Abstract
AIM: To identify plasma analytes using metabolomics that correlate with the diagnosis and severity of liver disease in patients with alcoholic hepatitis (AH).
METHODS: We prospectively recruited patients with cirrhosis from AH (n = 23) and those with cirrhosis with acute decompensation (AD) from etiologies other than alcohol (n = 25). We used mass spectrometry to identify 29 metabolic compounds in plasma samples from fasted subjects. A receiver operating characteristics analysis was performed to assess the utility of biomarkers in distinguishing acute AH from alcoholic cirrhosis. Logistic regression analysis was performed to build a predictive model for AH based on clinical characteristics. A survival analysis was used to construct Kaplan Meier curves evaluating transplant-free survival.
RESULTS: A comparison of model for end-stage liver disease (MELD)-adjusted metabolomics levels between cirrhosis patients who had AD or AH showed that patients with AH had significantly higher levels of betaine, and lower creatinine, phenylalanine, homocitrulline, citrulline, tyrosine, octenoyl-carnitine, and symmetric dimethylarginine. When considering combined levels, betaine and citrulline were highly accurate predictors for differentiation between AH and AD (area under receiver operating characteristics curve = 0.84). The plasma levels of carnitine [0.54 (0.18, 0.91); P = 0.005], homocitrulline [0.66 (0.34, 0.99); P < 0.001] and pentanoyl-carnitine [0.53 (0.16, 0.90); P = 0.007] correlated with MELD scores in patients diagnosed with AH. Increased levels of many biomarkers (carnitine P = 0.005, butyrobetaine P = 0.32, homocitrulline P = 0.002, leucine P = 0.027, valine P = 0.024, phenylalanine P = 0.037, tyrosine P = 0.012, acetyl-carnitine P = 0.006, propionyl-carnitine P = 0.03, butyryl-carnitine P = 0.03, trimethyl-lisine P = 0.034, pentanoyl-carnitine P = 0.03, hexanoyl-carnitine P = 0.026) were associated with increased mortality in patients with AH.
CONCLUSION: Metabolomics plasma analyte levels might be used to diagnose of AH or help predict patient prognoses.
Keywords: Metabolomics, Biomarkers, Liver disease, Model for end-stage liver disease, Cirrhosis, Alcoholic hepatitis, Liver biopsy
Core tip: The model for end-stage liver disease score, which is commonly used to predict outcomes in patients who have liver disease, is far from perfect. We report results from a study that uses metabolomics biomarkers as a means for assessing diagnosis and prognosis in patients who have liver disease. Plasma analytes from fasted subjects have provided information regarding 3 and 6 mo transplant free survival. This study is one of the first to employ the novel metabolomics approach as it relates to patient outcomes. These results can pave the way for future research that can enhance the way we assess patients with liver disease.
INTRODUCTION
Generally, clinical assessment is sufficient to generate a diagnosis of alcoholic hepatitis (AH). However, sole dependence on clinical signs and symptoms is not specific, and further confirmation is usually needed. Thus, the gold standard for the diagnosis of AH is liver biopsy. Liver biopsy is considered an expensive and invasive procedure, and 1%-5% of patients require post-procedural hospitalization[1]. In addition, sampling error and inter-observer variability contribute to the limitations of liver biopsy as a procedure[1]. Therefore, it behooves practitioners to utilize alternative non-invasive tools to diagnose AH. Hanouneh et al[1] have shown promise in the possibility of analyzing volatile compounds in breath samples as a useful diagnostic test in patients with AH. Consequently, a rapid, non-invasive, accurate, and precise test would greatly benefit AH diagnosis.
Furthermore, prognosis of AH is determined by several scoring systems, including the model for end-stage liver disease (MELD), which is primarily based on serum lab values and is one of the chief parameters in evaluation of long-term outcome and qualification for liver transplant. While the MELD score can detect short-term survival in patients with AH with good accuracy, its prediction of long-term survival is still debated[2]. Palaniyappan et al[2] evaluated several scoring systems and their ability to predict long-term outcome of AH and concluded that all scoring systems were uniformly poor in predicting long term survival beyond six months. In addition, the cut-off value for the MELD score in detecting severe AH has not been agreed upon, with various studies employing different values[2]. Therefore, MELD score may not accurately reflect the risk of death in some groups of patients with liver disease such as AH awaiting liver transplantation.
Metabolomics was originally defined as the detailed qualitative and quantitative analysis of the metabolites present in complex biological samples[3]. Metabolites are both the intermediate and end result of all the biological processes taking place in a cell, tissue, or organism, thereby serving as the most proximal reporters of the body’s response to a disease process or drug therapy[4]. By identifying and quantifying metabolites, one can gather a picture of the genetic variations and environmental influences (such as diet, lifestyle, drug use, and toxicological exposure) in a biological specimen. In more recent years physicians have been exploring the potential of metabolite profiling in providing diagnostic and prognostic information for many diseases, such as AH. For example, Rachakonda et al[5] demonstrated via metabolomics profiling that specific biomarkers could be used to determine disease prognosis in patients with severe AH. Thus, the potential of utilizing biomarkers in diagnosis of liver disease, assessing liver disease severity, and determining long-term survival in patients with AH is worth investigating; further exploration is warranted as there is limited information on this subject. Herein, we used a targeted metabolomics approach to identify plasma analytes that may provide improved diagnostic and prognostic value in patients with alcoholic hepatitis and end-stage liver disease.
MATERIALS AND METHODS
Patients
We recruited patients with liver cirrhosis awaiting liver transplantation from a single tertiary care center. The study population was divided between those with AH with cirrhosis (n = 25) and those with acute decompensated (AD) cirrhosis from etiologies other than alcohol (n = 23). The diagnosis of AH with cirrhosis was based on clinical and laboratory features: A patient with a history of heavy alcohol use, exclusion of other causes of liver disease, elevated aspartate aminotransferase that remained under < 300 IU/mL, a ratio of aspartate aminotransferase (AST) level to alanine aminotransferase (ALT) level that is > 2, total serum bilirubin level of > 5 mg/dL, an elevated international normalized ratio, and neutrophilia. Significant alcohol intake was defined as a consumption of > 2 drinks daily or > 6 drinks daily on weekends for the past 5 years. We used the definition of the American Association for the Study of Liver Disease guidelines of what constitutes a standard drink: 12 g of alcohol with range 9.3-13.2 g.
