2 deletion with the P250 kit. The use of only the experimental MLPA-kit did not enable us to delineate deletion sizes more accurately compared to the standard MLPA kit. It did however reveal deletions of the GGT3P gene in two patients. Therefore, we believe that the use of the experimental kit only has advantages if used together with the standard kit. Used together, delineating of the deletion size was more precisely in all samples: it revealed two smaller deletions. This confirms the hypothesis that the experimental MLPA kit gives extra information about deletion endpoints in 22q11DS, but only if it is used together with the standard kit. In all samples one of the two PRODH and TBX1 probes were deleted.
Statistical analysis between deletion size and IQ and of the deletion size with presence of psychotic problems, revealed no significant difference. This suggests that deletion size is not related to psychotic problems in 22q11DS nor with intelligence.
This study confirmed that the research MLPA kit gives extra information about deletion size, but in our opinion the preferable MLPA mixture, for use in scientific research focusing on deletion size, should be a combination of the standard and the experimental kit together. A more accurate definition of deletions can give valuable extra information on genotype-phenotype relations, but larger sample size is needed. High resolution microarray-CGH delineates deletion size more accurately and would in the same time give an idea of additional imbalances found at other locations in the genome. Therefore it can be seen as favourite method in clinical use. However, the combined MLPA kits can be seen as a cheaper and faster alternative for CGH-array, which may be more realistic when large samples need delineation of deletion size. The sample size in this cohort is too small to draw conclusions on the relation of deletion size and intelligence or psychopathology.
Strength and limitations
This is, to the best of our knowledge, the first study using the experimental MLPA P324 kit to establish deletion size and compared the results with the standard P250 MLPA kit. Although the sample size is relatively large for this specific group, it is still small from a methodological point of view. With the inclusion of 22q11DS patients that function at a lower intellectual level, we cover a broader range of this syndrome, however, it cannot be ruled out that low FSIQ is overrepresented. The use of two different methods to establish intelligence was unavoidable (IQ below 55 cannot be established by Wechsler instruments) but is a serious limitation in this study.
Supplementary Material
Acknowledgments
We thank the patients and their families for their participation in the study. Research reported in this publication was supported by the National Institute of Mental Health of the National Institutes of Health under Award Number U01MH101722. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Appendix A. Supplementary data
Supplementary data related to this article can be found at:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14:3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald-McGinn DM, Kirschner R, Goldmuntz E, Sullivan K, Eicher P, Gerdes M, Moss E, Solot C, Wang P, Jacobs I, Handler S, Knightly C, Heher K, Wilson M, Ming JE, Grace K, Driscoll D, Pasquariello P, Randall P, Larossa D, Emanuel BS, Zackai EH. The Philadelphia story: the 22q11.2 deletion: report on 250 patients. Genet Couns. 1999;10:11–24. [PubMed] [Google Scholar]
- 3.Shprintzen RJ, Higgins AM, Antshel K, Fremont W, Roizen N, Kates W. Velo-cardio-facial syndrome. Curr Opin Pediatr. 2005;17:725–730. doi: 10.1097/01.mop.0000184465.73833.0b. [DOI] [PubMed] [Google Scholar]
- 4.Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14:3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O’Leary LA, Wong LY, Elixson EM, Mahle WT, Campbell RM. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112:101–107. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- 6.Grati FR, Gomes D Molina, Ferreira JC, Dupont C, Alesi V, Gouas L, Kuitunen N Horelli, Choy KW, Conejero JA Martinez, Vega A Gonzalesdela, Piotrowski K, Genesio R, Queipo G, Malvestiti B, Herve B, Benzacken B, Novelli A, Vago P, Piippo K, Leung TY, Malvestiti F, Quibel T, Tabet AC, Simoni G, Vialard F. Prevalence of recurrent pathogenic microdeletions and microduplications in over 9,500 pregnancies. Prenat Diagn. 2015 doi: 10.1002/pd.4613. DOI. [DOI] [PubMed] [Google Scholar]
