Abstract
Hepatic steatosis is characterised by excessive triglyceride accumulation in the form of lipid droplets (LD); however, mechanisms differ in drug induced (DIS) and/or non-alcoholic fatty liver disease (NAFLD). Here we hypothesized distinct molecular circuits of microRNA/LD-associated target genes and searched for mechanistically linked serum and tissue biomarkers that would distinguish between DIS and human NAFLD of different grades. We analysed >800 rat hepatic whole genome data for 17 steatotic drugs and identified 157 distinct miRNAs targeting 77 DIS regulated genes. Subsequently, genomic data of N = 105 cases of human NAFLD and N = 32 healthy controls were compared to serum miRNA profiles of N = 167 NAFLD patients. This revealed N = 195 tissue-specific miRNAs being mechanistically linked to LD-coding genes and 24 and 9 miRNAs were commonly regulated in serum and tissue of advanced and mild NAFLD, respectively. The NASH serum regulated miRNAs informed on hepatic inflammation, adipocytokine and insulin signalling, ER-and caveolae associated activities and altered glycerolipid metabolism. Conversely, serum miRNAs associated with blunt steatosis specifically highlighted activity of FOXO1&HNF4α on CPT2, the lipid droplet and ER-lipid-raft associated PLIN3 and Erlin1. Altogether, serum miRNAs informed on the molecular pathophysiology of NAFLD and permitted differentiation between DIS and NAFLD of different grades.
The term hepatic steatosis refers to an intracellular accumulation of lipid droplets and is associated with an enlargement of the liver. Apart from drug induced steatosis over-nutrition is the most common cause of non-alcoholic fatty liver disease (NAFLD); it comprises diverse alterations in liver architecture and organ function and ranges from simple steatosis to non-alcoholic steatohepatitis (NASH), i.e. a condition accompanied by inflammation. If unresolved disease progression involves fibrosis, cirrhosis with risk for liver cancer1; however, mechanisms of disease progression are poorly understood. There is unmet need for non-invasive diagnostic markers that will permit disease monitoring and differentiation among different causes and grades of NAFLD.
Notably, based on current diagnostics, i.e. serum biochemistry, ultrasound or liver biopsies and by considering various patient populations and their associated co-morbidities a median prevalence for NAFLD of about 20% for the US and Europe has been reported. The incidence of NAFLD has substantially increased in recent years (http://www.bornnaturopathic.com/blog/health-articles/nonalcoholic-fatty-liver-disease-nafld-need-responsive/) and its progression to NASH is considered to be a “silent” killer2 with prognostic studies suggesting increased mortality in patients with NAFLD as compared to the general population3. Moreover, a strong link between NAFLD and cardiovascular disease has been established4. While the definitive diagnosis of NAFLD is based on histologic examination of liver biopsy inherent risks associated with the procedure limits its widespread use as a screening test (http://www.medscape.com/viewarticle/497001_5). Therefore, ultrasound and other imaging modalities have been encouraged for the diagnosis of NAFLD but apart from cost the procedures are cumbersome and cannot be used reliably for the grading of disease stages and differentiation between various causes of NAFLD including DIS.
Remarkably, the frequency of DIS is unknown and estimates of drug induced hepatic injury range considerably amongst different studies and countries with a crude incidence rate of 19.1/100,000 as was reported for the general population of Iceland5. Given that NAFLD has become an epidemic it would be highly desirable to have sensitive and specific serum markers for the rapid and cost effective screening of individuals at risk for NAFLD and/or DIS and to determine disease progression by minimal invasive or non-invasive means. Accordingly, therapeutic intervention strategies can be developed to prevent disease progression by actively screening patients with fatty liver disease. Although several risk factors for NAFLD have been identified and typically include type-2 diabetes & impaired glucose tolerance, clinical obesity, dyslipidaemia and eventually the metabolic syndrome, prolonged exposure of drugs can also be associated with DIS and NASH as reported for amidarone and other drugs6.
Importantly, drug induced hepatic steatosis is generally associated with more than one mechanism7 and either involves micro- or macrovesicular steatosis (e.g. valproic acid) and can be associated with ballooning degeneration of hepatocytes. Particularly at prolonged exposure to drugs cellular stress, as a result of impaired detoxification, and mitochondrial function becomes compromised thereby aggravating the condition. The main mechanism by which drugs cause steatosis were recently summarized and involve (a) inhibition of mitochondrial fatty acid beta oxidation (b) sequestration of CoA and/or L-Carnitine (c) inhibition of the mitochondrial respiration and modulation of the mitochondrial membrane potential (d) impairment of mitochondrial DNA replication (e) impaired peroxisome proliferator activated receptor (PPARα) transcriptional activity and other alterations in lipid homeostasis pathways8.
Due to the growing knowledge on the mechanisms of lipid droplet formation in hepatocytes and the possibilities to study mechanistically linked biomarkers we were particularly interested in studying molecular circuits and transcriptional responses of microRNA target genes in rat liver induced by steatotic drugs. The importance of non-coding RNAs (ncRNAs) in regulating lipid metabolism has just been unraveled with microRNAs (miRNAs) constituting a large family of small, approximately 21-nucleotide-long, non-coding RNAs that have emerged as key post-transcriptional regulators of gene expression9. By base pairing to mRNAs, miRNAs mediate translational repression or mRNA degradation10. In mammals, miRNAs are predicted to control the activity of approximately 30% of all protein-coding genes, and have been shown to participate in the regulation of almost every cellular process investigated so far10. Recent research highlighted the critical functions of miRNAs in regulating diverse biological processes including the immune system, cell proliferation, differentiation and development, cancer and cell cycle11,12,13 and several miRNAs function in a tissue-specific manner14,15 as determined for NAFLD16.
Altogether, the primary objective of our study was to define mechanistically linked serum miRNAs that would distinguish between DIS and NAFLD of different grades. We hypothesized distinct transcriptional regulations of co-expressed miRNA/target genes in rat liver induced by steatotic drugs and utilized our knowledge of about 200 genes being mechanistically involved in various pathophysiological processes during fatty liver disease as recently reported17. By interrogating the publically available Open TG-GATEs database the dose and time dependent effects of 17 DIS causing drugs could be investigated and by studying whole genome transcript expression data of human NAFLD co-expressed miRNA-target genes in DIS and NAFLD were ascertained. Importantly, the identification of mechanistically linked miRNA serum biomarkers for DIS and NAFLD of different grades provide opportunities for its early detection and the monitoring of disease progression and will aid drug selection in preclinical development programs.
Methods and Materials
Chemicals
Table 1 informs on drugs and chemicals used to induce hepatic steatosis.
Table 1. Summary information for N = 17 drugs/chemicals which induced hepatic steatosis in rat liver.
| Drug/chemical | CAS number | Pathology Image number* | Day | Dose (mg/kg) | Observation |
|---|---|---|---|---|---|
| carbon tetrachloride (CCL4) | 56-23-5 | 2663 | 6 hrs, Day 8, 29 | HD_300 mg | clear vacuolar formation |
| hydroxyzine (HYZ) | 68-88-2 | 19778 | Day 29 | HD_100 mg | clear vacuolar formation |
| imipramine (IMI) | 50-49-7 | 33959 | Day 29 | HD_100 mg | clear vacuolar formation |
| amitriptyline (AMT) | 50-48-6 | 34287 | Day 29 | HD_150 mg | clear vacuolar formation |
| ethinylestradiol (EE) | 57-63-6 | 33709 | Day 29 | HD_10 mg | clear vacuolar formation |
| methapyrilene hydrochloride (MP) | 91-80-5 | 46989 | Day 29 | HD_100 mg | clear vacuolar formation |
| coumarin (CMA) | 91-64-5 | 29057 | Day 29 | HD_150 mg | clear vacuolar formation |
| tetracycline (TC) | 60-54-8 | 11030 | 9 hr | HD_1000 mg | clear vacuolar formation |
| lomustine (LS) | 13010-47-4 | 32392 | Day 29 | HD_6 mg | clear vacuolar formation |
| vitamin A (VA) | 68-26-8 | 15733 | Day 29 | HD_100 mg | clear vacuolar formation |
| diltiazem (DIL) | 42399-41-7 | 22191 | Day 15 | HD_800 mg | clear vacuolar formation |
| disulfiram (DSF) | 97-77-8 | 47866 | Day 4 | HD_600 mg | clear vacuolar formation |
| colchicine (COL) | 64-86-8 | 41516 | 24 hr | HD_15 mg | clear vacuolar formation |
| ethionamide (ETH) | 536-33-4 | 46400 | Day 4 | HD_300 mg | clear vacuolar formation |
| ethanol (ETN) | 64-17-5 | 47867 | Day 29 | HD_4000 mg | clear vacuolar formation |
| adapin (ADP) | 1668-19-5 | 30832 | Day 29 | HD_100 mg | clear vacuolar formation |
| puromycin aminonucleoside (PAN) | 58-60-6 | 55728 | Day 8 | HD_40 mg | clear vacuolar formation |
*The image can be retrieved from http://toxico.nibio.go.jp/open-tggates/english/search.html
Ethical statement and animal experimentation
The animal studies conformed to the Guide for the Care and Use of Laboratory Animals (The National Academy Press, Washington, D.C., 1996) and were conducted by the Japanese National Institute of Health Science, the National Institute Biomedical Innovation and pharmaceutical companies after obtaining approval from the Ethics Review Committee for Animal Experimentation of the National Institute of Health Sciences, Japan18. Specifically, male Crj:CD Sprague Dawly (SD) rats at the age of 6 weeks were given either a single low, middle and high dose and were sacrificed at 3, 6, 9 and 24 hours or received repeated treatments for 3, 7, 14 and 28 days. At the end of the study the liver was removed and next to histopathology representative samples were stored at −80 °C for further analysis. Further information regarding administration routes and dosage details are available on the TG-GATE database (http://toxico.nibio.go.jp). In this study, only in vivo data were used, which included both single and repeated dosing experiments.
