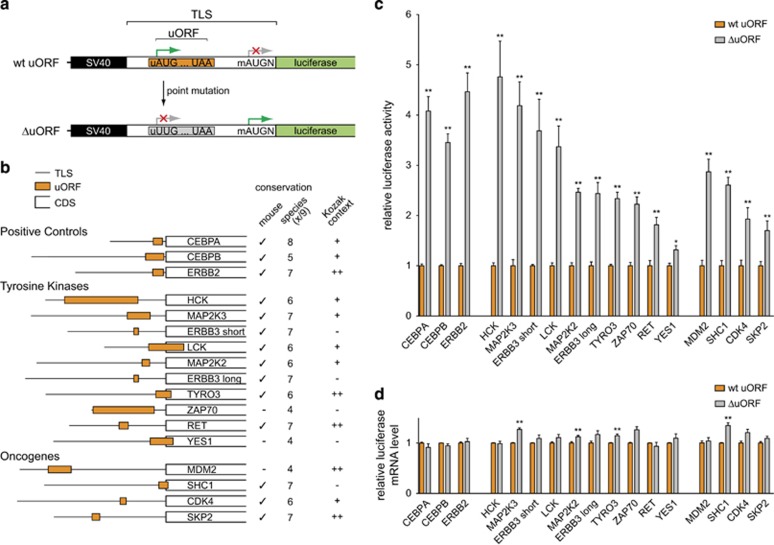
Figure 2.
uORFs repress downstream translation of TKs and other proto-oncogenes. (a) Graphic representation of the luciferase reporter construct. The plasmid allows the insertion of a complete TLS including the endogenous mAUG initiation codon plus the surrounding Kozak sequence including base +4 (N) to retain the gene-specific initiation context. For functional experiments, the wild-type (wt) uORF initiation codon was deleted by the insertion of a point mutation that creates a UUG codon instead (ΔuORF). To generate the reporter plasmid, an SV40 promoter originating from the pSG5 vector (Agilent Technologies) was cloned into the pGL3-Basic vector (Promega). Then, the SV40 promoter plus the remainder of the multiple cloning (MCS) site of the pGL3-Basic vector were PCR-amplified and cloned back into the KpnI and a newly generated SacII restriction site, obtained by mutating the Firefly luciferase initiation codon of pSG5 to GCG. This cloning strategy resulted in a reporter plasmid with the required structure: 5′-SV40 promoter–MCS–luciferase gene-3′. TLSs containing either the wt uORF start site or a respective uUUG mutation (ΔuORF) were synthesized by GeneArt Gene Synthesis (Life Technologies) and introduced in-frame with the luciferase coding sequence. (For details on restriction sites and inserted sequences, see Supplementary Figure 1 and Supplementary Table 3 and 4). (b) Schematic representation of the position and length of uORFs within the TLSs of three positive control transcripts, 10 selected TKs and 4 other proto-oncogenes. Conservation of the uAUG among human and mouse (check mark) and among a total of nine vertebrate species (human, rhesus, mouse, rat, cow, dog, elephant, chicken, zebrafish) is depicted. The Kozak context is indicated as strong (++, a purine base at −3 and a guanine base at +4), intermediate (+, one of the core Kozak bases matches) or weak (−, no core Kozak base matches). Note that SKP2, MDM2 and CDK4 contain additional potential uORFs that were excluded from display and functional analysis as they were not consistently translated in ribosome profiling studies. (c) Bar graph showing the relative reporter activity in the presence of wt uORF and ΔuORF containing TLSs of indicated proteins. For Luciferase assays, HeLa cells were seeded in duplicates and cultured under standard conditions. After 6 h, cells were transfected with 1 μg/12-well of the TLS luciferase reporter construct and 30 ng of Renilla luciferase reporter construct (pRL-CMV, Promega) using Metafectene transfection reagent (Biontex). After 42 h, Firefly and Renilla luciferase activities were determined following a published protocol.32 For each construct, Firefly luciferase signals were normalized to Renilla luciferase internal control signals. (d) Bar graph indicating the relative luciferase mRNA levels of wt uORF and ΔuORF reporter constructs for indicated TLSs. Quantitative real-time PCRs was performed after RNA extraction and cDNA synthesis following standard protocols and using custom primers that targeted Firefly and Renilla luciferase as an internal control (Supplementary Table 4). Error bars represent the s.e.m. of three independent experiments. Data were analyzed by using GraphPad Prism (Version 5.0a) and statistically evaluated by the two-tailed, nonparametric Mann–Whitney test. Asterisks indicate statistical significance (**P<0.01 and *P<0.05).