Abstract
The lack of an anti-capsular vaccine against serogroup B meningococcal disease has necessitated the exploration of alternative vaccine candidates, mostly proteins exhibiting varying degrees of antigenic variation. Analysis of variants of antigen-encoding genes is facilitated by publicly accessible online sequence repositories, such as the Neisseria PubMLST database and the associated Meningitis Research Foundation Meningococcus Genome Library (MRF-MGL). We investigated six proposed meningococcal vaccine formulations by deducing the prevalence of their components in the isolates represented in these repositories. Despite high diversity, a limited number of antigenic variants of each of the vaccine antigens were prevalent, with strong associations of particular variant combinations with given serogroups and genotypes. In the MRF-MGL and globally, the highest levels of identical sequences were observed with multicomponent/multivariant vaccines. Our analyses further demonstrated that certain combinations of antigen variants were prevalent over periods of decades in widely differing locations, indicating that vaccine formulations containing a judicious choice of antigen variants have potential for long-term protection across geographic regions. The data further indicated that formulations with multiple variants would be especially relevant at times of low disease incidence, as relative diversity was higher. Continued surveillance is required to monitor the changing prevalence of these vaccine antigens.
Introduction
Neisseria meningitidis, the meningococcus, a Gram-negative diplococcus, is a globally important causative agent of meningitis and septicaemia (severe sepsis), accounting for a significant amount of morbidity and mortality worldwide. However, it is frequently carried harmlessly in the human nasopharynx and can be considered part of the normal human commensal micro-biota. Currently, no comprehensive vaccine exists against meningococcal disease due in large part to the structural similarity of serogroup B polysaccharide to polysaccharides associated with the human neural cell adhesion molecule (NCAM). This is thought to account for the poor human immune response against group B polysaccharide and also raises safety concerns [1]. Many subcapsular vaccine antigen candidates, especially proteins, have therefore been investigated, with the intention of producing serogroup B substitute formulations. Several such antigens have been incorporated into vaccine formulations that are in various stages of development.
First developed in the 1980s, outer membrane vesicle (OMV) vaccines were created to counter higher levels of disease incidence caused by particular serogroup B meningococci. These OMV vaccines contained the respective epidemic antigen variants of the outer membrane protein PorA of these meningococci as major immunogens and were successfully deployed in Norway (MenBvac), Cuba (VA-MENGOC-BC), and New Zealand (MeNZB) [2-4]. In the last decade however, many high-income regions such as Europe and North America have experienced a period of relatively low incidence of serogroup B meningococcal disease [5,6]. In such periods, when disease incidence is lower, but caused by more diverse meningococci, vaccines should ideally contain several components in order to attain the widest possible strain coverage. An example is the proposed NonaMen (RIVM, the Netherlands) vaccine formulation comprising nine PorA variants corresponding to the most prevalent disease-associated strains [7]. Alternatively, Bexsero, developed by Novartis, is a supplemented OMV vaccine which contains four components: PorA P1.7–2, 4, fHbp subvariant 1.1, NHBA variant 2, and NadA-3.8 subvariant [8]. This was licenced in Europe in 2013 and in the United States (US) in 2015 and has been included in the infant immunisation schedule in the United Kingdom (UK) since September 2015. The rLP2086 vaccine, Trumenba developed by Pfizer, which was licenced by the Food and Drug Administration in the US in 2014, is a bivalent recombinant vaccine based on two fHbp antigens from subfamily A and B (subvariants 3.45 and 1.55 respectively) [9].
Over the past two decades, sequence-based molecular typing has become an intrinsic part of meningococcal disease surveillance and standardised typing methods and schemes have allowed for more comparability across reference and research laboratories in different countries [10-12]. For example, the European surveillance system (TESSy) of the European Centre for Disease Control (ECDC) (http://www.ecdc.europa.eu/en/activities/surveillance/Tessy) and the European Meningococcal Epidemiology in Real Time (EMERT) database (http://emgm.eu/emert/) include two typing antigens which are also vaccine candidates, PorA and FetA. Following the advent of whole genome sequencing (WGS) and its rapidly reducing costs, comprehensive investigations of the likely and actual impact of available or potential interventions may be made more easily. Publicly accessible online resources such as the Neisseria PubMLST database (http://pubmlst.org/neisseria/) and the Meningitis Research Foundation Meningococcus Genome Library (MRF-MGL) (http://www.meningitis.org/research/genome), which contain molecular typing information from single genes up to many hundreds, and for many thousands of isolates, allow fine-scaled analyses, including investigation of the distribution of vaccine components. In this study, the PubMLST and MRF-MGL databases were used in an investigation of the distribution of vaccine components in Bexsero, Trumenba, NonaMen, MenBvac, MeNZB and VA-MENGOC-BC.
Methods
This study made use of the public Neisseria PubMLST database http://pubmlst.org/neisseria/ and the MRF-MGL http://www.meningitis.org/research/genome which is hosted within it. The databases were accessed in August 2014. The MRF-MGL contained 1,344 N. meningitidis isolates which were all from England and Wales, covering all culture-confirmed cases of invasive meningococcal disease (IMD) from the epidemiological years 2010/11 to 2012/13. From the PubMLST database, we included 1,717 N. meningitidis isolates within the database which had assembled sequence data of at least 0.5 Mbp. This is the minimum amount of assembled sequence data that allows as complete an analysis of vaccine antigen distribution as possible. This multinational collection contained data on disease and carriage isolates from the years 1937 to 2014, from 53 countries, six continents and excluded MRF-MGL isolates (Table 1).
Table 1.