Histopathology
Formalin fixed paraffin-embedded tissue blocks were sectioned to approximately 3 μm thickness and stained with H and E. Hepatic steatosis was confirmed by board certified pathologists after evaluating randomly selected fields at 20X magnification.
Data processing
A total of 808 Affymetrix GeneChip Rat Genome 230 2.0 raw data (CEL files) of 17 steatotic drugs and matched controls were retrieved from TG-GATE (http://toxico.nibio.go.jp/english/index.html). The data were analyzed using the robust multi-array average (RMA) methodology in the Bioconductor, R package for background-adjusted, normalized, and log-transformed perfect matched values of individual probes from the Array. (http://www.bioconductor.org/packages/release/bioc/html/BufferedMatrixMethods.html).
The fold change for each compound was calculated by comparing treatment data and matched controls; the student t test with multiple testing corrections was employed to calculate p values for each gene. Differential expressed genes (DEGs) were considered for further analysis by using the criterion fold change more than 1.5 and Benjamini & Hochberg adjusted p value less than 0.05.
Genes involved in lipid droplet formation of hepatocytes
Based on our previous works we considered approximately 200 steatosis related genes17 which code for lipid transport & lipogenesis, lipid droplet associated proteins such as the perilipins, glucose & fatty acid metabolism and signalling events. Details are given in supplementary Table S1 and the gene symbols were mapped to Entrez Gene ID based on NCBI gene annotation (http://www.ncbi.nlm.nih.gov/gene/).
MiRNA-gene target predictions
MiRNA targets were predicted using the miRTarBase repository http://mirtar.mbc.nctu.edu.tw/human/index.php)19. The database defines experimentally proven miRNA-gene target relationships. DEGs for 17 steatotic drugs (see above) and data from N = 4 independent publically available genomic data sets of N = 105 cases of human liver NAFLD and N = 32 controls (see below “translational research”) were mapped to miRTarBase. A summary of the miRNA-gene relationships is given in supplementary Table S2 and the data were compared to N = 207 cases of experimentally verified serum miRNA profiles. Furthermore, consensus prediction of miRNA targets involved 12 different computational algorithms, i.e. DIANA-microTv4.0 [ http://diana.imis.athena-innovation.gr/DianaTools/index.php?r = microtv4/index], DIANA-microT-CDS [ http://diana.imis.athena-innovation.gr/DianaTools/index.php?r = microT_CDS/index], miRanda [ http://www.microrna.org/microrna/home.do], mirBridge [PMID:20385095], miRDB [ http://mirdb.org/miRDB], miRmap [ http://mirmap.ezlab.org], miRNAMap [ http://mirnamap.mbc.nctu.edu.tw], PicTar2 [ https://www.mdc-berlin.de/10440258/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/pictar], PITA [ http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html], RNA22 [ https://cm.jefferson.edu/rna22/Interactive/], RNAhybrid [ http://bibiserv.techfak.uni-bielefeld.de/rnahybrid] and TargetScan [ http://www.targetscan.org/].
Bona fide miRNA targets were defined when predicted by at least nine different algorithms.
KEGG pathway analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) was used to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) annotated pathways. The Benjamini-Hochberg (BH) adjusted p value of ≤0.05 was applied as a cut-off to determine significantly enriched pathways.
Protein–protein interaction
The STRING version 9.1 database was used to determine protein-protein interactions amongst steatosis regulated DEGs20. Only interactions with confidence scores >0.4 were considered.
Network analysis
Network construction of co-regulated miRNA-target genes induced by drugs was based on the MCODE method21. The parameters in MCODE were set as default with NodeScoreThreshold = 0.2, K-coreThreshold = 2 and MaxDepth = 100.
Chemical structure similarity
The structural similarity among steatosis inducing drugs/chemicals was determined by using functional class fingerprints (FCFPs) with a radius of FCFP_4 as chemical descriptors to calculate Tanimoto coefficients. The data were subsequently analyzed using the software Pipeline Pilot 8.0 (Accelrys, http://accelrys.com/).
Translational research
The genomics of human NAFLD involved whole genome data sets GSE48452, GSE63067, and GSE17470 that were retrieved from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Alike, NAFLD regulated serum miRNAs were studied by analyzing the data set GSE33857 and the results reported by Pirola et al.22 (Fig. 1 and Table 2). Furthermore, gene expression data from N = 72 NAFLD patients, i.e. N = 40 with mild NAFLD, fibrosis stage 0–1 and N = 32 with advanced NAFLD, fibrosis stage 3–4 were retrieved from the GEO database (GSE49541). Importantly, the study of Moylan et al.23 (GEO data set GSE49541) did not involve healthy control cases23. Therefore, DEGs were calculated by considering whole genome hepatic gene expression data of N = 7 healthy controls regained from the López-Vicario et al. study24. In their study (GEO data set GSE37031) NASH patients and healthy controls were analyzed with the same microarray platform as utilized in the study of Moylan et al.23. In order to eliminate a batch effect, the CEL files from the two studies were processed together using the robust multi-array average (RMA) methodology and DEGs for mild NAFLD and advanced NAFLD were generated with the R limma package. Altogether, genomic data sets of N = 105 cases of human liver NAFLD and N = 32 controls were considered and DEGs were generated with the GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) software with the filtering criteria fold change >1.5 and Benjamini & Hochberg adjusted p value < 0.05.
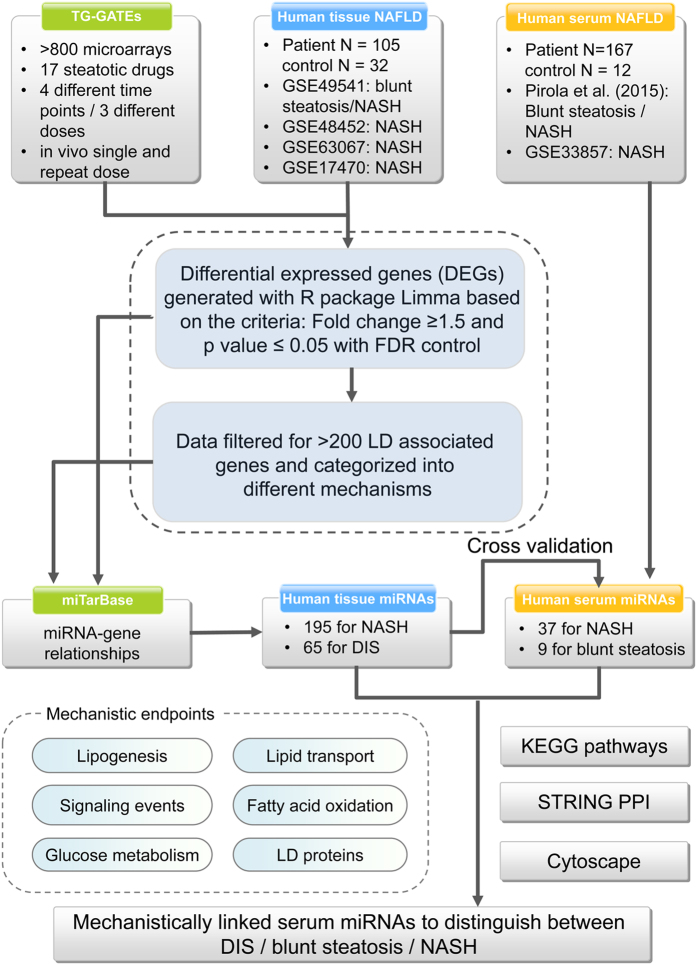
Figure 1. Flowchart of experimental strategies and associated data analysis.