The diagnosis of liver cirrhosis was based on the histologic features of cirrhosis on liver biopsy and/or a composite of clinical signs and findings of cirrhosis provided by laboratory tests, endoscopy, and radiologic imaging. AH was defined by the acute development of one major complication of liver disease including acute kidney injury, ascites, encephalopathy, or gastro-intestinal hemorrhage secondary to gastrointestinal varices or portal hypertensive gastropathy and enteropathy. Hepatic encephalopathy was assessed by a single individual using Conn score and asterixis grade. Acute kidney injury was defined as an abrupt (arbitrarily set at 48 h) reduction in kidney function manifested by an absolute increase in serum creatinine of 0.3 mg/dL or more, equivalent to a percentage increase in serum creatinine of 50% or more (1.5-fold from baseline)[6].
Among patients with acute decompensated liver cirrhosis, only those who remained abstinent from alcohol use for at least 6 mo before admission were included, whereas all patients with AH were (by definition) actively abusing alcohol before admission. The data was collected at the time of diagnosis and admission with alcoholic hepatitis - subjects were not drinking alcohol following admission. We also excluded all individuals with ongoing tobacco use. Patients with liver cancer or other malignancies were excluded, as were those with prior history of transplantation.
Data collection
Mass spectrometry identified and measured 29 metabolomics compounds related to amino acid and intermediary metabolism in plasma samples from fasted subjects. Samples and associated clinical data were collected from fasting subjects undergoing community health screens. All subjects gave written informed consent and the Institutional Review Board of the Cleveland Clinic approved all study protocols.
Quantification of plasma analytes by liquid chromatography/mass spectrometry/mass spectrometry
Stable isotope dilution liquid chromatography/mass spectrometry (MS)/MS was used to quantify plasma analytes. Four volumes of methanol containing isotope-labeled internal standards was added to one volume of plasma for protein precipitation. After centrifugation, supernatant was analyzed by injection onto a silica column that was interfaced with an atmospheric pressure ionization 4000 Q-TRAP mass spectrometer (AB SCIEX, Framingham, MA)[7]. A discontinuous gradient was generated to resolve analytes by mixing 0.1% propanoic acid in water with 0.1% acetic acid in methanol[7]. Analytes and the isotope-labeled internal standards were monitored in positive multiple reaction monitoring MS mode using characteristic precursor-product ion transitions (Table 1). Parameters for ion monitoring were optimized for each analyte. Various concentrations of analytes were spiked into a control plasma sample to prepare calibration curves for quantification of analytes.
Table 1.
Characteristic precursor-product transitions
| Name | Precursor | Product | |
| Analytes | Trimethylamine N-oxide | 76 | 58 |
| Choline | 104 | 60 | |
| Betaine | 118 | 59 | |
| Valine | 118 | 72 | |
| Leucine | 86 | 43 | |
| Isoleucine | 86 | 56 | |
| Ornithine | 133 | 70 | |
| Crotonobetaine | 144 | 59 | |
| Butyrobetaine | 146 | 60 | |
| Lysine | 147 | 84 | |
| Methyl-lysine | 161 | 84 | |
| Carnitine | 162 | 60 | |
| Phenylalanine | 166 | 120 | |
| Arginine | 175 | 70 | |
| Citrulline | 176 | 70 | |
| Tyrosine | 182 | 136 | |
| Methyl-arginine | 189 | 70 | |
| Symmetric dimethyl-arginine | 203 | 70 | |
| Asymmetric dimethylarginine | 203 | 70 | |
| Acetyl-carnitine | 204 | 85 | |
| Propionyl-carnitine | 218 | 85 | |
| Butyryl-carnitine | 232 | 85 | |
| Pentanoyl-carntine | 246 | 85 | |
| Hexanoyl-carnitine | 260 | 85 | |
| Octenoyl-carnitine | 286 | 85 | |
| Internal standard | Trimethylamine N-oxide-d9 | 85 | 66 |
| Choline-trimethyl-d9 | 113 | 69 | |
| Betaine-trimethyl-d9 | 127 | 68 | |
| Valine-13C5, 15N1 | 124 | 77 | |
| Leucine-13C6, 15N1 | 139 | 92 | |
| Ornithine 3, 3, 4, 4, 5, 5-d6 | 139 | 76 | |
| Crotonobetaine-trimethyl-d9 | 153 | 68 | |
| Butyrobetaine-trimethyl-d9 | 155 | 69 | |
| Lysine-u-13C6, 15N2 | 155 | 90 | |
| Phenylalanine-13C6 | 172 | 126 | |
| Citrulline 2, 3, 4, 5-d4 | 180 | 74 | |
| Arginine-13C6 | 181 | 74 | |
| Tyrosine-u-13C9, 15N1 | 192 | 145 | |
| Asymmetric dimethylarginine 2, 3, 3, 4, 4, 5, 5-d7 | 210 | 77 | |
| Acetyl-carnitine-d3 | 207 | 85 | |
| Propionyl-carnitine-d3 | 221 | 85 | |
| butyryll-carnitine-d3 | 235 | 85 | |
| Pentanoyl-carnitine-d9 | 246 | 85 | |
| Hexanoyl-carnitine-d3 | 263 | 85 |
Statistical analysis
Data are presented as mean ± SD, median (25th, 75th percentiles) or n (%). Univariable analysis was performed to compare clinical characteristics and biomarker levels between the two groups. Analysis of variance or the non-parametric Kruskal-Wallis test were used to assess differences in continuous variables and Pearson’s χ2 tests or Fisher’s exact tests were used for categorical factors. Analysis of covariance was used to assess differences in biomarker levels while adjusting for MELD; the logarithm of each compound was modeled as the outcome variable with group and MELD as the independent variables. Receiver operating characteristics (ROC) analysis was performed to assess the utility of biomarkers in distinguishing acute alcoholic hepatitis from alcoholic cirrhosis; the area under the ROC curves [area under receiver operating characteristics curve (AUC)] and corresponding 95%CI are presented.
We used various statistical analyses to compare clinical characteristics and plasma levels of compounds among groups and to test the correlation between levels of compounds and severity of liver disease. Correlations between 0.0-0.3 are considered low, between 0.3-0.5 are considered moderate, and between 0.5-0.7 are considered high, and between 0.7-1.0 are considered very high. Spearman’s correlation coefficients were also used to assess correlations between biomarkers and severity of liver disease for each group separately. Finally, logistic regression analysis was performed to build a predictive model for AH.