- 7.Miller KA. FISH Diagnosis of 22q11.2 Deletion Syndrome. Newborn Infant Nurs Rev. 2008;8:e11–q19. [Google Scholar]
- 8.Jalali GR, Vorstman JA, Errami A, Vijzelaar R, Biegel J, Shaikh T, Emanuel BS. Detailed analysis of 22q11.2 with a high density MLPA probe set. Hum Mutat. 2008;29:433–440. doi: 10.1002/humu.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bittel DC, Yu S, Newkirk H, Kibiryeva N, Holt A, 3rd, Butler MG, Cooley LD. Refining the 22q11.2 deletion breakpoints in DiGeorge syndrome by aCGH. Cytogenet Genome Res. 2009;124:113–120. doi: 10.1159/000207515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niklasson L, Gillberg C. The neuropsychology of 22q11 deletion syndrome. A neuropsychiatric study of 100 individuals. Res Dev Disabil. 2010;31:185–194. doi: 10.1016/j.ridd.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Antshel KM, Fremont W, Kates WR. The neurocognitive phenotype in velo-cardio-facial syndrome: a developmental perspective. Dev Disabil Res Rev. 2008;14:43–51. doi: 10.1002/ddrr.7. [DOI] [PubMed] [Google Scholar]
- 12.Henry JC, van Amelsvoort T, Morris RG, Owen MJ, Murphy DG, Murphy KC. An investigation of the neuropsychological profile in adults with velo-cardio-facial syndrome (VCFS) Neuropsychologia. 2002;40:471–478. doi: 10.1016/s0028-3932(01)00136-1. [DOI] [PubMed] [Google Scholar]
- 13.van Amelsvoort T, Henry J, Morris R, Owen M, Linszen D, Murphy K, Murphy D. Cognitive deficits associated with schizophrenia in velo-cardio-facial syndrome. Schizophr Res. 2004;70:223–232. doi: 10.1016/j.schres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Chow EW, Watson M, Young DA, Bassett AS. Neurocognitive profile in 22q11 deletion syndrome and schizophrenia. Schizophr Res. 2006;87:270–278. doi: 10.1016/j.schres.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Smedt B, Devriendt K, Fryns JP, Vogels A, Gewillig M, Swillen A. Intellectual abilities in a large sample of children with Velo-Cardio-Facial Syndrome: an update. J Intellect Disabil Res. 2007;51:666–670. doi: 10.1111/j.1365-2788.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- 16.Evers LJ, Smulders CE De Die, Smeets EE, Clerkx MG, Curfs LM. The velo-cardio-facial syndrome: the spectrum of psychiatric problems and cognitive deterioration at adult age. Genet Couns. 2009;20:307–315. [PubMed] [Google Scholar]
- 17.Devriendt K, Thienen MN, Swillen A, Fryns JP. Cerebellar hypoplasia in a patient with velo-cardio-facial syndrome. Dev Med Child Neurol. 1996;38:949–953. doi: 10.1111/j.1469-8749.1996.tb15052.x. [DOI] [PubMed] [Google Scholar]
- 18.Iascone MR, Vittorini S, Sacchelli M, Spadoni I, Simi P, Giusti S. Molecular characterization of 22q11 deletion in a three-generation family with maternal transmission. Am J Med Genet. 2002;108:319–321. [PubMed] [Google Scholar]
- 19.Kozma C. On cognitive variability in velocardiofacial syndrome: profound mental retardation and autism. Am J Med Genet. 1998;81:269–270. doi: 10.1002/(sici)1096-8628(19980508)81:3<269::aid-ajmg12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 20.Swillen A, Devriendt K, Legius E, Eyskens B, Dumoulin M, Gewillig M, Fryns JP. Intelligence and psychosocial adjustment in velocardiofacial syndrome: a study of 37 children and adolescents with VCFS. J Med Genet. 1997;34:453–458. doi: 10.1136/jmg.34.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evers LJ, van Amelsvoort TA, Candel MJ, Boer H, Engelen JJ, Curfs LM. Psychopathology in adults with 22q11 deletion syndrome and moderate and severe intellectual disability. J Intellect Disabil Res. 2014;58:915–925. doi: 10.1111/jir.12117. [DOI] [PubMed] [Google Scholar]