Table 2. Publically available human miRNAs and mRNAs data sets associated with fatty liver disease.
| GEO ACESSION number | Species | Diseases | Number of patients | Number of healthy controls | References |
|---|---|---|---|---|---|
| miRNA data sets | |||||
| GSE33857 | Homo sapiens (serum) | non-alcoholic steatohepatitis (NASH) | 7 | 12 | 25 |
| – | Homo sapiens (serum) | non-alcoholic fatty liver disease (NAFLD) | 66 benign steatosis | – | 22 |
| 94 NASH | |||||
| Total cases | 167 | 12 | |||
| mRNA data sets | |||||
| GSE48452 | Homo sapiens (tissue) | non-alcoholic steatohepatitis (NASH) | 18 | 14 | 48 |
| GSE63067 | Homo sapiens (tissue) | non-alcoholic steatohepatitis (NASH) | 9 | 7 | – |
| GSE17470 | Homo sapiens (tissue) | non-alcoholic steatohepatitis (NASH) | 6 | 4 | 49 |
| GSE49541 | Homo sapiens (tissue) | non-alcoholic fatty liver disease (NAFLD) | 40 mild NAFLD | 7 control from GSE37031 | 23 |
| 32 advanced NAFLD | |||||
| Total cases | 105 | 32 | |||
Tissue regulated miRNA were predicted from whole genome hepatic gene expression changes by querying the miRTarBase repository (see above). Additionally, the whole genome DEG data were filtered for N = 200 genes mechanistically involved in lipid droplet formation of hepatocytes (supplementary Table S1 and [18]) and both approaches were subsequently mapped to miRTarBase to identify associated miRNAs. Moreover, prediction of gene targets was based on miRNA consensus among 9 out of 12 different algorithms (see above).
The tissue regulated miRNA were validated by considering experimentally verified serum miRNA profiles obtained from N = 12 healthy controls and N = 167 NAFLD patients samples. Note, for all patients liver biopsies were obtained to permit an assessment of grades of fatty liver disease and included N = 66 cases of benign steatosis and N = 94 NASH cases of different grades22. A further independent data set of serum microRNA expression in human NAFLD (GEO Accession: GSE33857) encompassing N = 12 controls with normal liver function and N = 7 NASH cases was considered25.
Results
The study design and work flow is depicted in Fig. 1 and statistically significant differentially expressed genes (DEGs) were identified in liver tissue of rats after single and repeated treatment with 17 steatotic drugs for up to 28 days. Subsequently, the data were filtered for genes known to be mechanistically linked to lipid droplet formation in hepatocytes (see supplementary Table S1) and miRNAs targeting DIS regulated genes were obtained by interrogating the experimentally verified miRTar database. Thus, the data analysis focused on (1) an identification of miRNAs targeting LD associated genes in hepatic steatosis; (2) time/dose effects in drug induced steatosis (3) an identification of commonly regulated tissue and serum miRNAs for its utility as serum biomarker and (4) serum miRNAs to distinguish DIS and human NAFLD of different grades.
Prediction of tissue regulated miRNA in drug induced steatosis
Computational analysis of 17 steatotic drugs involved >800 whole genome microarrays and the subsequent filtering of DEGs for 200 hepatic steatosis related genes (supplementary Table S2). This revealed 409 miRNA-gene associations and consisted of 157 distinct miRNAs and 77 genes. The mapping of significant DEGs to different processes in lipid droplet formation (Fig. 2) discovered >20 genes to be regulated in the process of lipogenesis while KEGG pathways analysis highlighted significant enrichment for glycerol lipid and glycerophospholipid metabolism (Table 3) as exemplified by the significant regulation of DGAT1, i.e. a key enzyme catalyzing the terminal step in triacylglycerol synthesis as well as AGPAT isoforms involved in the metabolism of lysophosphatidic acid to phosphatidic acid (PA). The latter are important signalling molecules but also function as building block intermediates in lipid biosynthesis. Other highly regulated genes include fatty acid synthase as well as lecithin cholesterol acyltransferase, the desaturase FADS1, FADS2, the transcription factors PPARγ, SREBF1 and the SREBP cleavage protein SCAP, the glucosylceramide synthase and various annexins to influence Ca2+ dependent phospholipid binding to modify cytoskeletal dynamics.
Figure 2. Co-regulation of miRNA and gene targets in drug induced steatosis.
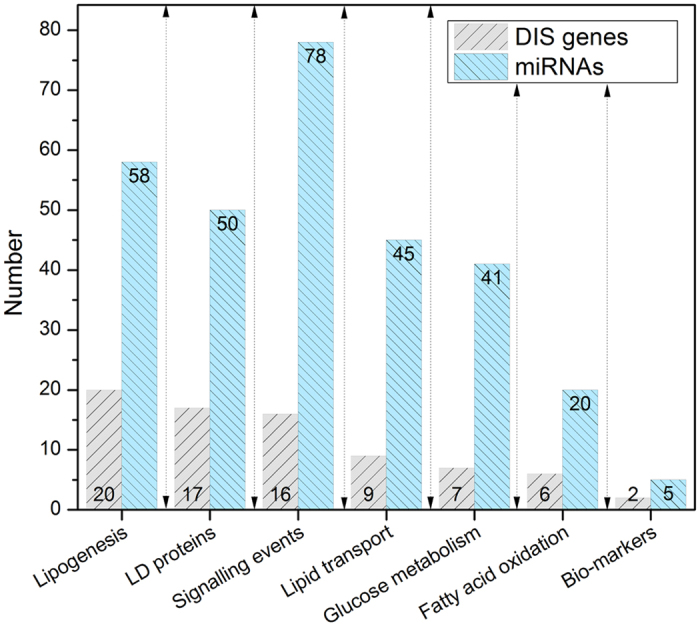
The histogram depicts the number of miRNAs involved in the regulation of N = 77 DIS gene targets. The data are grouped according to distinct processes in the pathophysiology of lipid droplet formation in hepatocytes. The DIS signature genes are given in supplementary Table S3 and are mechanistically linked to fatty liver disease.
Table 3. KEGG pathways analysis of DIS genes involved in lipogenesis.
| KEGG ID: Pathways | Count | Genes | adjusted p values |
|---|---|---|---|
| hsa01040:Biosynthesis of unsaturated fatty acids | 4 | ELOVL5, FADS1, ELOVL2, FADS2, | 7.7E-4 |
| hsa00561:Glycerolipid metabolism | 3 | DGAT1, AGPAT3, AGPAT2 | 9.1E-2 |
| hsa00564:Glycerophospholipid metabolism | 3 | LCAT, AGPAT3, AGPAT2 | 1.3E-1 |
The frequency by which individual miRNAs target steatosis related genes was considered; Fig. 3a depicts hsa-mir-335-5p and has-miR-124-3p as top ranking. These are involved in the regulation of genes coding for lipogenesis (n = 6/3), signalling events (n = 3/2), lipid transport (n = 1/3), lipid droplet associated proteins (n = 1/5), fatty acid oxidation (n = 4/1) and glucose metabolism (1/2). The network of LD-associated gene regulations among the top 10 ranking miRNAs is given in Fig. 3b and Table 4 summarizes independent studies to confirm the top ranking miRNAs as significantly regulated in animal and human NAFLD.
Figure 3. DIS associated gene regulations by miRNAs.
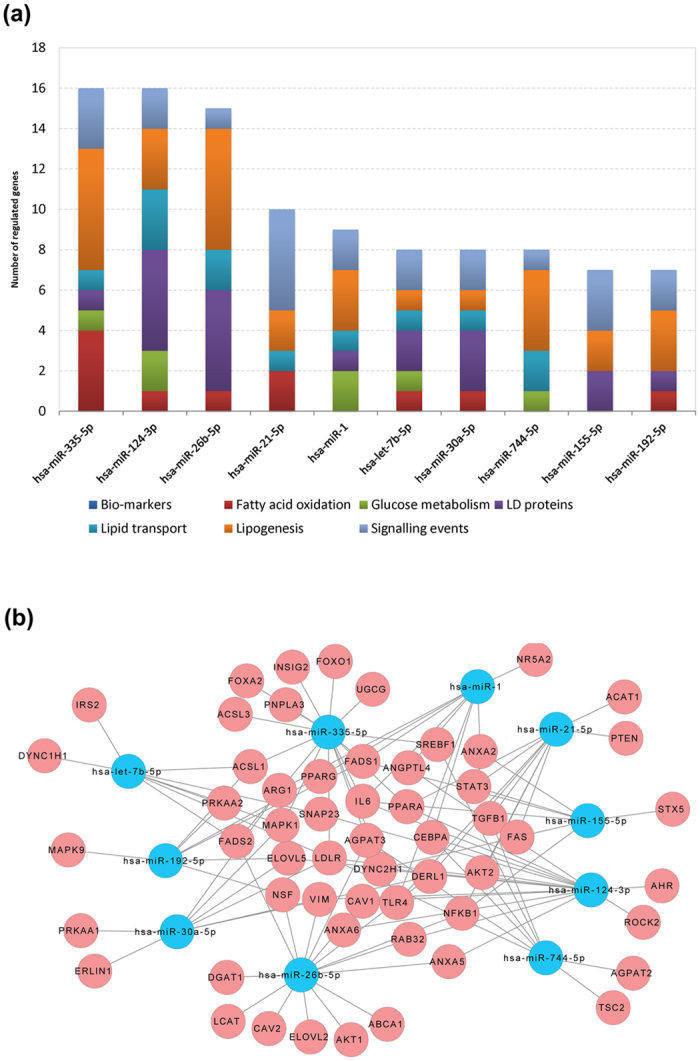
(a) The histogram depicts the top 10 ranked miRNAs and their distribution amongst different lipid droplet associated processes; (b) Cytoscape network analysis of mechanistically linked LD-associated gene regulations of the top 10 ranking miRNAs. The blue and pink colored nodes refer to miRNAs and LD-associated genes, respectively.