Characteristics of the PubMLST global Neisseria meningitidis collection, 1937–2014 (n = 1,717)
| Sample source |
Number of isolates |
Country | Number of isolates |
Year of isolation |
Number of isolates |
Clonal complex |
Number of isolates |
|---|---|---|---|---|---|---|---|
| Disease | 926 | United Kingdom | 1,143 | Unknown | 319 | ST-11cc | 677 |
| Carriage | 112 | South Africa | 178 | 1937 | 1 | ST-41/44cc | 154 |
| Unknown | 679 | Czech Republic | 53 | 1940 | 1 | ST-269cc | 102 |
| Greece | 47 | 1941 | 1 | ST-32cc | 88 | ||
| Serogroup | Number of isolates | Norway | 36 | 1947 | 1 | ST-23cc | 79 |
| Unknown | 596 | United States | 25 | 1962 | 1 | ST-213cc | 52 |
| W | 413 | Canada | 22 | 1963 | 5 | ST-1cc | 47 |
| B | 278 | Ireland | 20 | 1964 | 3 | ST-22cc | 46 |
| C | 256 | Unknown | 15 | 1965 | 1 | ST-60cc | 44 |
| A | 82 | Spain | 15 | 1966 | 5 | ST-53cc | 35 |
| Y | 46 | Niger | 13 | 1967 | 3 | ST-1157cc | 28 |
| NG | 40 | Burkina Faso | 12 | 1968 | 2 | ST-35cc | 22 |
| E | 2 | Germany | 12 | 1969 | 3 | ST-5cc | 21 |
| X | 2 | The Netherlands | 12 | 1970 | 8 | ST-174cc | 19 |
| Z | 2 | France | 11 | 1971 | 1 | ST-8cc | 19 |
| China | 11 | 1973 | 1 | ST-167cc | 18 | ||
| Continent | Number of isolates | Cameroon | 8 | 1974 | 2 | ST-162cc | 15 |
| Europe | 1,373 | Malta | 7 | 1975 | 9 | ST-4cc | 14 |
| Africa | 234 | Turkey | 6 | 1976 | 6 | ST-198cc | 10 |
| North America | 49 | Brazil | 5 | 1977 | 4 | ST-865cc | 10 |
| Asia | 30 | Chile | 4 | 1978 | 4 | ST-18cc | 9 |
| Unknown | 15 | Denmark | 4 | 1979 | 5 | ST-254cc | 8 |
| South America | 10 | New Zealand | 4 | 1980 | 4 | ST-103cc | 6 |
| Oceania | 6 | Algeria | 3 | 1981 | 1 | ST-4821cc | 5 |
| Chad | 3 | 1982 | 1 | ST-1136cc | 4 | ||
| Gambia | 3 | 1983 | 3 | ST-1117cc | 3 | ||
| Italy | 3 | 1984 | 6 | ST-178cc | 3 | ||
| Mali | 3 | 1985 | 20 | ST-334cc | 3 | ||
| Morocco | 3 | 1986 | 10 | ST-364cc | 3 | ||
| Portugal | 3 | 1987 | 9 | ST-461cc | 3 | ||
| Russia | 3 | 1988 | 19 | ST-181cc | 2 | ||
| Senegal | 3 | 1989 | 6 | ST-282cc | 2 | ||
| Australia | 2 | 1990 | 5 | ST-37cc | 2 | ||
| Cuba | 2 | 1991 | 4 | ST-613cc (lactamica) | 2 | ||
| Finland | 2 | 1992 | 5 | ST-175cc | 1 | ||
| Ghana | 2 | 1993 | 58 | ST-212cc | 1 | ||
| Iceland | 2 | 1994 | 7 | ST-231cc | 1 | ||
| India | 2 | 1995 | 1 | ST-4240/6688cc | 1 | ||
| Japan | 2 | 1996 | 39 | ST-750cc | 1 | ||
| Philippines | 2 | 1997 | 35 | Unassigned STs | 157 | ||
| Argentina | 1 | 1998 | 62 | ||||
| Austria | 1 | 1999 | 51 | ||||
| Djibouti | 1 | 2000 | 100 | ||||
| Israel | 1 | 2001 | 101 | ||||
| Ivory Coast | 1 | 2002 | 34 | ||||
| Pakistan | 1 | 2003 | 52 | ||||
| Poland | 1 | 2004 | 56 | ||||
| Saudi Arabia | 1 | 2005 | 38 | ||||
| Sudan | 1 | 2006 | 36 | ||||
| Switzerland | 1 | 2007 | 41 | ||||
| Thailand | 1 | 2008 | 29 | ||||
| 2009 | 42 | ||||||
| 2010 | 33 | ||||||
| 2011 | 168 | ||||||
| 2012 | 167 | ||||||
| 2013 | 69 | ||||||
| 2014 | 19 | ||||||
cc: clonal complex.
The PubMLST database is a publicly-accessible repository of isolate and typing information for several species including N. meningitidis. It contains a large set of global records of Neisseria genus isolates (33,019 in March 2014) spanning 100 years. It is a complete catalogue of known genotypic and phenotypic variation, date and location of isolation, and permits an estimation of the minimum lifespan of particular genotypes and deduced antigenic types. While the collection is not an epidemiologically coherent sample set as a whole, it does include several such datasets (including the MRF-MGL), and permits several types of investigation into the evolution and population biology of N. meningitidis.