Lastly, a survival analysis was done to evaluate transplant-free survival. Kaplan-Meier product-limit estimates were used to assess transplant-free survival. Follow-up time was defined as time from sample collection to death and subjects were censored at time of orthotopic liver transplantation (OLT), if applicable, or last follow-up visit. Cox regression was used to assess associations between biomarker levels and transplant-free survival. In addition, inverse probability of censoring weighting estimation of cumulative/dynamic time-dependent ROC curve was used to assess the role of novel biomarkers in prediction of 3 and 6-mo LT-free survival[8,9]. Each marker was compared to the MELD score and markers with AUC of at least 0.70 were further assessed to see if any of these improved prediction of survival in combination with MELD. A P < 0.05 was considered statistically significant. A 95%CI encompassing 0.5 was considered to indicate no significant predictive value. SAS (version 9.2, the SAS Institute, Cary, NC) and R (version 3.0.3, the R Foundation for Statistical Computing) were used to perform all analyses. The statistical methods of this study were reviewed by Rocio Lopez from the Cleveland Clinic Foundation.
RESULTS
Baseline characteristics
Table 2 presents a summary of patient demographic and clinical characteristics. A total of 45 subjects were included in the analysis. The average age was 53 ± 10 years, 54% were male, and 75% were Caucasian. The mean MELD score was 18.0 ± 9.3. MELD score was comparable between subjects with AH and those with AD.
Table 2.
Patient characteristics
| Factor |
Cirrhosis with acute decompensation from etiologies other than alcohol (n = 23) |
Alcoholic hepatitis (n = 25) |
P-value | ||
| n | Summary | n | Summary | ||
| Age (yr) | 14 | 53.8 ± 9.8 | 20 | 51.5 ± 10.4 | 0.511 |
| Male | 21 | 13 (61.9) | 23 | 10 (43.5) | 0.223 |
| Caucasian | 10 | 10 (100.0) | 19 | 16 (84.2) | 0.534 |
| AST | 23 | 40.0 (33.0, 75.0) | 25 | 138.0 (88.0, 161.0) | < 0.0012b |
| ALT | 23 | 21.0 (15.0, 30.0) | 25 | 51.0 (42.0, 71.0) | < 0.0012b |
| Bilirubin | 23 | 3.8 (1.4, 6.0) | 25 | 9.4 (6.8, 21.7) | 0.0052b |
| Albumin | 23 | 2.8 ± 0.66 | 25 | 2.8 ± 0.68 | 0.861 |
| INR | 23 | 1.5 ± 0.41 | 25 | 1.7 ± 0.59 | 0.161 |
| PT | 23 | 16.4 ± 4.5 | 25 | 19.1 ± 6.1 | 0.0951 |
| Creatinine | 23 | 0.94 (0.74, 1.5) | 25 | 0.72 (0.57, 1.04) | 0.0622 |
| MELD score | 23 | 16.4 ± 8.8 | 25 | 20.5 ± 10.0 | 0.141 |
| Maddrey's score | 16 | 22.2 (16.0, 37.0) | 17 | 43.5 (34.0, 60.6) | 0.0282a |
| Ascites | 23 | 25 | 0.142 | ||
| None | 9 (39.1) | 14 (56.0) | |||
| Small | 4 (17.4) | 6 (24.0) | |||
| Large | 9 (39.1) | 4 (16.0) | |||
| Severe | 1 (4.3) | 1 (4.0) | |||
| HE | 23 | 25 | 0.942 | ||
| None | 2 (8.7) | 6 (24.0) | |||
| Mild | 12 (52.2) | 7 (28.0) | |||
| Severe | 9 (39.1) | 12 (48.0) | |||
| Steroids | 23 | 2 (8.7) | 25 | 13 (52.0) | 0.0013b |
| Trental | 23 | 3 (13.0) | 25 | 9 (36.0) | 0.0673 |
| OLT | 23 | 2 (8.7) | 25 | 1 (4.0) | 0.604 |
| Deceased | 23 | 6 (26.1) | 25 | 8 (32.0) | 0.653 |
P-values were calculated using the test corresponding to superscript characters:
ANOVA;
Kruskal-Wallis test;
Pearson’s χ2 test;
Fisher’s exact test.
P < 0.05 and
P < 0.01. AST: Aspartate aminotransferase; ALT: Alanine aminotransferase; MELD: Model for end-stage liver disease; OLT: Orthotopic liver transplantation; PT: Prothrombin time; INR: International normalized ratio; HE: Hepatic encephalopathy.
Metabolomics biomarkers of alcoholic hepatitis
Table 3 presents a summary of MELD-adjusted biomarker levels in the two study groups. Betaine, creatinine, homocitrulline and citrulline, tyrosine, phenylalanine, octenoyl carnitine, and symmetric dimethylarginine (SDMA) were significantly higher in patients with AH compared to those with AD.
Table 3.