Table 4. The top 10 ranked drug-induced steatosis regulated miRNAs and their links to NAFLD.
| miRNAs | Species | Tissue | Serum | Associated disease | Experimental evidence | References |
|---|---|---|---|---|---|---|
| hsa-miR-335-5p | mouse | liver | lipid metabolism | The up-regulated expressions of miR-335 in liver and white adipose tissue of obese mice might contribute to the pathophysiology of obesity. | 27 | |
| rat | liver | hepatic fibrosis | miR-335 restoration inhibited HSC migration, at least in part, via down-regulation the TNC expression, which accounts for the increased numbers of activated HSCs in areas of inflammation during hepatic fibrosis. | 50 | ||
| hsa-miR-124-3p | human | liver | cholestatic liver disease | Down-regulation of miR-124 and up-regulation of miR-200 collaboratively promote bile duct proliferation through the IL-6-STAT3 pathway. | 51 | |
| – | MIN6 cells | lipid and glucose process | Over-expression of miR124a leads to exaggerated hormone release under basal conditions and a reduction in glucose-induced secretion. | 52 | ||
| hsa-miR-26b-5p | human | liver | hepatocellular carcinoma (HCC) | The miR-26 expression status is associated with survival and response to adjuvant therapy with interferon-α in liver cancer patients. | 53 | |
| rat | liver | NASH | miR-26b was up-regulated with fold change = 8 in differentiating steatohepatitis from steatosis | 54 | ||
| hsa-miR-155-5p | mouse/human | liver | NASH-induced hepatocarcinogenesis | In a microarray analysis conducted on mice fed a choline-deficient L-aminoacid-defined diet, miR-155 was found to be upregulated at early stages of NASH-induced hepatocarcinogenesis, as well as in primary human HCCs compared to matching liver tissues. | 55 | |
| human | liver | A gradual ascension of miR-155 expression in cirrhotic and HCC tissues, compared with low expression levels detected in normal liver tissues. | 56 | |||
| human | liver | Hepatitis C virus - hepatocellular carcinoma | The up-regulated expression of miR-155 induced by inflammation after HCV infection promotes hepatocyte proliferation and tumorigenesis by activating Wnt signalling. | 57 | ||
| mouse | liver | Hepatic steatosis | Increased expression of miR-155 in models of NAFLD likely plays a critical homeostatic role designed to prevent excessive lipid accumulation in livers that can ultimately lead to liver damage. | 58 | ||
| hsa-miR-30a-5p | zebra fish | liver | hepatic organogenesis | Demonstrated that miR-30a is required for biliary development in hepatic organogenesis in zebra fish. | 59 | |
| hsa-miR-1 | human | Serum | hepatocellular carcinoma | Serum miR-1 is a new independent parameter of OS in HCC patients and may therefore improve the predictive value of classical HCC staging scores. | 60 | |
| hsa-let-7b-5p | rat | liver | NASH | Let-7b was found to be upregulated in steatohepatitis compared to steatosis samples. | 54 | |
| hsa-miR-744-5p | human | liver | hepatocellular carcinoma | miR-744 was suggested as an independent predictor of survival in HCC patients after LT and may therefore be a potential biomarker for their prognosis. | 61 | |
| human | serum | chronic hepatitis B/NASH | Serum levels of miR-122, -572, -575, -638 and -744 are deregulated in patients with CHB or NASH. The levels of these miRNAs may serve as potential biomarkers for liver injury caused by CHB and NASH. | 62 | ||
| hsa-miR-192-5p | human | serum | drug-induced liver injury | microRNAs (miR-122 and miR-192) are promising biomarkers of acetaminophen-induced acute liver injury (APAP-ALI). | 63 | |
| house | liver | serum | drug-induced liver injury | We have demonstrated that specific microRNA species, such as mir-122 and mir-192, both are enriched in the liver tissue and exhibit dose- and exposure duration-dependent changes in the plasma that parallel serum aminotransferase levels and the histopathology of liver degeneration, but their changes can be detected significantly earlier. | 64 | |
| hsa-miR-21-5p | human | serum | hepatic steatosis | Serum level of miR-21 is higher in the NAFLD patients compared to the control participants. | 65 | |
| human | liver | hepatic fibrosis | MicroRNA-21 is important in hepatic fibrosis development, but the mechanism is unclear. | 66 |
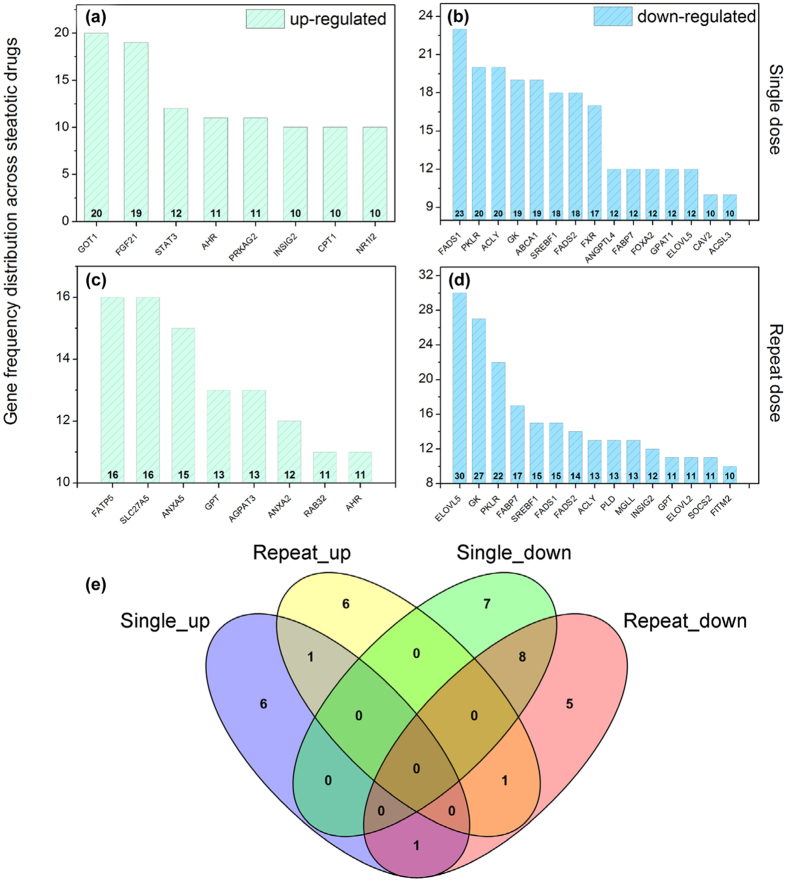
Moreover, the drug induced dose and time dependent regulations were considered and as shown in Fig. 4 the higher dose majorly affected repression rather than induction of gene transcription after single and repeated drug treatment. In an effort to define commonly regulated genes (frequency >10) the different drug, dose and time constellations were considered (Fig. 5a–d) and compared by constructing a Venn diagram to obtain information on the overlap of genes among the various considerations (Fig. 5e). This defined the aryl hydrocarbon receptor (AhR) as commonly up-regulated after single and repeat dosing with 17 different drugs while the genes FADS1, PKLR, ACLY, GK, SREBF1, FADS2, FABP7, and ELOVL5 were commonly repressed in expression. Note, the molecular functions of the aryl hydrocarbon receptor and the liver X receptor in the regulation of metabolic homeostasis is the subject of intense research; evidence was obtained for the aryl hydrocarbon receptor to induce hepatic steatosis via the up-regulation of the fatty acid transporter CD3618,26. Additional miRNAs involved in the regulation of the AhR include has-mir-26a-5p, hsa-mir-130b-3p, has-mir-124-3p, has-miR-625-5p and has-miR-98-5p with proven experimental evidence for their participation in the regulation of genes coding for lipid transport most notable CD36, fatty acid binding proteins FABP1, FAB6, FAB7, low density lipoprotein receptor, RXRß and others based on miRTarBase data analysis and PubMed searches.
Figure 4. Cumulative DIS associated gene regulations after single and repeated treatment of rats with N = 17 steatotic drugs.