The presence of components of each of the fHbp-containing vaccines, were analysed in the collections [8,9]. Before the development of a unified nomenclature scheme in which each unique allele is assigned a unique numerical identifier [13], separate schemes were developed which divided fHbp into either two subfamilies (subfamily A and B) or three variant families (variant families 1, 2 and 3) according to nomenclature system [14,15]. These schemes can be cross-referenced online (http://pubmlst.org/neisseria/fHbp/). Briefly, subfamily B is equivalent to variant family 1 and subfamily A incorporates both variant families 2 and 3. Peptides are then numbered with the variant family/subfamily name e.g. fHbp 1.1 is variant family 1 peptide 1 [13]. This is the fHbp nomenclature used throughout this paper. The recombinant vaccine Trumenba contains two fHbp antigen subvariants: peptide 45 (Pfizer nomenclature A05, subfamily A/variant family 3) and peptide 55 (Pfizer nomenclature B01, subfamily B/variant family 1). As well as Bexsero and Trumenba, other vaccine formulations analysed were: NonaMen, which contains nine PorA antigens variants (P1.7,16; P1.5–1,2–2; P1.19,15–1; P1.5–2,10; P1.12–1,13; P1.7–2,4, P1.22,14; P1.7–1,1; P1.18–1,3), MenBvac, (P1.7,16), MeNZB (P1.7–2,4) and VA-MENGOC-BC (P1.19,15) [4,7,16,17]. Analysis of the distribution of vaccine components among clonal complexes and patient age groups in the MRF-MGL was based on an exact match of deduced peptide sequences (‘sequence match’) to at least one component of each vaccine formulation investigated. The analysis was carried out on isolates of all genogroups (organisms with a cps region, encoding a capsule) and genogroup B isolates only (those containing a cps region encoding the group B polysaccharide capsule). For the purposes of this analysis is was assumed that such meningococci either had the capacity to express a capsule or had a very closely related ancestor which could [18]. It should be noted that OMV vaccines include other potentially immunogenic proteins not assessed here, although WGS allows for such analyses.
Simpson’s index of diversity (D) was used to determine the diversity of each Bexsero vaccine antigen by age group in the MRF-MGL. The value of the index ranged from 0 to 1, with values nearer to 1 indicating greater diversity. Calculation of the index was performed as described previously [19]. The 95% confidence intervals (CIs) for the index were calculated as described previously [20].
Results
Current vaccine antigens in the MRF-MGL collection: invasive meningococcal disease isolates from England and Wales, 2010/11 to 2012/13
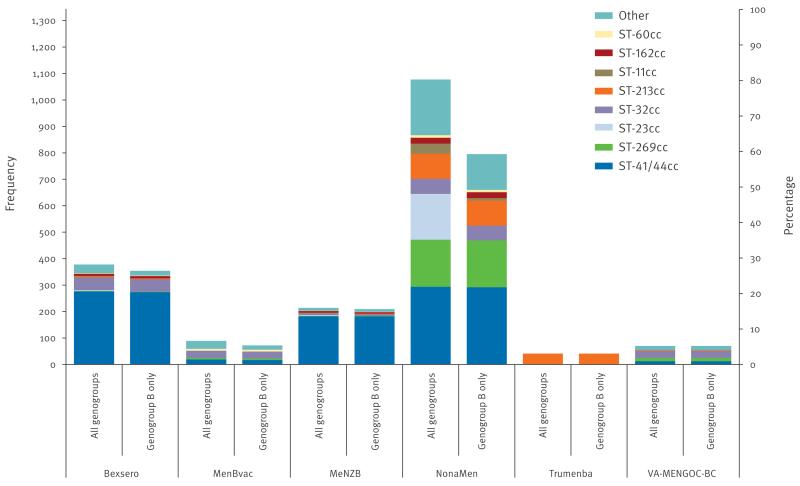
Within the MRF-MGL collection, NadA was significantly the least diverse peptide antigen examined (D = 0.39; 95% CI: 0.36–0.42) compared with: PorA variable region 1 (VR1) (D = 0.87; 95% CI: 0.86–0.88), PorA VR2 (D = 0.92; 95% CI: 0.92–0.93), fHbp (D = 0.91; 95% CI: 0.91–0.92) and NHBA (D = 0.91; 95% CI: 0.91–0.92). However, a total of 1,025 (76.3%) isolates were inferred to be missing the nadA gene as it was not found in their genomes (Table 2). A total of 1,102 of 1,344 (82.0%) isolates contained exact peptide sequence matches to components of any of the vaccines investigated (Figure 1). Isolates belonging to the two most prevalent clonal complexes (cc) in the MRF-MGL, ST-41/44cc and ST-269cc, had most sequence matches (302 and 181 instances, respectively). Exact matches to NonaMen components were found in 20 different clonal complexes. In total there were 1,077 of 1,344 isolates (80.1%) with genomes that encoded the PorA VR1 or VR2 antigen variants included in the NonaMen formulation. ST-41/44cc isolates comprised 294 of these 1,077 (27.3%), a consequence of their association with PorA subtype P1.7–2,4.
Table 2.