Model for end-stage liver disease-adjusted average biomarker levels
| Biomarker (μmol/L) | Cirrhosis with acute decompensation from etiologies other than alcohol (n = 23) | Alcoholic hepatitis (n = 25) | P-value |
| Choline | 5.8 (4.7, 7.2) | 7.0 (5.7, 8.6) | 0.22 |
| TMAO | 0.74 (0.34, 1.6) | 0.87 (0.42, 1.8) | 0.76 |
| Carnitine | 37.2 (32.1, 43.1) | 32.6 (28.3, 37.5) | 0.2 |
| Betaine | 83.6 (64.7, 108.2) | 134.0 (104.7, 171.5) | 0.012a |
| Butyrobetaine | 1.6 (1.3, 1.8) | 1.7 (1.4, 2.0) | 0.46 |
| Crotonobetaine | 0.12 (0.10, 0.15) | 0.14 (0.12, 0.18) | 0.35 |
| Creatinine | 92.0 (75.0, 112.9) | 59.3 (48.8, 72.1) | 0.003b |
| Ornithine | 71.6 (59.1, 86.9) | 62.0 (51.5, 74.6) | 0.29 |
| Lysine | 131.3 (112.7, 153.1) | 134.8 (116.4, 156.2) | 0.81 |
| Methyl-lysine | 4.0 (2.9, 5.6) | 3.4 (2.5, 4.7) | 0.46 |
| Argine | 61.7 (52.0, 73.2) | 61.8 (52.4, 72.8) | 0.99 |
| Citrulline | 40.2 (33.2, 48.6) | 23.7 (19.7, 28.5) | < 0.001b |
| MMA | 0.24 (0.20, 0.27) | 0.21 (0.19, 0.24) | 0.32 |
| Homocitrulline | 0.73 (0.55, 0.97) | 0.37 (0.28, 0.48) | 0.001b |
| Leucine | 52.8 (44.0, 63.4) | 48.8 (40.9, 58.1) | 0.54 |
| Iso-leucine | 27.1 (21.7, 33.8) | 28.0 (22.6, 34.6) | 0.83 |
| Valine | 115.4 (100.4, 132.7) | 101.8 (89.1, 116.4) | 0.2 |
| Phenylalanine | 90.4 (78.0, 104.7) | 60.2 (52.3, 69.3) | < 0.001b |
| Tyrosine | 166.0 (126.8, 217.3) | 107.4 (83.0, 139.1) | 0.025a |
| Acetyl-carnitine | 17.8 (14.9, 21.1) | 16.9 (14.3, 20.0) | 0.69 |
| Propionyl-carnitine | 1.02 (0.76, 1.4) | 1.2 (0.88, 1.5) | 0.5 |
| Butyryl-carnitine | 2.0 (1.6, 2.5) | 2.2 (1.8, 2.7) | 0.58 |
| Trimethyl-Lysine | 1.00 (0.81, 1.2) | 1.1 (0.92, 1.4) | 0.42 |
| SDMA | 0.81 (0.65, 1.02) | 0.58 (0.47, 0.72) | 0.042a |
| Dimethyl-Lysine | 0.92 (0.72, 1.2) | 0.74 (0.59, 0.94) | 0.22 |
| ADMA | 0.90 (0.78, 1.03) | 0.82 (0.72, 0.93) | 0.32 |
| Pentanoyl-carnitine | 0.25 (0.20, 0.31) | 0.27 (0.22, 0.34) | 0.48 |
| Hexanoyl-carnitine | 0.69 (0.56, 0.85) | 0.61 (0.50, 0.74) | 0.38 |
| Octenoyl-carnitine | 0.05 (0.02, 0.12) | 0.01 (0.00, 0.02) | 0.009b |
Values presented as mean (95%CI) and P-values obtained from analysis of covariance. The natural logarithm of each biomarker was modeled as the outcome variable with disease group and MELD as the independent variables.
P < 0.05,
P < 0.01. ADMA: Asymmetric dimethylarginine; SDMA: Symmetric dimethylarginine; MMA: Monomethylarginine; TMAO: Trimethylamine N-oxide.
Table 4 presents AUC data using ROC analysis, where values greater than 0.7 are strongly predictive for differentiation between AH and AD. Citrulline, betaine, and tyrosine were all notable for their values in differentiating AH from AD. Using a combination of citrulline and betaine provided the greatest AUC, at 0.835 with a 95%CI between 0.747 and 0.978. Other significant biomarkers include homocitrulline, SDMA, octenoyl-carnitine, creatinine, and phenylalanine. The remaining biomarkers were insignificant.
Table 4.
Utility of biomarkers in differentiating cirrhosis with alcoholic hepatitis from cirrhosis with acute decompensation from etiologies other than alcohol: Receiver operating characteristics analysis
| Biomarker | AUC (95%CI) |
| Citrulline and betaine | 0.835 (0.747, 0.978) |
| Betaine and phenylalanine | 0.810 (0.684, 0.937) |
| Citrulline and phenylalanine | 0.758 (0.609, 0.907) |
| Citrulline | 0.758 (0.610, 0.907) |
| Betaine | 0.732 (0.588, 0.877) |
| Phenylalanine | 0.715 (0.567, 0.863) |
| Crotonobetaine | 0.663 (0.498, 0.827) |
| Tyrosine | 0.650 (0.484, 0.817) |
| Butyrobetaine | 0.649 (0.489, 0.808) |
| Creatinine | 0.645 (0.486, 0.805) |
| Octenoyl-carnitine | 0.631 (0.488, 0.774) |
| Propionyl-carnitine | 0.620 (0.453, 0.787) |
| Homocitrulline | 0.618 (0.457, 0.779) |
| Trimethyl-lysine | 0.616 (0.447, 0.784) |
| Butyryl-carnitine | 0.615 (0.451, 0.778) |
| Pentanoyl-carnitine | 0.613 (0.455, 0.771) |
| Choline | 0.597 (0.432, 0.762) |
| Valine | 0.588 (0.424, 0.752) |
| Lysine | 0.559 (0.393, 0.725) |
| Methyl-lysine | 0.552 (0.383, 0.721) |
| Iso-leucine | 0.548 (0.379, 0.716) |
| Acetyl-carnitine | 0.543 (0.375, 0.710) |
| Hexanoyl-carnitine | 0.529 (0.362, 0.696) |
| TMAO | 0.521 (0.347, 0.695) |
| Argine | 0.507 (0.339, 0.675) |
| ADMA | 0.481 (0.314, 0.647) |
| Leucine | 0.477 (0.309, 0.644) |
| Carnitine | 0.470 (0.301, 0.638) |
| MMA | 0.453 (0.286, 0.620) |
| SDMA | 0.447 (0.277, 0.617) |
| Dimethyl-lysine | 0.417 (0.243, 0.592) |
| Ornithine | 0.351 (0.186, 0.517) |
AUC: Area under receiver operating characteristics curve; ADMA: Asymmetric dimethylarginine; SDMA: Symmetric dimethylarginine; MMA: Monomethylarginine; TMAO: Trimethylamine N-oxide.
Table 5 presents the correlations between biomarkers and liver disease severity for alcoholic hepatitis. There was moderate to strong correlation between several biomarkers and both MELD and Maddrey’s scores. Correlations between 0.0-0.3 are considered trivial/low, 0.3-0.5 are considered moderate, 0.5-0.7 are considered high and 0.7-1.0 are considered very high/strong.
Table 5.