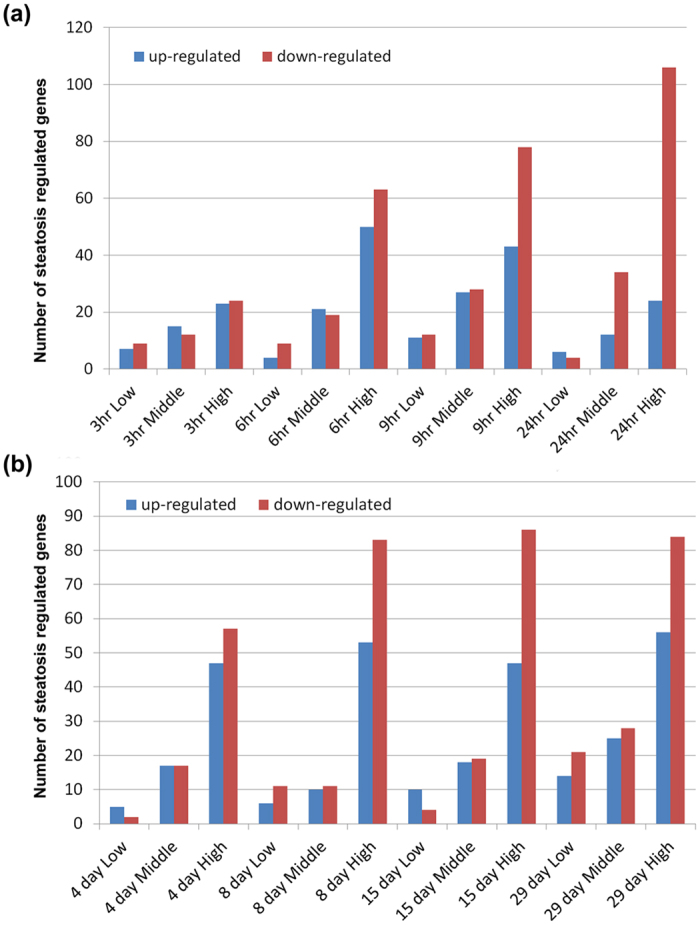
(a) Single dose treatment. (b) Repeated dose treatment for up to 28 days. Details regarding the animal protocol are given in the Material and Method section and in18.
Figure 5. Frequency distribution of top 10 ranked DIS signature genes.
Panels (a,b) depict the frequency distribution of up- or down regulated genes after single treatment of rats with N = 17 steatotic drugs; Panels (c,d) describe the frequency distribution of up- or down regulated genes after repeated treatment of rats with N = 17 steatotic drugs for 28 days; (e) Venn diagram of common regulated genes among the different treatment conditions (see panels a–d). Details regarding the animal protocol are given in the Material and Method section and in18.
As depicted in Fig. 5e eight genes were commonly down regulated after single and repeated treatment of rats with 17 steatotic drugs and included FADS1, PKLR, ACLY, GK, SREBF1, FADS2, FABP7 and ELOVL5.
We also considered the chemical structure similarity amongst 17 steatotic drugs by calculating the Tanimoto coefficient and found it to be insignificant. Nonetheless, when the frequency of DIS associated miRNAs were considered hsa-miR-335 was most frequently regulated across a wide range of dissimilar drugs.
Lastly, when miRNAs obtained from consensus predictions were compared with results obtained from miRTarBase queries a total of 35 miRNAs were in common (supplementary Fig. S1).
Molecular circuits of co-expressed miRNA-gene targets in DIS
Based on the built miRNA- steatosis and drug-steatosis gene interactions, we considered drug, dose and time associations (supplementary Table S3) and subsequently constructed protein-protein-interaction networks by probing the STRING PPI database version 9.1. As a result, 128 and 162 PPIs corresponding to 48 and 50 genes were obtained after single and repeated drug treatment of rats with 17 steatotic drugs (supplementary Table S4). Furthermore, by utilizing the MCODE algorithm the endoplasmic reticulum localized fatty acid desaturases FADS1, FADS2 and the fatty acid elongases ELOVL2 and ELOVL5 were identified as commonly regulated after single and repeat drug treatment. Consistent with previous findings expression of these genes is repressed and confer functional diversity through changes in chain length and degree of unsaturation in the process of lipogenesis.
Alike, individual networks for top regulated miRNAs were considered and as an example Fig. 6 depicts hsa-miR-124-3p and hsa-miR-335 regulated DIS genes after single and repeated treatment of rats with 17 drugs for up to 28 days. Transcriptional regulation involved 12, 10, 9 and 11 repressed genes after single and repeated treatments, respectively and the LDL receptor was commonly targeted amongst these miRNAs irrespective of the treatment conditions. Note, up-regulation of liver tissue specific hsa-miR-335 was reported for murine models of NAFLD, e.g. the leptin deficient ob/ob, the leptin-receptor-deficient db/db and KKAy44 mice. It is of considerable importance that hsa-miR-335 is a major regulator for lipid accumulation and adipose tissue differentiation in mice27 and the transcription factors FOXO1 and FOXA2, the insulin induced gene 2 and the low density lipoprotein receptor LDLR were equally regulated by miR-335. For its frequent regulation by different drugs and its important role in lipid homeostasis has-miR-335 should be considered as a DIS biomarker in drug safety evaluation. Similarly, among the miR-124 target genes was the Rho-associated, coiled-coil containing protein kinase 2. Recent evidence suggests repression of ROCK proteins to prevent hepatic steatosis by reducing lipid synthesis while pharmacological inhibition of ROCK prevented severe ischemia/reperfusion injury in steatotic fatty liver allografts28. The observed repression of ROCK transcripts can therefore be considered as an adaptive response to drug induced steatosis.
Figure 6. Transcriptional repression of lipid droplet associated genes by has-miR-3355p and has-miR-124-3p.
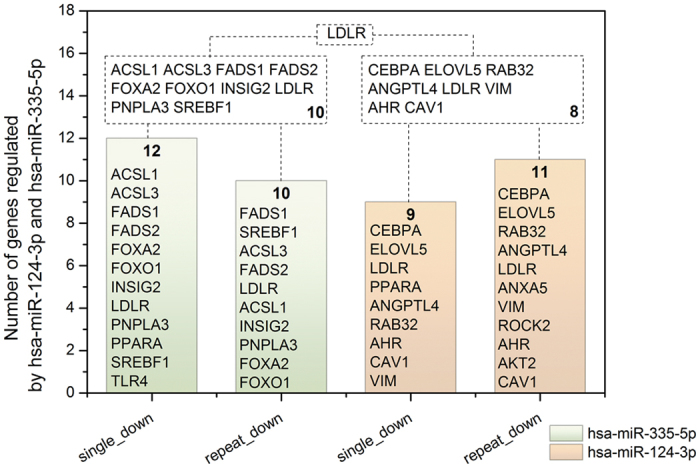
Green and orange colored bars refer to transcriptional repression of genes induced by has-miR-335-5p and hsa-miR-124-3p after single and repeat treatment of rats with N = 17 steatotic drugs.
Translational research
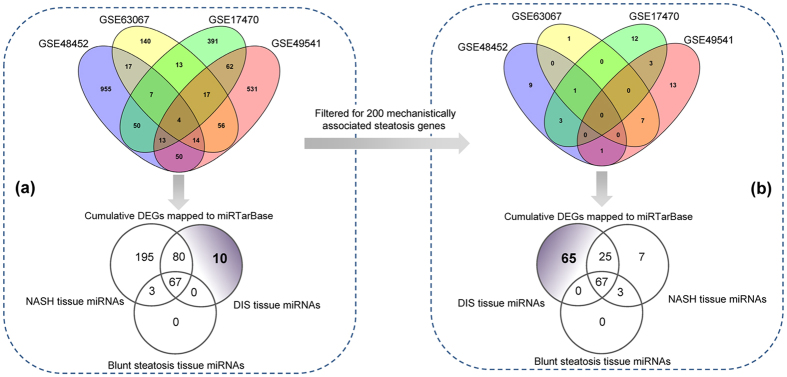
As depicted in Fig. 1 N = 4 independent studies were retrieved from the GEO database. Next to determining differentially expressed genes (DEGs) the data were mapped to miRTarBase and the results were compared with predicted DIS regulated miRNAs for 17 drugs after single and repeated treatment of rats for up to 28 days. Among 157 DIS regulated miRNAs 147 were regulated in common with human NAFLD (Fig. 7a). Alike, when human NAFLD data were filtered for 200 genes mechanistically linked to hepatic steatosis (Fig. 7b) 92 DIS regulated miRNAs were commonly regulated; vice versa 10 (see Fig. 7a) or 65 (see Fig. 7b) miRNA were predicted to be DIS tissue specific. Altogether, a total of 195 or 7 NASH specific miRNAs were identified by either considering whole genome human liver tissue DEGs or by filtering DEGs for 200 mechanistically linked genes. This analysis also revealed the lack of blunt steatosis specific tissue regulated miRNAs.
Figure 7. Venn diagram of differentially expressed genes in DIS and NAFLD.
(a) The publically available human NAFLD gene expression data (GEO accession number: GSE48452, GSE63067, GSE17470 and GSE49541) were analysed as described in the Material and Method section. DEGs were mapped to the miRTar database and compared to DIS associated miRNAs after single and repeated treatment of rats with N = 17 steatotic drugs for up to 28 days. (b) DEGs obtained from the publically available data sets GSE48452, GSE63067, GSE17470 and GSE49541 were filtered for N = 200 mechanistically linked LD-associated gene regulations (see supplementary Table S1) and mapped to the miRTar database. The data were compared with DIS associated miRNA regulations.