Allelic diversity of Bexsero vaccine antigens per patient age group, MRF-MGL invasive meningococcal disease isolate genomes, England and Wales, 2010/11–2012/13 (n = 1,344)
| Age (years) | NadA D (95% CI) | NHBA D (95% CI) | FHBP D (95% CI) | PorA VR1 D (95% CI) | PorA VR2 D (95% CI) |
|---|---|---|---|---|---|
| < 1 (n = 300) | 0.31 (0.24–0.38) | 0.88 (0.86–0.91) | 0.89 (0.87–0.91) | 0.82 (0.78–0.85) | 0.88 (0.86–0.90) |
| 1–4 (n = 315) | 0.39 (0.33–0.46) | 0.88 (0.86–0.90) | 0.90 (0.88–0.92) | 0.84 (0.81–0.87) | 0.92 (0.91–0.94) |
| 5–14 (n = 99) | 0.33 (0.21–0.44) | 0.91 (0.88–0.94) | 0.91 (0.88–0.94) | 0.88 (0.85–0.92) | 0.91 (0.89–0.94) |
| 15–24 (n = 168) | 0.42 (0.33–0.52) | 0.90 (0.88–0.92) | 0.90 (0.88–0.92) | 0.88 (0.85–0.90) | 0.93 (0.92–0.95) |
| 25–39 (n = 73) | 0.50 (0.36–0.64) | 0.92 (0.90–0.95) | 0.93 (0.91–0.95) | 0.84 (0.79–0.90) | 0.93 (0.91–0.96) |
| 40–64 (n = 173) | 0.42 (0.33–0.51) | 0.91 (0.89–0.93) | 0.90 (0.88–0.92) | 0.87 (0.84–0.90) | 0.92 (0.91–0.94) |
| ≥ 65 (n = 207) | 0.41 (0.32–0.49) | 0.87 (0.84–0.90) | 0.85 (0.81–0.89) | 0.82 (0.78–0.86) | 0.91 (0.89–0.92) |
| Total (n = 1,344) | 0.39 (0.36–0.42) | 0.91 (0.91–0.92) | 0.91 (0.91–0.92) | 0.87 (0.86–0.88) | 0.92 (0.92–0.93) |
CI: confidence interval; MRF-MGL: Meningitis Research Foundation Meningococcus Genome Library.
Diversity determined by Simpson’s Diversity Index
Figure 1.
Distribution of clonal complexes in meningococcal disease isolates in the MRF-MGL collection with exact peptide matches to at least one antigen of various vaccines, England and Wales, 2010/11–2012/13 (n = 1,344)
MRF-MGL: Meningitis Research Foundation Meningococcus Genome Library.
Invasive meningococcal disease isolates from England and Wales from epidemiological years 2010/11 to 2012/13 inclusive of all genogroups or genogroup B only. ‘Other’ includes uncommon ccs and unassigned STs. Secondary y-axis indicates percentage of total number of isolates.
Unlike the other vaccine formulations, exact matches to Trumenba vaccine components were found in a single clonal complex (ST-213cc). Exact amino acid sequence matches to at least one of the four Bexsero vaccine antigen variants were present in 378 of 1,344 (28.1%) isolates, which belonged to 10 clonal complexes. Again, ST-41/44cc contained the majority of matches (276 isolates) due to its association with NHBA peptide 2 and PorA VR2 P1.4, but only two isolates of ST-269cc contained an exact match to any Bexsero antigen peptide variant. None of the major hyperinvasive clonal complexes, except for a single genogroup A ST-5cc isolate, contained an exact match to NadA subvariant 8, which was found in 13 ST-174cc isolates. However, 65.7% of ST-32cc isolates contained Bexsero fHbp variant family 1 peptide 1, consistent with derivation of this component from MC58, an isolate of the same complex [21].
In total, 37.0% of genogroup B isolates contained at least one exact match to any Bexsero vaccine component. For the other vaccines, the proportion of isolates with at least one exact match to vaccine antigen variants were NonaMen: 82.7%, MeNZB: 21.8%, MenBvac: 7.5% and VA-MENGOC-BC: 7.3%. When cross-protection among NadA peptide alleles belonging to variants NadA-1 and NadA-2/3 and among fHbp peptide alleles belonging to family 1 was assumed, the overall number of isolates containing an exact match to Bexsero antigen variants more than doubled to 847 isolates, or 63.0% of the MRF-MGL.
Vaccine antigen distribution by age group
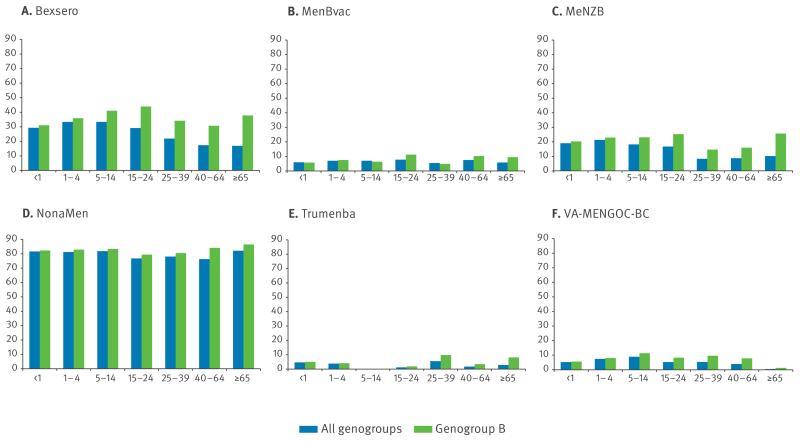
Vaccine peptide diversity and distribution in the MRF-MGL were assessed by age group (Table 2, Figure 2). For all antigens, peptide diversity was lowest among meningococci isolated from patients younger than one year or 65 years and older. Significant differences in peptide diversity, those where the 95% CIs did not overlap, were seen. For example, fHbp diversity (D = 0.93; 0.91–0.95) was significantly greater in isolates from patients aged 25 to 29 years than in isolates from patients 65 years and older (D = 0.85; 0.81–0.89). Apart from isolates from patients 65 years and older, where PorA P1.7–2 predominated, and those from patients 40 to 64 years of age, where NHBA and PorA P1.7–2 were equally prevalent (15 occurrences), NHBA-2 was the most frequently identified Bexsero antigen peptide in all age categories (data not shown). PorA P1.7–2 was the second most prevalent vaccine subvariant in age groups 25 to 39 years and younger. The fHbp 1.1 subvariant was found at the lowest proportions in patients younger than one year and in those 65 years and older. NadA-3.8 was absent from isolates from patients aged one to four years and 15 to 24 years, and was identified in less than 2% of isolates from all other age groups except the group 65 years and older, where it was present in 3.9% of isolates. Thus, isolates possessing at least one of the four Bexsero antigen peptide variants were found at the highest proportions in patients aged one to four and five to 14 years (114/315 (36.2%) and 34/99 (34.3%) isolates, respectively), and at the lowest proportions in patients aged 65 years and older (37/207 (17.9%) isolates). The greatest proportion of genogroup B isolates with at least one exact amino-acid sequence match was identified in patients aged 15 to 24 years (48/107 (44.9%) genogroup B isolates), and the least was in patients younger than one year (88/277 (31.8%) genogroup B isolates). The association of ST-41/44cc and less common lineages with patients younger than one year meant that 30.3% of all isolates contained at least one exact peptide match. Isolates possessing at least NadA-1 or NadA-2/3, fHbp variant family 1, PorA P1.4, or NHBA-2 allele were found at the highest proportion in patients aged five to 14 years (72/99 (72.7%) isolates). Of the vaccine formulations assessed, NonaMen antigen peptide variants were the most frequently identified in isolates of all age groups (Figure 2), ranging from 76.3% of patients aged 40 to 64 years to 82.1% of patients 65 years and older. A total of 81.7% of all isolates and 82.3% of genogroup B isolates from patients younger than one year possessed at least one NonaMen PorA variant. The Trumenba vaccine antigen peptide subvariants fHbp 3.45 and fHbp 1.55 were not found in isolates from patients aged five to 14 years (n = 99).