Correlations between biomarkers and model for end-stage liver disease and Maddrey’s score in patients with alcoholic hepatitis
| Biomarker |
Alcoholic hepatitis |
|||
|
MELD |
Maddrey’s score |
|||
| rho (95%CI) | P-value | rho (95%CI) | P-value | |
| Choline | 0.28 (-0.13, 0.69) | 0.18 | 0.02 (-0.53, 0.57) | 0.95 |
| TMAO | -0.25 (-0.67, 0.17) | 0.23 | 0.29 (-0.24, 0.82) | 0.26 |
| Carnitine | 0.54 (0.18, 0.91) | 0.005b | 0.48 (-0.01, 0.96) | 0.054 |
| Betaine | 0.24 (-0.18, 0.65) | 0.26 | -0.00 (-0.55, 0.55) | 0.99 |
| Butyrobetaine | 0.30 (-0.11, 0.71) | 0.15 | 0.29 (-0.24, 0.81) | 0.26 |
| Crotonobetaine | 0.07 (-0.36, 0.50) | 0.74 | -0.03 (-0.58, 0.52) | 0.91 |
| Creatinine | 0.44 (0.05, 0.83) | 0.027a | 0.48 (-0.00, 0.96) | 0.052 |
| Ornithine | 0.28 (-0.13, 0.70) | 0.17 | 0.26 (-0.27, 0.79) | 0.32 |
| Lysine | 0.37 (-0.03, 0.77) | 0.068 | 0.27 (-0.25, 0.80) | 0.29 |
| Methyl-lysine | 0.09 (-0.34, 0.52) | 0.67 | -0.01 (-0.56, 0.54) | 0.96 |
| Argine | 0.00 (-0.43, 0.44) | 0.98 | 0.42 (-0.08, 0.92) | 0.095 |
| Citrulline | 0.09 (-0.34, 0.52) | 0.66 | 0.05 (-0.50, 0.60) | 0.86 |
| MMA | 0.34 (-0.07, 0.74) | 0.098 | 0.39 (-0.11, 0.90) | 0.12 |
| Homocitrulline | 0.66 (0.34, 0.99) | < 0.001b | 0.59 (0.14, 1.00) | 0.014a |
| Leucine | 0.03 (-0.40, 0.46) | 0.89 | 0.50 (0.02, 0.97) | 0.043 |
| Iso-leucine | 0.11 (-0.32, 0.54) | 0.59 | 0.29 (-0.24, 0.82) | 0.26 |
| Valine | 0.20 (-0.22, 0.62) | 0.34 | 0.54 (0.08, 1.00) | 0.025a |
| Phenylalanine | 0.34 (-0.06, 0.75) | 0.092 | 0.56 (0.11, 1.00) | 0.018a |
| Tyrosine | 0.30 (-0.11, 0.71) | 0.14 | 0.44 (-0.05, 0.94) | 0.074 |
| Acetyl-carnitine | 0.49 (0.11, 0.86) | 0.014a | 0.50 (0.03, 0.98) | 0.04a |
| Propionyl-carnitine | 0.40 (0.01, 0.80) | 0.046a | 0.28 (-0.25, 0.81) | 0.28 |
| Butyryl-carnitine | 0.48 (0.10, 0.86) | 0.016a | 0.25 (-0.29, 0.78) | 0.34 |
| Trimethyl-lysine | 0.48 (0.11, 0.86) | 0.014a | 0.38 (-0.13, 0.89) | 0.14 |
| SDMA | 0.38 (-0.02, 0.78) | 0.064 | 0.55 (0.09, 1.00) | 0.023a |
| Dimethyl-lysine | 0.31 (-0.10, 0.72) | 0.13 | 0.18 (-0.37, 0.72) | 0.5 |
| ADMA | 0.42 (0.03, 0.81) | 0.037a | 0.60 (0.16, 1.00) | 0.011a |
| Pentanoyl-carnitine | 0.53 (0.16, 0.90) | 0.007b | 0.31 (-0.22, 0.83) | 0.23 |
| Hexanoyl-carnitine | 0.49 (0.12, 0.87) | 0.013a | 0.40 (-0.11, 0.90) | 0.11 |
| Octenoyl-carnitine | 0.45 (0.07, 0.84) | 0.024a | 0.23 (-0.30, 0.77) | 0.37 |
Values presented as mean (95%CI) and P-values obtained from analysis of covariance. The natural logarithm of each biomarker was modeled as the outcome variable with disease group and MELD as the independent variables. A superscript of a indicates
P < 0.05,
P < 0.01. rho: Spearman’s correlation; MELD: Model for end-stage liver disease; ADMA: Asymmetric dimethylarginine; SDMA: Symmetric dimethylarginine; MMA: Monomethylarginine; TMAO: Trimethylamine N-oxide.
The objective of this study was to detect patterns in biomarkers or hypothesis generation. In addition, adjustments for multiple comparisons are typically somewhat conservative and it would be possible to miss many potential associations that should be further explored. Holm-Bonferroni adjustment is quite conservative when the number of tests is large or the tests are not independent[10]. Despite this, we performed the Holm-Bonferroni adjustment to provide a more complete set of data (Table 6). In this case only citrulline, phenylalanine, and homocitrulline remain significantly different between the groups.
Table 6.