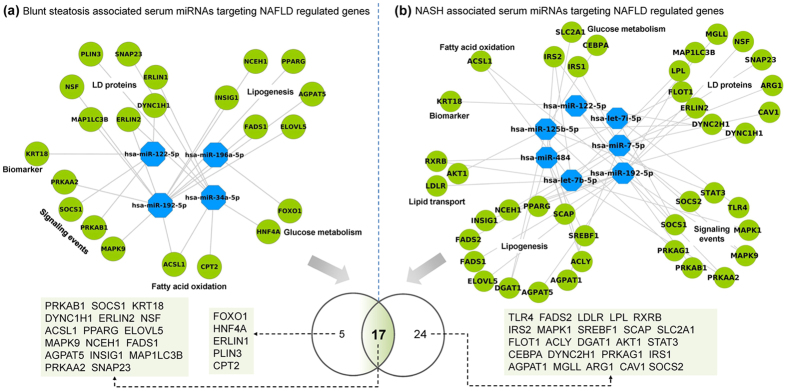
Serum biomarker candidates
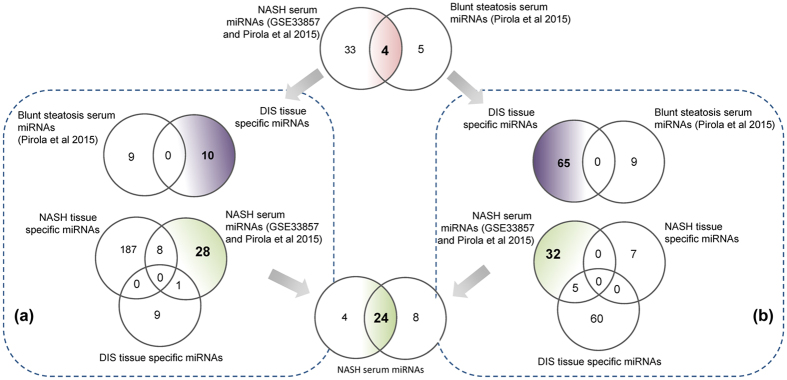
Based on whole genome gene expression profiling and by considering commonalities amongst independent publically available studies NASH and DIS specific miRNAs were analyzed. Their regulation was compared to experimentally verified serum miRNA profiling studies and as shown in Fig. 8 a total of 37 and 9 miRNAs were significantly changed in serum miRNA profiling studies of NASH and cases of blunt steatosis, respectively (Fig. 8 upper panel). Subsequently, DIS tissue specific miRNAs were compared to blunt steatosis regulated serum miRNAs. This revealed none of the miRNA to be in common thus evidencing specificity of the performed analysis (Fig. 8a,b). Additionally, miRNAs commonly regulated in liver tissue amongst 4 independent NASH studies were compared with DIS tissue and serum regulated miRNAs. Such an analysis discovered 28 and 32 NASH serum specific miRNAs by either considering total DEGs (approach A) or DEGs filtered for 200 mechanistically linked hepatic steatosis genes (approach B). The Venn diagram shown in the lower panel of Fig. 8 informs on 24 common miRNAs to demonstrate that the hypothesis driven approach (see panel b) yielded 65% of experimentally verified serum regulated miRNAs in NAFLD.
Figure 8. Translational biomarkers.
(a) Upper panel: Venn diagram of blunt steatosis serum regulated miRNAs and DIS tissue regulated miRNAs. Note, the DIS tissue specific miRNAs are based on the data given in Fig. 7a. Lower panel: Comparison of NASH tissue and NASH serum regulated miRNAs with DIS tissue specific miRNAs. (b) Upper panel: Venn diagram of DIS tissue and blunt steatosis serum regulated miRNAs. Note, the DIS tissue specific miRNAs are based on the data given in Fig. 7b (i.e. the whole genome data was filtered for N = 200 mechanistically linked LD-associated gene regulations (see supplementary Table S1). Lower panel: Comparison of NASH and DIS tissue miRNAs with NASH serum regulated miRNAs.
Our study highlights 8 common miRNAs when independent NASH tissue and serum profiling studies are compared and includes the miRNAs: hsa-let-7c-5p, hsa-let-7d-5p, hsa-let-7g-5p, hsa-miR-326, hsa-miR-150-5p, has-miR-885-5p, has-miR-486-5p and has-miR-126-5p. The network of regulated genes by these miRNAs is depicted in Fig. 9 and the results given in Fig. 10 evidence differentiation between NAFLD of different grades. Specifically, miRNAs regulated in blunt steatosis were analysed for target genes by mapping them to miRTarBase; the resultant target genes were compared to DEGs determined in hepatic tissue of NAFLD patients and to the list of 200 genes mechanistically involved in fatty liver disease (supplementary Table S2). Accordingly, a network was constructed and visualised with Cytoscape and 4 miRNAs were found to influence transcriptional expression of 22 steatosis related genes. This included the lipid droplet associated protein PLIN3, the transcription factors FOXO1, HNF4a and PPARγ, the ER lipid raft associated ERLIN1 and 2, the lysophosphatidic acid acyltransferase, protein and mitogen activated kinases and SNAP23 involved in the lipid fusion process. Using the same approach NASH regulated serum miRNAs were analysed and a total of 7 miRNAs were identified to target 41 fatty liver disease regulated genes. Here, significant enrichment for adipocytokine (i.e. AKT1, IRS2, ACSL1, RXRB, PRKAG1, SLC2A1, PRKAB1, MAPK9, PRKAA2, IRS1, STAT3) and insulin signalling pathways (SREBF1, AKT1, MAPK1, IRS2, SOCS2, PRKAG1, SOCS1, FLOT1, PRKAB1, MAPK9, PRKAA2, IRS1), regulation of suppressor of cytokine (SOCS1 & SOCS2) and Toll-like receptor 4 signalling were important findings. Further notable findings were miRNAs involved in the regulation of the LDL receptor, various transcription factors (RXRB, STAT3), calveole and fusion associated proteins CAV1, SNAP23, Flotilin, the monoglyceride lipase 1 and the microtubule-activated ATPase dynein 1 which functions in vesicle transport. Additionally, a total of 17 genes were regulated in common when serum miRNAs regulated in blunt steatosis and/or NASH were compared (Fig. 10 lower panel). Taken collectively, circulating miRNAs provide opportunities for disease diagnostic that may be extended to drug therapeutic monitoring. Table 4 compiles independent experimental evidence for the top ten DIS regulated miRNAs in serum or tissue of animal and human NAFLD. For instance, the anti-fibrotic miR-335-5p as well as miR-30a-5p are known to repress lipid biosynthesis, lipoprotein secretion and autophagasome formation and were significantly regulated as was mir-21 and mir-335 which are mechanistically linked to hepatic triglyceride and cholesterol metabolism.
Figure 9. Commonly regulated tissue and serum miRNAs in NASH.
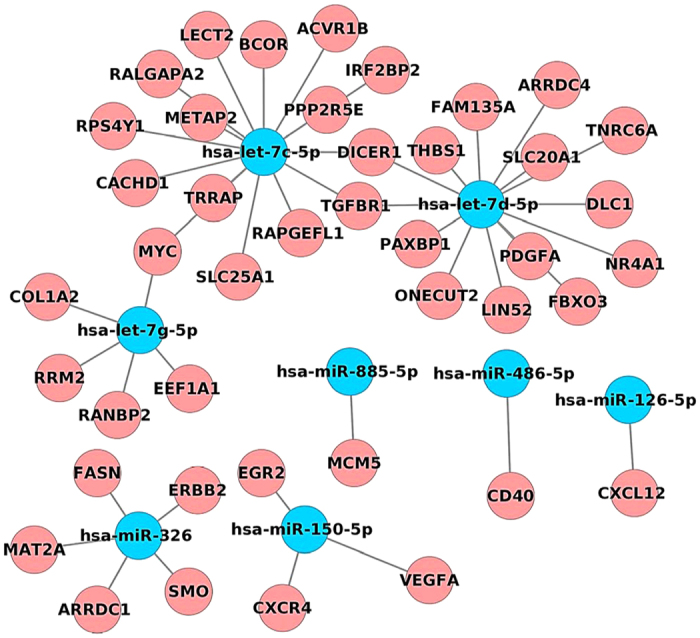
Eight miRNAs were identified as commonly regulated when independent NASH tissue and serum profiling studies are compared.
Figure 10. Serum regulated miRNAs distinguish between blunt steatosis and NASH.
Significantly regulated serum miRNAs inform on pathophysiological processes in fatty liver disease and permit differentiation between NAFLD of different grades.
Discussion
A recent white paper by the American Association for the Study of Liver Diseases, the American College of Gastroenterology and the American Gastroenterological Association provided guidance for the diagnosis and the management of NAFLD29. In this report the current limitations in the diagnosis of NAFLD are summarized with serum biochemistries frequently failing to be informative as they can be within normal range or are insufficiently sensitive to reflect grades of disease; alike liver ultrasound imaging requires skilled healthcare professionals and might be obscured by gross morphologic changes other than NAFLD. Furthermore, ultrasound imaging does not inform on some important diagnostic and prognostic factors in NAFLD. Therefore, the diagnosis and assessment of progressive fatty liver disease remains challenging; nonetheless the discoveries of molecular disease markers carry the hope to replace liver biopsies by novel blood and/or urinary tests.