Figure 2.
Percentage distribution of age groups in meningococcal disease isolates in the MRF-MGL collection with exact peptide matches to at least one antigen of various vaccines, England and Wales, 2010/11–2012/13 (n = 1,344)
MRF-MGL: Meningitis Research Foundation Meningococcus Genome Library.
Invasive meningococcal disease isolates from England and Wales from epidemiological years 2010/11 to 2012/13 inclusive of all genogroups and genogroup B only.
Vaccine candidate antigen variants in PubMLST/neisseria database
There were 38 isolates in the PubMLST Neisseria collection with the fHbp 3.45 subvariant and none with the 1.55 subvariant (Table 3). A total of 36 of the 38 isolates that possessed the 3.45 subvariant were ST-213cc and were all from the UK. Those which had a date of isolation recorded in the database were from between 1999 and 2012. The Bexsero subvariant fHbp 1.1 was found in 73 isolates in the dataset. Of these, 66 were ST-32cc and 47 of 73 were serogroup B. They were found over a time span of 40 years from 1969 to 2009 and on all continents. NadA-3.8 was found in 41 isolates. Of those, 17 were ST-174cc and 16 were ST-5cc; 16 were serogroup Y, 16 serogroup A and five were serogroup B. They were found across 49 years from 1963 to 2012 and on all continents except Oceania. NHBA variant 2 was found in 121 isolates and 98 of these were ST-41/44cc. Of the 121 isolates for which serogroup data were recorded, 66 were serogroup B, 12 were serogroup C and 11 were non-groupable. NHBA variant 2 was found on all continents except Asia and Africa and across 37 years from 1976 to 2013.
Table 3.
Characteristics of Bexsero and Trumenba antigens in PubMLST Neisseria database
| Antigen | Number of isolates | Dominant clonal complex(es) n (%) | Dominant serogroup(s) n (%) | Minimum lifespan (years) | Observed time period | Continents found |
|---|---|---|---|---|---|---|
| fHbp subvariant 3.45 (A05)a | 38 | ST-213: 36 (94.7) | Unassigned: 36 (94.7) | 13 | 1999–2012 | Europe |
| fHbp subvariant 1.55 (B01)a | 0 | NA | NA | NA | NA | NA |
| fHbp variant family 1/subfamily B | 873 | ST-11: 349 (40.0), ST-41/44: 98 (11.2), Unassigned: 92 (7.4) |
W: 241 (27.6), B: 159 (18.2), C: 113 (12.9), A: 76 (8.7) |
77 | 1937–2014 | Africa, Asia, Europe, North America, Oceania, South America |
| fHbp subvariant 1.1 | 73 | ST-32: 66 (90.4) | B: 47 (64.4) | 40 | 1969–2009 | Africa, Asia, Europe, North America, Oceania, South America |
| NadA-3.8 subvariant | 41 | ST-174: 17 (41.5), ST-5: 16 (39.0) |
Y: 16 (39.0), A: 16 (39.0), B: 5 (12.2) |
49 | 1963–2012 | Africa, Asia, Europe, North America, South America |
| NadA-1,2,3 variants | 709 | ST-11: 568 (80.1) | W: 368 (51.9), C: 135 (19.0) |
51 | 1963–2014 | Africa, Asia, Europe, North America, Oceania, South America |
| NHBA variant 2 | 121 | ST-41/44: 98 (80.9) | B: 66 (54.6), C: 12 (9.9), NG: 11 (9.1) |
37 | 1976–2013 | Europe, North America, Oceania, South America |
NA: not available; NG: non-groupable.
A05, B01: Pfizer nomenclature.
Cross-protection among fHbp variant family 1 and NadA-1, NadA-2 and NadA-3 variant family members has been described [22,23], prompting an analysis of their distribution. There were 873 fHbp variant family 1 peptide variants and these were found across the whole time span of the collection (77 years) from 1937 to 2014 and on all continents. Of these, 349 (40.0%) were ST-11cc, 98 (11.2%) were ST-41/44cc, and unassigned sequence types (STs) accounted for 92 (7.4%) of the isolates. Of the 873, 241 were serogroup W, 159 were serogroup B, 113 were serogroup C and 76 were serogroup A. There were 709 isolates that were NadA-1, 2 and 3 variant family members. These spanned 51 years of the collection from 1963 to 2014 and were found on all continents. The majority were ST-11cc (n = 568; 80.1%), 368 were serogroup W and 135 were serogroup C.