- 22.Lindsay EA. Chromosomal microdeletions: dissecting del22q11 syndrome. Nat Rev Genet. 2001;2:858–868. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- 23.Michaelovsky E, Frisch A, Carmel M, Patya M, Zarchi O, Green T, Basel-Vanagaite L, Weizman A, Gothelf D. Genotype-phenotype correlation in 22q11.2 deletion syndrome. BMC Med Genet. 2012;13:122. doi: 10.1186/1471-2350-13-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rauch A, Zink S, Zweier C, Thiel CT, Koch A, Rauch R, Lascorz J, Huffmeier U, Weyand M, Singer H, Hofbeck M. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radoeva PD, Coman IL, Salazar CA, Gentile KL, Higgins AM, Middleton FA, Antshel KM, Fremont W, Shprintzen RJ, Morrow BE, Kates WR. Association between autism spectrum disorder in individuals with velocardiofacial (22q11.2 deletion) syndrome and PRODH and COMT genotypes. Psychiatr Genet. 2014;24:269–272. doi: 10.1097/YPG.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carmel M, Zarchi O, Michaelovsky E, Frisch A, Patya M, Green T, Gothelf D, Weizman A. Association of COMT and PRODH gene variants with intelligence quotient (IQ) and executive functions in 22q11.2DS subjects. J Psychiatr Res. 2014;56:28–35. doi: 10.1016/j.jpsychires.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 27.Zarchi O, Carmel M, Avni C, Attias J, Frisch A, Michaelovsky E, Patya M, Green T, Weinberger R, Weizman A, Gothelf D. Schizophrenia-like neurophysiological abnormalities in 22q11.2 deletion syndrome and their association to COMT and PRODH genotypes. J Psychiatr Res. 2013;47:1623–1629. doi: 10.1016/j.jpsychires.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Koning MB de, Boot E, Bloemen OJ, Duin ED van, JA B, Abel KM, Amelsvoort TA van. PRODH RS450046 and proline × COMT Val158Met Interaction effects on Intelligence and Startle in Adults with 22q11 Deletion Syndrome. Psychopharmacology. 2015 doi: 10.1007/s00213-015-3971-5. in press. [DOI] [PubMed] [Google Scholar]
- 29.Evers LJM, van Amelsvoort AMJ, Bakker JA, de Koning MB, Drukker M, Curfs LMG. Glutamatergic markers, age, intellectual functioning and psychosis in 22q11 deletion syndrome. Psychopharmacology. 2015 doi: 10.1007/s00213-015-3979-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao S, Li X, Amendt BA. Understanding the role of Tbx1 as a candidate gene for 22q11.2 deletion syndrome. Curr Allergy Asthma Rep. 2013;13:613–621. doi: 10.1007/s11882-013-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 32.Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, Sparks C, Choi CH, Oghalai J, Curran S, Murphy KC, Monks S, Williams N, O’Donovan MC, Owen MJ, Scambler PJ, Lindsay E. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci U S A. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiroi N, Takahashi T, Hishimoto A, Izumi T, Boku S, Hiramoto T. Copy number variation at 22q11.2: from rare variants to common mechanisms of developmental neuropsychiatric disorders. Mol Psychiatry. 2013;18:1153–1165. doi: 10.1038/mp.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiroi N, Hiramoto T, Harper KM, Suzuki G, Boku S. Mouse Models of 22q11.2-Associated Autism Spectrum Disorder. Autism Open Access. 2012;(Suppl 1):001. doi: 10.4172/2165-7890.S1-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wechsler D. WAIS-III, nederlandstalige bewerking: Technische handleiding. Lisse. 2001 [Google Scholar]
- 36.Kraijer DW, Plas JJ. Handboek psychodiagnostiek en beperkte begaafdheid. 4. Harcourt Assesment B.V.; Amsterdam: 2006. [Google Scholar]
- 37.Prosser H, Moss S, Costello H, Simpson N, Patel P, Rowe S. Reliability and validity of the Mini PAS-ADD for assessing psychiatric disorders in adults with intellectual disability. J Intellect Disabil Res. 1998;42(Pt 4):264–272. doi: 10.1046/j.1365-2788.1998.00146.x. [DOI] [PubMed] [Google Scholar]
- 38.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, Hergueta T, Baker R, Dunbar GC. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 20):22–33. quiz 34–57. [PubMed] [Google Scholar]
- 39.Coffa J, Berg J van de. Analysis of MLPA Data Using Novel Software Coffalyser.NET by MRC-Holland. In: Eldin DAB, editor. Modern Approaches To Quality Control, InTech. 2011. [Google Scholar]
- 40.StataCorp. Stata Statistical Software. StataCorparation; Texas: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.