The significant role of miRNAs in regulating lipid metabolism was the subject of several reviews30; however, only a few studies investigated the regulation of miRNAs in drug induced steatosis31. As of today it remains uncertain whether miRNAs known to regulate cholesterol and fatty acid metabolism and lipid droplet formation are the same in DIS and NAFLD.
In the present study whole genome transcript expression changes were examined to investigate tissue regulated miRNAs and to permit construction of miRNA-gene networks specific for fatty liver disease and DIS. Based on diverse computational approaches distinct circuits of co-expressed microRNA target genes in NAFLD and DIS were postulated and validated by comparing tissue with serum regulated miRNAs. This enabled an identification of mechanistically linked serum biomarker candidates for DIS and NAFLD of different grades. Table 5 compiles a summary of regulated miRNAs in human NAFLD (Table 5A) and in animal models of NAFLD (Table 5B).
Table 5. miRNAs in human (A) and animal models (B) of NAFLD of different grades.
| miRNAs | Regulation | Species | Liver tissue | Serum | Diseases | Description | References |
|---|---|---|---|---|---|---|---|
| (A) A summary of regulated miRNAs in human NAFLD of different grades | |||||||
| Blunt steatosis | |||||||
| miR-15b | up-regulated | homo sapiens | serum | hepatic steatosis | The expression of miR-15b was also significantly elevated in the serum of fatty liver disease patients compared with healthy subjects | 67 | |
| miR-21 miR-34a miR-122 miR-451 | up-regulated | homo sapiens | serum | hepatic steatosis | Serum levels of circulating miRNAs, miR-21, miR-34a, miR-122 and miR-451 are associated with nonalcoholic fatty liver disease | 65 | |
| miR-101 | up-regulated | homo sapiens | human THP-1-derived macrophages and HepG2 hepatoblastoma cells | blunt steatosis | miR-101 promotes intracellular cholesterol retention under inflammatory conditions through suppressing ABCA1 expression and suggests that the miR-101-ABCA1 axis may play an intermediary role in the development of NAFLD and vascular atherosclerosis | 68 | |
| miR-103 | up-regulated | homo sapiens | serum | blunt steatosis | Compared with the normal control group, higher serum levels of miR-103 were expressed in the NAFLD group (8.18 ± 0.73 vs 4.23 ± 0.81, P = 0.000). | 69 | |
| miR-199a-5p | up-regulated | homo sapiens | HepG2 and AML12 cells | hepatic steatosis | Upregulated miR199a-5p in hepatocytes may contribute to impaired FA β-oxidation in mitochondria and aberrant lipid deposits, probably via CAV1 and the PPARα pathway | 70 | |
| miR-122 | down-regulated | homo sapiens | liver tissue | hepatic steatosis | miR-122 is significantly under-expressed in NAFLD subjects. | 71 | |
| NASH | |||||||
| pri-miR-7-1 | up-regulated | homo sapiens | liver tissue | NASH | Histologic NASH correlated positively with the expression levels of pri-miR-16-2 and pri-miR-7-1. | 72 | |
| pri-miR-16-2 | |||||||
| hsa-miR-125b | down-regulated | Both NASH and ballooning degeneration of hepatocytes correlated negatively with the expression levels of hsa-miR-125b. | |||||
| miR-21 | down-regulated | homo sapiens | serum | NAFLD | serum levels of miR-21 were lower in patients with NAFLD compared with the healthy controls | 73 | |
| hsa-miR-28-3p hsa-miR-132 hsa-miR-150 hsa-miR-433 hsa-miR-511 hsa-miR-517a hsa-miR-671 | homo sapiens | visceral adipose tissue | NASH | Seven hsa-miR-132, hsa-miR-150, hsa-miR-433, hsa-miR-28-3p, hsa-miR-511, hsa-miR-517a, hsa-miR-671 differentially expressed between NASH patients and non-NASH patients (P < 0.05 after multiple test correction) | 74 | ||
| miR-30c miR-331-3p | up-regulated | homo sapiens | serum | NAFLD | miR-331-3p and miR-30c were differentially expressed between NAFLD and non-NAFLD groups | 75 | |
| miR-194 | down-regulated | homo sapiens | human THP-1 cells | NAFLD | miR-194 negatively regulates the TLR4 signal pathway which is activated by PA through directly negative TRAF6 expression, which is related to inflammatory in NAFLD and obesity | 76 | |
| miR-296 | down-regulated | homo sapiens | liver tissue | NASH | miR-296-5p was reduced in liver samples from nonalcoholic steatohepatitis (NASH) patients compared with patients with simple steatosis (SS) or controls | 77 | |
| miRNAs linked to NAFLD and other hepatic disorders | |||||||
| miR-16 miR-21 miR-34a miR-122 | down-regulated | homo sapiens | serum | chronic hepatitis C and liver fibrosis | Serum levels of miR-34a and miR-122 may represent novel, noninvasive biomarkers of diagnosis and histological disease severity in patients with CHC or NAFLD | 78 | |
| hsa-miR-17 hsa-miR-186 hsa-miR-219a-2 hsa-miR-373 hsa-miR-376b hsa-miR-378c hsa-miR-378i hsa-miR-590 hsa-miR-1286 hsa-miR-3611 hsa-miR-5699 | down-regulated | homo sapiens | liver tissue | blunt steatosis/fibrosis or cirrhosis | A total of 75 miRNAs showing statistically significant evidence for differential expression between the NAFLD VS. Non-NAFLD, including 30 upregulated and 45 downregulated miRNAs | 79 | |
| hsa-miR-31 hsa-miR-92b hsa-miR-150 hsa-miR-182 hsa-miR-183 hsa-miR-200a hsa-miR-224 hsa-miR-708 hsa-miR-766 hsa-miR-3613 | up-regulated | ||||||
| hsa-miR-27b-3p hsa-miR-122-5p hsa-miR-192-5p hsa-miR-1290 | up-regulated | homo sapiens | serum | blunt steatosis/NASH | a serum microRNA panel (hsa-miR-122-5p, hsa-miR-1290, hsa-miR-27b-3p, and hsa-miR-192-5p) with considerable clinical value in NAFLD diagnosis | 80 | |
| miR-33a miR-224 | up-regulated | homo sapiens | liver tissue | steatotic chronic hepatitis C | he expression of miR-33a and miR-224 were elevated in CHC-Steatosis and Steatosis in comparison to control tissue (P < 0.01) | 81 | |
| miR-122 | up-regulated | homo sapiens | serum | hepatic steatosis and fibrosis | The hepatic and serum miR-122 levels were associated with hepatic steatosis and fibrosis. The serum miR-122 level can be a useful predictive marker of liver fibrosis in patients with NAFLD | 82 | |
| (B) A summary of regulated miRNAs in animal models of NAFLD of different grades | |||||||
| Blunt steatosis | |||||||
| miR-10b | down-regulated | L02 cells | hepatic steatosis | The established miRNA profile of the steatotic L02 cell model and the novel effect of miRNA-10b in regulating hepatocyte steatosis may provide a new explanation of the pathogenesis of NAFLD | 83 | ||
| miR-21 | down-regulated | C57BL/6J mice | liver tissue | hepatic steatosis | miR-21 expression was decreased in livers from high-fat diet-fed mice and Hepa 1-6 cells treated with SA. | 84 | |
| miR-27 miR-122 miR-451 | down-regulated | rat | liver tissue | hepatic steatosis | The miRNAs analysis showed the significant down regulation of three miRNAs (miR-122, miR-451 and miR-27) and the up regulation of miR-200a, miR-200b and miR-429 in HFD, SD-HF and HFD-HF rats. | 85 | |
| miR-200a miR-200b miR-429 | up-regulated | ||||||
| miR-34a miR-122 miR-181a miR-192 miR-200b | up-regulated | Mouse | serum | hepatic steatosis | The levels of circulating miR-34a, miR-122, miR-181a, miR-192, and miR-200b miRNAs were significantly correlated with a severity of NAFLD-specific liver pathomorphological features, with the strongest correlation occurring with miR-34a. | 86 | |
| miR-34a | up-regulated | mouse | ob/ob mouse liver | hepatic steatosis | Up regulation of miR-34a and down regulation of miR-122 was found in livers of STZ-induced diabetic mice. | 87 | |
| miR-122 | down-regulated | ||||||
| miR-122 | up-regulated | rat | serum | blunt steatosis | serum miR-122 level is indeed useful for assessing early NAFLD and might be superior to clinical markers traditionally used to monitor hepatic disease | 88 | |
| miR-122 miR-192 | up-regulated | mouse | serum | blunt steatosis | the liver appears to be an important source of circulating EVs in NAFLD animals as evidenced by the enrichment in blood with miR-122 and 192 | 89 | |
| miR-125b | up-regulated | mouse | primary mouse hepatocytes | blunt steatosis | Estrogen protects against hepatic steatosis in female mice via up-regulating miR-125b expression | 90 | |
| miR-146a miR-146b miR-152 miR-200a miR-200b miR-200c | up-regulated | rat | HepG2 cells and human hepatocytes | blunt steatosis | miR-200a, miR-200b, miR-200c, miR-146a, miR-146b and miR-152 were up-regulated both in vitro and vivo. | 91 | |
| miR-155 | up-regulated | mouse | liver tissue | lipid metabolism | miR-155 gain of function the altered lipid metabolism and provide new insights into the metabolic state of the liver in Rm155LG/Alb-Cre mice | 92 | |
| miR-155 miR-200b | up-regulated | rat | NAFLD rats and in the free fatty acid-treated HepG2 | blunt steatosis | The pharmacological inhibition of EZH2 by 3-Deazaneplanocin A (DZNep) significantly reduces EZH2 expression/activity, while it increases lipid accumulation, inflammatory molecules and miR-200b/155 | 93 | |