Discussion
Since its introduction in the 1990s, sequence-based molecular typing has established a role in the clinical microbiology laboratory, replacing or complementing existing phenotypic typing methods. WGS is the latest sequencing technology and, as costs continue to decrease, will become more commonplace in clinical and reference laboratories [24-26]. WGS provides definitive sequence-level resolution with widespread applications including molecular epidemiology, surveillance, vaccine design and vaccine implementation monitoring [27]. To be useable by physicians and public health specialists, databases will need to use uniform nomenclature and be interoperable and compatible with other databases such as those that contain phenotypic information [28].
As WGS databases such as PubMLST are generic and scalable, they enable detailed deduction of potential coverage and preliminary assessment of the impact of given vaccine formulations on the meningococcal population, thus informing further work such as phenotypic assays [29]. This requires the assembly of representative collections of meningococcal isolate genomes. The MRF-MGL is an exemplar of a representative, contemporary, curated, publicly accessible database containing many hundreds of genomes and was expressly established as a resource for the meningococcal research and public health communities. It is embedded within the PubMLST database, which has been running for many years as a community resource and repository for isolate and characterisation data. The MRF-MGL is the most comprehensive epidemiological sample of meningococcal WGS currently available, allowing an assessment of vaccine antigen distribution among disease cases.
Molecular epidemiology using phenotypic or genotypic data has been used to inform vaccine design: tailor-made OMV vaccines were designed to contain the respective outbreak strain PorA variant [2-4]; and the broad-spectrum multivalent Hexamen/Nonamen formulation was based on the most prevalent PorA serosubtypes documented in the Netherlands at the time of its development [30]. One of the earliest uses of genome data in vaccine design was in the discovery of novel meningococcal vaccine candidates by ‘reverse vaccinology’ based on a single isolate genome [31]. Several of these genome-derived antigens are components of vaccines at various stages of clinical development and deployment at the time of writing [8,9].
The temporal and geographic spread of antigen variants and combination of variants, with some existing for long time periods and across several continents, demonstrated the stability of antigen clonal complex relationships and therefore the potential longevity of appropriate vaccine formulations [32,33]. Previous work using the PubMLST database collection [32] demonstrated the longevity of PorA VR types and their association with clonal complex which we here extend to other antigens available from WGS. For example, the most frequent strain type (PorA:FetA:cc) P1.5,2:F3–6:cc11 had a minimum lifespan of 49 years and was found on three continents (Europe, North America and South America) [32]. In the present analysis, multicomponent vaccines exhibited more potential to protect against isolates represented in the MRF-MGL than vaccine formulations containing one or a few components, although this did not take into account any potential cross-protection that may be offered by any particular vaccine antigen. Europe and North America are experiencing low rates of meningococcal disease at present [5,6], and in the absence of a dominant epidemic clone accounting for disease, a multicomponent vaccine formulation would be required to cover most disease [34,35]. Since a differential distribution of clonal complex and antigens has been demonstrated among different age groups and those at highest risk (the under one year-olds) are less at risk from lineages that affect the next peak of disease incidence, late adolescents, multicomponent vaccines are likely to be most appropriate, especially in a period of low incidence [36]. Therefore, comprehensive molecular epidemiology and surveillance is required in order to maximise the coverage of a given vaccine formulation. Continuous surveillance will be required to track changes in epidemiology that may need vaccine reformulation.
While genotypic data can provide valuable information on the potential utility of vaccines, the evaluation of antigen expression and potential cross-reactivity is fundamental to gauging the actual success of a given formulation. Assays have been developed and expression studies carried out that attempt to predict the coverage of various meningococcal vaccine antigens in the population [14,37]. One assay, the ELISA-based meningococcal antigen typing system (MATS) was developed to predict the strain coverage of the Bexsero vaccine [37]. Based on a panel of invasive serogroup B-associated meningococcal isolates from several European countries it was estimated that it could protect against 78% of serogroup B cases and against a panel of serogroup B invasive isolates from Greece up to 90% [38,39]. One of the features of the PubMLST database is that phenotypic information such as MATS data can be added to isolate or allele data so that phenotypic and genotypic information may be associated allowing further analyses.
Conclusion
Highly variable pathogens require detailed characterisation to appropriately tailor clinical and public health responses such as treatment, immunisation, outbreak control, and novel vaccine design. This is especially true for organisms such as meningococci, in particular those that express the serogroup B polysaccharide, given that a universal capsular vaccine is unavailable. Well-characterised isolate collections can easily be investigated for any number of vaccine formulations and vaccine candidates when they are housed within databases embedded with analysis tools which can handle phenotype and genotype data including WGS. This high level of characterisation and molecular epidemiology provides a foundation for further phenotypic analyses so that a fuller picture of potential vaccine effectiveness can be seen. Detailed characterisation and monitoring is particularly relevant in periods of low incidence, such as experienced in high-income regions at present, as multivalent vaccines may be most appropriate and also most adaptable should changes in the meningococcal population occur. This rationale for vaccine formulation using molecular epidemiology may be applied to any pathogen and will become more readily applicable as well characterised datasets like the MRF-MGL and PubMLST become increasingly available. A combination of detailed genotypic characterisation and phenotypic investigation offer the best hope of producing vaccines with the widest possible coverage.
Acknowledgements
This publication made use of the Meningitis Research Foundation Meningococcus Genome Library (http://www.meningitis.org/research/genome) developed by Public Health England, the Wellcome Trust Sanger Institute and the University of Oxford as a collaboration. The project is funded by Meningitis Research Foundation. It also made use of the Neisseria PubMLST website (http://pubmlst.org/neisseria/) developed by Keith Jolley and sited at the University of Oxford (BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11(1):595). The development of this site has been funded by the Wellcome Trust and European Union.