Model for end-stage liver disease-adjusted average biomarker levels
| Biomarker (μmol/L) | Alcoholic cirrhosis (n = 23) | Alcoholic hepatitis (n = 25) | Holm-bonferroni corrected P-value |
| Citrulline | 40.2 (33.2, 48.6) | 23.7 (19.7, 28.5) | 0.009b |
| Phenylalanine | 90.4 (78.0, 104.7) | 60.2 (52.3, 69.3) | 0.009b |
| Homocitrulline | 0.73 (0.55, 0.97) | 0.37 (0.28, 0.48) | 0.029a |
| Creatinine | 92.0 (75.0, 112.9) | 59.3 (48.8, 72.1) | 0.087 |
| Octenoyl-carnitine | 0.05 (0.02, 0.12) | 0.01 (0.00, 0.02) | 0.26 |
| Betaine | 83.6 (64.7, 108.2) | 134.0 (104.7, 171.5) | 0.35 |
| Tyrosine | 166.0 (126.8, 217.3) | 107.4 (83.0, 139.1) | 0.73 |
| SDMA | 0.81 (0.65, 1.02) | 0.58 (0.47, 0.72) | 0.99 |
| Carnitine | 37.2 (32.1, 43.1) | 32.6 (28.3, 37.5) | 0.99 |
| Valine | 115.4 (100.4, 132.7) | 101.8 (89.1, 116.4) | 0.99 |
| Choline | 5.8 (4.7, 7.2) | 7.0 (5.7, 8.6) | 0.99 |
| Dimethyl-lysine | 0.92 (0.72, 1.2) | 0.74 (0.59, 0.94) | 0.99 |
| Ornithine | 71.6 (59.1, 86.9) | 62.0 (51.5, 74.6) | 0.99 |
| MMA | 0.24 (0.20, 0.27) | 0.21 (0.19, 0.24) | 0.99 |
| ADMA | 0.90 (0.78, 1.03) | 0.82 (0.72, 0.93) | 0.99 |
| Crotonobetaine | 0.12 (0.10, 0.15) | 0.14 (0.12, 0.18) | 0.99 |
| Hexanoyl-carnitine | 0.69 (0.56, 0.85) | 0.61 (0.50, 0.74) | 0.99 |
| Trimethyl-lysine | 1.00 (0.81, 1.2) | 1.1 (0.92, 1.4) | 0.99 |
| Butyrobetaine | 1.6 (1.3, 1.8) | 1.7 (1.4, 2.0) | 0.99 |
| Methyl-lysine | 4.0 (2.9, 5.6) | 3.4 (2.5, 4.7) | 0.99 |
| Pentanoyl-carnitine | 0.25 (0.20, 0.31) | 0.27 (0.22, 0.34) | 0.99 |
| Propionyl-carnitine | 1.02 (0.76, 1.4) | 1.2 (0.88, 1.5) | 0.99 |
| Leucine | 52.8 (44.0, 63.4) | 48.8 (40.9, 58.1) | 0.99 |
| Butyryl-carnitine | 2.0 (1.6, 2.5) | 2.2 (1.8, 2.7) | 0.99 |
| Acetyl-carnitine | 17.8 (14.9, 21.1) | 16.9 (14.3, 20.0) | 0.99 |
| TMAO | 0.74 (0.34, 1.6) | 0.87 (0.42, 1.8) | 0.99 |
| Lysine | 131.3 (112.7, 153.1) | 134.8 (116.4, 156.2) | 0.99 |
| Iso-leucine | 27.1 (21.7, 33.8) | 28.0 (22.6, 34.6) | 0.99 |
| Argine | 61.7 (52.0, 73.2) | 61.8 (52.4, 72.8) | 0.99 |
Values presented as mean (95%CI) and P-values obtained from analysis of covariance. The natural logarithm of each biomarker was modeled as the outcome variable with disease group and MELD as the independent variables. A superscript of a indicates
P < 0.05,
P < 0.01. ADMA: Asymmetric dimethylarginine; SDMA: Symmetric dimethylarginine; MMA: Monomethylarginine; TMAO: Trimethylamine N-oxide.
Metabolomics biomarkers of severity of liver disease
Patients were followed over 12.5 (P25, P75: 4.3, 14.1) mo during which three subjects received a liver transplant and a total of 24% of subjects expired. As seen in Figure 1, tyrosine was strongly associated with transplant-free survival outcome in patients with liver cirrhosis [AUC for 3-mo OLT-free survival AUC = 0.91 (0.74-1.0)]. Combined MELD scores and tyrosine levels provided the best accuracy for 3-mo transplant-free survival AUC = 0.92 (0.76-1.0). Evidently these biomarkers can be used to predict OLT-free survival with reasonable sensitivity and specificity.
Figure 1.
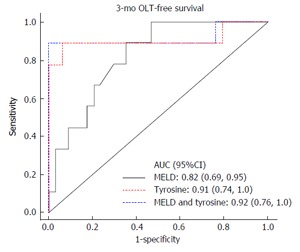
Tyrosine predicts 3-mo liver transplant-free survival in patients with end-stage liver disease. Results are presented as AUC (P25, P75). AUC: Area under receiver operating characteristics curve; OLT: Orthotopic liver transplantation; MELD: Model for end-stage liver disease.
Figure 2 shows the same analysis with similar results, except for 6-mo OLT-free survival. MELD provided an AUC of 0.77, tyrosine provided an AUC of 0.90, and MELD and tyrosine together provided an AUC of 0.89. Tyrosine alone as well as tyrosine in combination with MELD provided better AUC values than MELD alone, suggesting its utility in predicting OLT-free survival.
Figure 2.
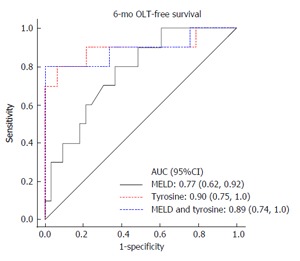
Tyrosine predicts 6-mo liver transplant-free survival in patients with end-stage liver disease. AUC: Area under receiver operating characteristics curve; OLT: Orthotopic liver transplantation; MELD: Model for end-stage liver disease.
A multivariable Cox regression analysis was used to adjust for MELD, the most important predictor of mortality, and tyrosine remained significantly associated with mortality [HR = 1.02 (1.01, 1.04) for a one unit increase in tyrosine; P = 0.002]. In Figures 1 and 2 it can also be seen that tyrosine performs better than MELD for prediction of 3- and 6-mo mortality, and the combination of MELD and tyrosine (in a multivariable analysis) performs more or less the same as the compound by itself.
Time-dependent ROC analysis (Table 7) shows that phenylalanine [AUC = 0.77 (0.56, 0.97)], carnitine [AUC = 0.73 (0.53, 0.93)], asymmetric dimethylarginine (ADMA) [AUC = 0.72 (0.49, 0.96)] and monomethylarginine (MMA) [AUC = 0.71 (0.47, 0.94)] all provide excellent predictive value for transplant-free survival in patients with liver cirrhosis, but there was no evidence to suggest that they were significantly better than MELD.
Table 7.
Utility of biomarkers in predicting transplant-free survival
| Biomarker | 3-mo survival | 6-mo survival |
| (μmol/L) | AUC (95%CI) | AUC (95%CI) |
| MELD | 0.82 (0.69, 0.95) | 0.77 (0.62, 0.92) |
| Tyrosine | 0.91 (0.74, 1.0) | 0.89 (0.74, 1.0) |
| Phenylalanine | 0.77 (0.56, 0.97) | 0.79 (0.61, 0.98) |
| Carnitine | 0.73 (0.53, 0.93) | 0.74 (0.56, 0.93) |
| ADMA | 0.72 (0.49, 0.96) | 0.70 (0.48, 0.92) |
| MMA | 0.71 (0.47, 0.94) | 0.70 (0.49, 0.91) |
AUC: Area under receiver operating characteristics curve; MELD: Model for end-stage liver disease; ADMA: Asymmetric dimethylarginine; MMA: Monomethylarginine.