Footnotes
Conflict of interest
DH was supported by the Meningitis Research Foundation. RB does contract research on behalf of his employer, Public Health England, for manufacturers of meningococcal vaccines including Baxter Biosciences, GlaxoSmithKline, Novartis Vaccines, Pfizer, and Sanofi Pasteur. MM has received grants and personal fees from Novartis outside the submitted work.
The other authors declare no competing interests.
References
- 1.Finne J, Leinonen M, Mäkelä PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;322(8346):355–7. doi: 10.1016/S0140-6736(83)90340-9. PMID: 6135869. [DOI] [PubMed] [Google Scholar]
- 2.Bjune G, Høiby EA, Grønnesby JK, Arnesen O, Fredriksen JH, Halstensen A, et al. Effect of outer membrane vesicle vaccine against group B meningococcal disease in Norway. Lancet. 1991;338(8775):1093–6. doi: 10.1016/0140-6736(91)91961-S. PMID: 1682541. [DOI] [PubMed] [Google Scholar]
- 3.O’Hallahan J, Lennon D, Oster P. The strategy to control New Zealand’s epidemic of group B meningococcal disease. Pediatr Infect Dis J. 2004;23((12) Suppl):S293–8. PMID: 15597072. [PubMed] [Google Scholar]
- 4.Sierra GVG, Campa CH, Garcia LI, Sotolongo FP, Izquierdo LP, Valcarcel MN, et al. Efficacy evaluation of the Cuban vaccine VA-MENGOC-BC against disease caused by serogroup B Neisseria meningitidis. In: Mea Achtman., editor. Niesseria 1990. Walter de Gruyter; Berlin: 1991. pp. 129–134. [Google Scholar]
- 5.European Centre for Disease Prevention and Control (ECDC) Reporting on 2011 surveillance data and 2012 epidemic intelligence data. ECDC; Stockholm: 2013. Annual epidemiological report 2013. Available from: http://ecdc.europa.eu/en/publications/Publications/annual-epidemiological-report-2013.pdf. [Google Scholar]
- 6.Castelblanco RL, Lee M, Hasbun R. Epidemiology of bacterial meningitis in the USA from 1997 to 2010: a population-based observational study. Lancet Infect Dis. 2014;14(9):813–9. doi: 10.1016/S1473-3099(14)70805-9. PMID: 25104307. [DOI] [PubMed] [Google Scholar]
- 7.Kaaijk P, van Straaten I, van de Waterbeemd B, Boot EP, Levels LM, van Dijken HH, et al. Preclinical safety and immunogenicity evaluation of a nonavalent PorA native outer membrane vesicle vaccine against serogroup B meningococcal disease. Vaccine. 2013;31(7):1065–71. doi: 10.1016/j.vaccine.2012.12.031. PMID: 23273968. [DOI] [PubMed] [Google Scholar]
- 8.Bambini S, Piet J, Muzzi A, Keijzers W, Comandi S, De Tora L, et al. An analysis of the sequence variability of meningococcal fHbp, NadA and NHBA over a 50-year period in the Netherlands. PLoS ONE. 2013;8(5):e65043. doi: 10.1371/journal.pone.0065043. PMID: 23717687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissen MD, Marshall HS, Richmond PC, Jiang Q, Harris SL, Jones TR, et al. A randomized, controlled, phase 1/2 trial of a Neisseria meningitidis serogroup B bivalent rLP2086 vaccine in healthy children and adolescents. Pediatr Infect Dis J. 2013;32(4):364–71. doi: 10.1097/INF.0b013e31827b0d24. PMID: 23114369. [DOI] [PubMed] [Google Scholar]
- 10.Brehony C, Jolley KA, Maiden MC. Multilocus sequence typing for global surveillance of meningococcal disease. FEMS Microbiol Rev. 2007;31(1):15–26. doi: 10.1111/j.1574-6976.2006.00056.x. PMID: 17168997. [DOI] [PubMed] [Google Scholar]
- 11.Jolley KA, Brehony C, Maiden MC. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol Rev. 2007;31(1):89–96. doi: 10.1111/j.1574-6976.2006.00057.x. PMID: 17168996. [DOI] [PubMed] [Google Scholar]
- 12.Trotter CL, Chandra M, Cano R, Larrauri A, Ramsay ME, Brehony C, et al. A surveillance network for meningococcal disease in Europe. FEMS Microbiol Rev. 2007;31(1):27–36. doi: 10.1111/j.1574-6976.2006.00060.x. PMID: 17168995. [DOI] [PubMed] [Google Scholar]
- 13.Brehony C, Wilson DJ, Maiden MC. Variation of the factor H-binding protein of Neisseria meningitidis. Microbiology. 2009;155(Pt 12):4155–69. doi: 10.1099/mic.0.027995-0. PMID: 19729409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72(4):2088–100. doi: 10.1128/IAI.72.4.2088-2100.2004. PMID: 15039331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197(6):789–99. doi: 10.1084/jem.20021911. PMID: 12642606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Hallahan J, Lennon D, Oster P, Lane R, Reid S, Mulholland K, et al. From secondary prevention to primary prevention: a unique strategy that gives hope to a country ravaged by meningococcal disease. Vaccine. 2005;23(17-18):2197–201. doi: 10.1016/j.vaccine.2005.01.061. PMID: 15755594. [DOI] [PubMed] [Google Scholar]
- 17.Bjune G, Grønnesby JK, Høiby EA, Closs O, Nøkleby H. Results of an efficacy trial with an outer membrane vesicle vaccine against systemic serogroup B meningococcal disease in Norway. NIPH Ann. 1991;14(2):125–30. discussion 130-2. PMID: 1812425. [PubMed] [Google Scholar]