DISCUSSION
The purpose of this study was two-fold. First, the utility of metabolomics as an un-invasive diagnostic tool for AH was assessed. The diagnosis of AH is usually a clinical one, based on severe liver dysfunction in the context of excessive alcohol consumption, excluding other causes of acute and chronic liver disease (CLD)[1]. However, this method of diagnosis is not steadfast, as some studies that have included a liver biopsy in all patients with clinically suspected AH have shown histologic confirmation in only 70%-80% of patients[1]. Thus, liver biopsy remains the gold standard in diagnosing AH patients; however it is invasive, expensive, and burdensome for the patient. The utilization of metabolic biomarkers as an alternative, objective, un-invasive diagnostic tool is promising.
Our results demonstrated that AH patients have a specific metabolome that can be employed for diagnostic purposes. AH patients had higher levels of betaine, and lower levels of creatinine, citrulline, homocitrulline, tyrosine, phenylalanine, octenoyl-carnitine, and SDMA. Most importantly, betaine and citrulline provided excellent prediction accuracy in distinguishing AH from AD. Figure 3 shows the sensitivity and specificity of citrulline and betaine for diagnosis and acute decompensation from non-alcohol-related etiologies. Alcohol consumption in patients with alcoholic liver disease results in bacterial overgrowth and increases gut permeability and translocation of bacteria-derived lipopolysaccharides from the gut to the liver[1]. This could explain the altered levels of amino acids in these patients.
Figure 3.
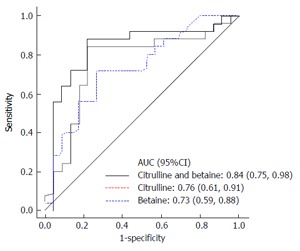
Citrulline and betaine serve as diagnostic biomarkers in patients with alcoholic hepatitis. AUC: Area under receiver operating characteristics curve.
Betaine is a molecule involved in transmethylation reactions in biological systems. S-adenosylmethionine (SAM), a critical methylating agent, is crucial to maintaining the integrity of the liver. One important function of SAM is its conversion of phosphatidylethanolamine to phosphatidylcholine, the latter of which constitutes lipoproteins involved in transporting fat away from the liver, thereby preventing hepatic fat infiltration and subsequent liver injury[11]. Betaine plays a significant role in this pathway as a methylating agent in the liver. Betaine transfers a methyl group to homocysteine via betaine-homocysteine methyl transferase (BHMT) in order to form methionine, which then goes on to form SAM and methylate biological molecules to protect the liver. Thus, betaine is protective against harmful fatty deposits in the liver due to alcohol abuse. While acute alcohol ingestion induces BMHT activity so that SAM levels can remain physiologically normal, chronic alcohol abuse leads to diminished SAM levels due to exhaustion of this system[11]. Consequently, this lead to increased betaine levels in the serum of AH patients, as the hepatocytes cannot compensate and regenerate SAM via the BHMT pathway. Furthermore, other studies have shown that dietary supplementation with betaine generated increased SAM in the liver and protected against ethanol-induced steatosis in rats[12]. However, with chronic alcohol abuse and dysfunction of the BHMT pathway, betaine cannot be metabolized.
Citrulline, in particular, is a biomarker of intestinal functionality[13]. Consequently, changes in intestinal flora due to liver disease can lead to imbalances in citrulline. Furthermore, it has been demonstrated that portal hypertension secondary to cirrhosis stimulates nitric oxide synthase (NOS)[14]. NOS converts arginine and oxygen into citrulline and nitric oxide (NO), resulting in vasodilation and increased blood flow. Pârvu et al[13] found that CLD patients had an increased serum citrulline concentration, indicating increased NO production to counter the mechanisms of portal hypertension[1]. Therefore, it makes sense that acute exacerbations of liver function seen in AH patients would deplete citrulline stores in an effort to produce NO and increase blood flow to an acute on chronic liver injury.
The second goal of this study was to assess the utility of metabolomics as a marker for the prognosis of liver disease. One particularly noteworthy result is the association between tyrosine and transplant-free survival outcome in patients with liver cirrhosis. The MELD score and tyrosine level, considered together, provided the greatest sensitivity and specificity for predicting 3-mo transplant-free survival. Tyrosine and phenylalanine are aromatic amino acids whose metabolism can become impaired as a consequence of liver injury, as the enzymes that metabolize these compounds are produced by the liver. Concentrations of aromatic amino acids are increased in patients with chronic liver disease who experience an acute inflammatory event such as acute alcoholic hepatitis, gastrointestinal bleeding, infection, or encephalopathy[15]. Liver cirrhosis with a superimposed liver injury will lead to systemic inflammation resulting in elevated tyrosine levels. Systemic inflammation in the context of acute on chronic liver failure exacerbates the patient’s health through the release of various pro-inflammatory cytokines[15]. Therefore, plasma tyrosine levels can be used to estimate the degree and severity of this inflammation, and provide novel information on prognosis and outcome.
Other analytes such as phenylalanine, carnitine, ADMA, and MMA were all found to be accurate predictors of transplant-free survival in patients with liver cirrhosis. It should be noted that there was no evidence to suggest that tyrosine, phenylalanine, carnitine, ADMA, and MMA were significantly better than MELD. However, despite the lack of statistical significance, in terms of the AUC even a small jump of 0.01-0.02 is promising as it denotes clinical significance. This study was an exploratory analysis designed to assess usefulness of metabolites and given the small sample size of 45 patients, no strong conclusions can be reached. However, the promising results of this study indicate the need for future studies with larger sample sizes to evaluate and corroborate the usefulness of these analytes in predicting transplant-free survival. Future studies can also explore more complex combinations of metabolites in predicting OLT-free survival. More data can help determine if plasma analytes are superior or inferior to the MELD score, or if they should be used in combination with the MELD score as a prognostic tool.
Limitations of this study include the small number of patients in the sample size. Furthermore, no power calculations were done to determine optimum sample size. Given no power calculations and a small sample size, we are only capable of generating sufficient power for large differences and the false negative rate may be high. Further research must validate the findings from this study utilizing larger patient populations. Another limitation was the lack of control group in this study. A control group is an essential part of any experiment that seeks to find a significant difference among populations. While this project had no control group, other research has corroborated the results from this study with control groups[7]. Lastly, this study was limited in that liver biopsy was not performed in all patients to confirm the diagnosis of AH; it was only performed in a subset of patients. One final limitation of this study is the lack of biopsy confirmation of AH as a diagnosis. Since liver biopsy is considered the gold standard in diagnosing AH, it cannot be said with absolute certainty that all patients were diagnosed with AH. Further research in this area might involve standardized biopsy evaluation alongside metabolomic correlations to liver disease.