- 18.Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis. 2013;19(4):566–73. doi: 10.3201/eid1904.111799. PMID: 23628376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. PMID: 3069867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001;39(11):4190–2. doi: 10.1128/JCM.39.11.4190-4192.2001. PMID: 11682558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287(5459):1809–15. doi: 10.1126/science.287.5459.1809. PMID: 10710307. [DOI] [PubMed] [Google Scholar]
- 22.Brunelli B, Del Tordello E, Palumbo E, Biolchi A, Bambini S, Comanducci M, et al. Influence of sequence variability on bactericidal activity sera induced by Factor H binding protein variant 1.1. Vaccine. 2011;29(5):1072–81. doi: 10.1016/j.vaccine.2010.11.064. PMID: 21130753. [DOI] [PubMed] [Google Scholar]
- 23.Comanducci M, Bambini S, Caugant DA, Mora M, Brunelli B, Capecchi B, et al. NadA diversity and carriage in Neisseria meningitidis. Infect Immun. 2004;72(7):4217–23. doi: 10.1128/IAI.72.7.4217-4223.2004. PMID: 15213166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter S, Ellington MJ, Cartwright EJ, Köser CU, Török ME, Gouliouris T, et al. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med. 2013;173(15):1397–404. doi: 10.1001/jamainternmed.2013.7734. PMID: 23857503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10(2):e1001387. doi: 10.1371/journal.pmed.1001387. PMID: 23424287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price JR, Didelot X, Crook DW, Llewelyn MJ, Paul J. Whole genome sequencing in the prevention and control of Staphylococcus aureus infection. J Hosp Infect. 2013;83(1):14–21. doi: 10.1016/j.jhin.2012.10.003. PMID: 23164609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45(6):656–63. doi: 10.1038/ng.2625. PMID: 23644493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolley KA, Maiden MC. Automated extraction of typing information for bacterial pathogens from whole genome sequence data: Neisseria meningitidis as an exemplar. Euro Surveill. 2013;18(4):20379. doi: 10.2807/ese.18.04.20379-en. PMID: 23369391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bratcher HB, Bennett JS, Maiden MCJ. Evolutionary and genomic insights into meningococcal biology. Future Microbiol. 2012;7(7):873–85. doi: 10.2217/fmb.12.62. PMID: 22827308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Ley P, van der Biezen J, Poolman JT. Construction of Neisseria meningitidis strains carrying multiple chromosomal copies of the porA gene for use in the production of a multivalent outer membrane vesicle vaccine. Vaccine. 1995;13(4):401–7. doi: 10.1016/0264-410X(95)98264-B. PMID: 7793138. [DOI] [PubMed] [Google Scholar]
- 31.Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3(5):445–50. doi: 10.1016/S1369-5274(00)00119-3. PMID: 11050440. [DOI] [PubMed] [Google Scholar]
- 32.Watkins ER, Maiden MC. Persistence of hyperinvasive meningococcal strain types during global spread as recorded in the PubMLST database. PLoS ONE. 2012;7(9):e45349. doi: 10.1371/journal.pone.0045349. PMID: 23028953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell JE, Urwin R, Gray SJ, Fox AJ, Feavers IM, Maiden MC. Molecular epidemiology of meningococcal disease in England and Wales 1975-1995, before the introduction of serogroup C conjugate vaccines. Microbiology. 2008;154(Pt 4):1170–7. doi: 10.1099/mic.0.2007/014761-0. PMID: 18375809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law DKS, Zhou J, Deng S, Hoang L, Tyrrell G, Horsman G, et al. Determination of serotyping antigens, clonal analysis and genetic characterization of the 4CMenB vaccine antigen genes in invasive Neisseria meningitidis from Western Canada, 2009 to 2013. J Med Microbiol. 2014;63(Pt 11):1490–9. doi: 10.1099/jmm.0.079921-0. PMID: 25165123. [DOI] [PubMed] [Google Scholar]
- 35.Holst J, Comanducci M, Bambini S, Muzzi A, Comandi S, Oksnes J, et al. Variability of genes encoding surface proteins used as vaccine antigens in meningococcal endemic and epidemic strain panels from Norway. Vaccine. 2014;32(23):2722–31. doi: 10.1016/j.vaccine.2014.02.068. PMID: 24631075. [DOI] [PubMed] [Google Scholar]
- 36.Brehony C, Trotter CL, Ramsay ME, Chandra M, Jolley KA, van der Ende A, et al. Implications of differential age distribution of disease-associated meningococcal lineages for vaccine development. Clin Vaccine Immunol. 2014;21(6):847–53. doi: 10.1128/CVI.00133-14. PMID: 24695776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch C, et al. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci USA. 2010;107(45):19490–5. doi: 10.1073/pnas.1013758107. PMID: 20962280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel U, Taha MK, Vazquez JA, Findlow J, Claus H, Stefanelli P, et al. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: a qualitative and quantitative assessment. Lancet Infect Dis. 2013;13(5):416–25. doi: 10.1016/S1473-3099(13)70006-9. PMID: 23414709. [DOI] [PubMed] [Google Scholar]
- 39.Tzanakaki G, Hong E, Kesanopoulos K, Xirogianni A, Bambini S, Orlandi L, et al. Diversity of Greek meningococcal serogroup B isolates and estimated coverage of the 4CMenB meningococcal vaccine. BMC Microbiol. 2014;14(1):111. doi: 10.1186/1471-2180-14-111. PMID: 24779381. [DOI] [PMC free article] [PubMed] [Google Scholar]