This is the first study that profiles plasma metabolites in patients with AH and CLD. Investigation of the human metabolome in disease states can be very useful in generating diagnostic markers and understanding the pathophysiology of those disease states. However, this study is limited in its relatively small sample size. Future larger studies are needed to confirm the diagnostic value of biomarkers in AH and CLD.
Metabolomics plasma analyte levels could help diagnose AH and determine the prognosis of patients with liver cirrhosis awaiting liver transplantation. Specifically, combined citrulline and betaine plasma levels yield a highly sensitive and specific discriminatory test of AH vs AD. Tyrosine, in combination with MELD score, provides even greater sensitivity and specificity for predicting 3 mo OLT-free survival than the MELD score on its own.
In conclusion, metabolomics plasma analyte levels could aid in diagnosing AH or in determining potential patient prognosis.
COMMENTS
Background
Liver biopsy remains the gold standard for the diagnosis of alcoholic hepatitis (AH). Herein, the authors use a novel metabolomics approach to identify plasma analytes that may correlate with the diagnosis of AH and the severity of liver disease in patients with AH.
Research frontiers
Metabolomics represents the analysis of metabolites present in biological samples. By identifying and quantifying metabolites, one can gather a picture of the genetic variations and environmental influences (such as diet, lifestyle, drug use, and toxicological exposure) in a biological specimen. The authors use metabolomics to assess prognostic and diagnostic factors in patients with liver disease with the hopes of developing more accurate measures of patient outcomes.
Innovations and breakthroughs
In this study, several metabolites were found to be associated with survival in patients with liver disease.
Applications
These findings could potentially be used to develop more robust measures to provide a diagnosis and prognosis in patients with liver disease. The model for end-stage liver disease score and liver biopsy, which are currently used, are imperfect; a less invasive and more accurate measure is needed.
Peer-review
This is a very important paper and presents impact on health system. It is very well elaborated.
Footnotes
Supported by In part by NIH grant R01 HL122283 (Brown JM).
Institutional review board statement: The study was reviewed and approved by the Cleveland Clinic Foundation Institutional Review Board.
Institutional animal care and use committee statement: No animals were involved in this study.
Conflict-of-interest statement: The authors have no conflict of interest to report.
Data sharing statement: Technical appendix, statistical code, and dataset available from the corresponding author at mxa256@case.edu. Participants gave informed consent for data sharing.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 16, 2015
First decision: October 28, 2015
Article in press: March 14, 2016
P- Reviewer: da Silva NMO, Fan L S- Editor: Gong XM L- Editor: A E- Editor: Liu SQ
References
- 1.Hanouneh IA, Zein NN, Cikach F, Dababneh L, Grove D, Alkhouri N, Lopez R, Dweik RA. The breathprints in patients with liver disease identify novel breath biomarkers in alcoholic hepatitis. Clin Gastroenterol Hepatol. 2014;12:516–523. doi: 10.1016/j.cgh.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palaniyappan N, Subramanian V, Ramappa V, Ryder SD, Kaye P, Aithal GP. The utility of scoring systems in predicting early and late mortality in alcoholic hepatitis: whose score is it anyway? Int J Hepatol. 2012;2012:624675. doi: 10.1155/2012/624675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumas ME, Davidovic L. Metabolic Profiling and Phenotyping of Central Nervous System Diseases: Metabolites Bring Insights into Brain Dysfunctions. J Neuroimmune Pharmacol. 2015;10:402–424. doi: 10.1007/s11481-014-9578-5. [DOI] [PubMed] [Google Scholar]
- 4.Lewis GD. The emerging role of metabolomics in the development of biomarkers for pulmonary hypertension and other cardiovascular diseases (2013 Grover Conference series) Pulm Circ. 2014;4:417–423. doi: 10.1086/677369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rachakonda V, Gabbert C, Raina A, Bell LN, Cooper S, Malik S, Behari J. Serum metabolomic profiling in acute alcoholic hepatitis identifies multiple dysregulated pathways. PLoS One. 2014;9:e113860. doi: 10.1371/journal.pone.0113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angeli P, Gines P, Wong F, Bernardi M, Boyer TD, Gerbes A, Moreau R, Jalan R, Sarin SK, Piano S, et al. Diagnosis and management of acute kidney injury in patients with cirrhosis: revised consensus recommendations of the International Club of Ascites. Gut. 2015;64:531–537. doi: 10.1136/gutjnl-2014-308874. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Levison BS, Hazen JE, Donahue L, Li XM, Hazen SL. Measurement of trimethylamine-N-oxide by stable isotope dilution liquid chromatography tandem mass spectrometry. Anal Biochem. 2014;455:35–40. doi: 10.1016/j.ab.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung H, Chiang C. Estimation methods for time-dependent AUC with survival data. Can J Stat. 2010;38:8–26. [Google Scholar]
- 9.Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32:5381–5397. doi: 10.1002/sim.5958. [DOI] [PubMed] [Google Scholar]
- 10.Barak AJ, Beckenhauer HC, Tuma DJ. Betaine, ethanol, and the liver: a review. Alcohol. 1996;13:395–398. doi: 10.1016/0741-8329(96)00030-4. [DOI] [PubMed] [Google Scholar]
- 11.Barak AJ, Beckenhauer HC, Badakhsh S, Tuma DJ. The effect of betaine in reversing alcoholic steatosis. Alcohol Clin Exp Res. 1997;21:1100–1102. [PubMed] [Google Scholar]
- 12.Crenn P, Coudray-Lucas C, Thuillier F, Cynober L, Messing B. Postabsorptive plasma citrulline concentration is a marker of absorptive enterocyte mass and intestinal failure in humans. Gastroenterology. 2000;119:1496–1505. doi: 10.1053/gast.2000.20227. [DOI] [PubMed] [Google Scholar]
- 13.Pârvu AE, Negrean V, Pleşca-Manea L, Cosma A, Drăghici A, Uifălean A, Moldovan R. Nitric oxide in patients with chronic liver diseases. Rom J Gastroenterol. 2005;14:225–230. [PubMed] [Google Scholar]
- 14.Amathieu R, Triba MN, Nahon P, Bouchemal N, Kamoun W, Haouache H, Trinchet JC, Savarin P, Le Moyec L, Dhonneur G. Serum 1H-NMR metabolomic fingerprints of acute-on-chronic liver failure in intensive care unit patients with alcoholic cirrhosis. PLoS One. 2014;9:e89230. doi: 10.1371/journal.pone.0089230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–46. [PubMed] [Google Scholar]