| miR-302a | down-regulated | mouse | liver tissue | hepatic steatosis | miR-302a may prove to be a valuable therapeutic target in the regulation of hepatic fatty acid utilization and insulin resistance. | 94 | |
| miR-467b | down-regulated | mouse | liver tissue | hepatic steatosis | Down regulation of miR-467b is involved in the development of hepatic steatosis by modulating the expression of its target, LPL. | 95 | |
| NASH | |||||||
| miR-21 | up-regulated | mouse | liver tissue | NASH | The studies show the novel role of leptin-NADPH oxidase induction of miR21 as a key regulator of TGF-β signalling and fibrogenesis in experimental and human NASH | 96 | |
| miR-34a | up-regulated | rat | liver tissue | NASH | A link between liver cell apoptosis and miR-34a/SIRT1/p53 signalling, specifically modulated by UDCA, and NAFLD severity. | 97 | |
| miR-122 | down-regulated | mouse | HCC tissue | NASH | Silencing of miR-122 is an early event during hepatocarcinogenesis from NASH, and that miR-122 could be a novel molecular marker for evaluating the risk of HCC in patients with NASH | 98 | |
| miR-122 | down-regulated | mouse | liver and hepatocyte | NASH | Decreased liver miR-122 contributes to up regulation of modulators of tissue remodelling (HIF-1α, vimentin and MAP3K3) and might play a role in NASH-induced liver fibrosis | 99 | |
| miR-122 | up-regulated | mouse | serum | NASH | Serum levels of miR-122 can potentially be used as a sensitive biomarker for the early detection of hepatotoxicity and can aid in monitoring the extent of NAFLD-associated liver injury in mouse efficacy models | 100 | |
| miR-199a-5p miR-219-5p | mouse | liver tissue | obesity-induced steato-hepatitis | RvD1 acts as a facilitator of the hepatic resolution process by reducing the inflammatory component of obesity-induced NASH, which was regulated by miR-219-5p and miR-199a-5p | 101 | ||
| miR-199a-5p | up-regulated | mouse | C57BL/6J mice | NASH | miR-199a-5p plays a key role in the progression of NASH through inhibition of NCOR1 translation, and provide novel insights into NASH pathogenesis | 102 | |
| miR-451 | down-regulated | mouse | Liver tissue | NASH | The negative regulation of miR-451 in fatty acid-induced inflammation via the AMPK/AKT pathway and demonstrate potential therapeutic applications for miR-451 in preventing the progression from simple steatosis to severely advanced liver disease | 103 | |
| miRNAs linked to NAFLD and other hepatic disorders | |||||||
| miR-24 | up-regulated | mouse | liver tissue | hepatic lipid accumulation and hyperlipidemia | miR-24 promotes hepatic lipid accumulation and hyperlipidemia by repressing Insig1, and suggest the use of miR-24 inhibitor as a potential therapeutic agent for NAFLD and/or atherosclerosis | 104 | |
| miR-34a | down-regulated | mouse | Liver tissue | Hepatic ischemia/reperfusion (I/R) injury | The miR-34a/SIRT1 pathway may represent a therapeutic target for hepatic injury. | 105 | |
| miR-200a miR-223 | up-regulated | mouse | male A/J, 129S1/SvImJ and WSB/EiJ mice | liver fibrosis | Downregulation of IRP1 was linked to an increased expression of microRNAs miR-200a and miR-223, which was negatively correlated with IRP1. The results of this study demonstrate that the interstrain variability in the extent of fibrogenesis was associated with a strain-dependent deregulation of hepatic iron homeostasis. | 106 | |
The present study highlights the importance of mechanistically linked lipid droplet associated gene regulations to inform on the pathophysiological state of fatty liver disease with serum miRNA profiling representing a rapid and cost effective means for an identification of diagnostic biomarkers.
Importantly, fatty liver disease is a clinico-pathologic feature of drug induced liver injury (DILI) and is typically seen with drugs such as amiodarone, perhexiline, carbamazepine amongst others. It results from an imbalance between xenobiotic defense and adaptation leading to ER/mitochondrial stress, unfolded protein response and liver injury. Recently, whole genome transcript profiling of drug induced steatosis was reported and comprehensive information on mechanistically linked gene expression changes was obtained for a wide range of drugs causing either steatosis and/or phospholipidosis while an identification of signature genes encourage their use in preclinical screening assays18.
Unfortunately, commonly used markers of liver injury lack specificity and do not distinguish between DIS and NAFLD of different grades but rely primarily on serum biochemistries, for instance by assaying serum alanine and aspartate aminotransferase activities as a means to examine the metabolic competence of the liver. In recent years molecular biomarkers of hepatotoxicity and their potential to improve understanding and management of DILI have been intensely researched as summarized in a recent research highlight32,33.
To overcome current limitations mechanistically based biomarkers have been advocated and await translational research to better predict prognosis and outcome than currently used clinical biochemistry parameters34. Serum miRNAs may therefore be employed to predict individuals at risk for developing fatty liver disease and in our previous research we identified serum acute phase reactants to define healthy individuals at risk for acetaminophen induced liver injury prior to drug treatment35, while the study of Yamaura and co-workers31 identified miRNAs indicative of the pattern of liver injury that is steatosis versus NASH, cholestatic disease and acute versus chronic hepatocellular injury using a range of drugs and dietary animal models. Indeed, when the data of the Yamaura’s study were compared with findings of the present study several miRNAs were regulated in common and included for blunt steatosis miR-10b and miR-183; similarly with NASH the miRNAs miR-17, miR148b-5p and miR-197 were commonly regulated thus providing independent evidence for their diagnostic utility in animal studies.
In the present study diverse genomic data sets were examined to define regulatory miRNA gene networks in DIS and NAFLD of different grades and unique miRNAs targeting LD-associated genes were identified to provide mechanistic information on the onset and progression (micro- versus macrovesicular steatosis) of fatty liver disease. The network analysis revealed 29 out of 56 DIS genes to be commonly regulated by the top 10 regulated miRNAs (Fig. 3a,b) and >70% of all DIS regulated genes (56 over 77 DIS genes) are targeted by these miRNAs. Apart from common DIS regulated genes the transcription factors FOXO1 and FOXA2 and the insulin induced gene 2 are specific targets of miR-335-5p; note polymorphisms of INSIG2 are associated with severe obesity. Furthermore, the patatin-like phospholipase domain-containing protein 3 is a target of miR-335-5p and a single nucleotide polymorphism of this gene is strongly associated with NAFLD of different grades36. Other genes regulates by this miRNA are the UDP-Glucose Ceramide Glucosyltransferase (UCGC) and acyl-CoA synthetase long-chain family member 3. Besides, regulation of syntaxin 5 by miR-155-5p is of critical importance. This protein facilitates vesicle docking and its fusion and is part of the SNARE complex. Alike, miR-26b-5p influences translation of caveolin 2 and diacylglycerol O-acyltransferase 1, i.e. a key enzyme in hepatic steatosis in addition to lecithin-cholesterol acyltransferase which catalyzes its esterification for cholesterol transport. For its role in the regulation of the Ah-receptor miR-124-3p is of great importance. Specifically, AhR sustained activation results in induction of fatty acid transport proteins and enhanced lipid uptake via activity of the fatty acid transporter CD36 to eventually cause accumulation of triglycerides leading to hepatic steatosis. Likewise, it is of great significance that AhR suppresses fatty acid oxidation and export of triglycerides from the liver by hampering VLDL synthesis and secretion. Lastly, the AhR engages in a vicious cycle whereby serum triacylglycerides levels are increased through peripheral fat mobilization thus perpetuating hepatic lipid uptake from the systemic circulation37.
Regulation of 1-acylglycerol-3-phosphate O-acyltransferase 2 by miR-744-5p is also of key importance. This ER localized protein catalyzes the conversion of lysophosphatidic acid to phosphatidic acid, e.g. a building block in phospholipid synthesis and essential second messenger that mediates cellular functions through different modes of action. Noteworthy is also the regulation of the phosphatase and tensin homolog PTEN by miR21-5p and recent research identified Maf1 as a novel target of PTEN and PI3K signalling to negatively regulate carcinogenesis and lipid metabolism by repressing intracellular lipid accumulation and de novo lipogenesis38. Alike, this miRNA targets fatty acid synthase and the enzyme catalyze the synthesis of long-chain saturated fatty acids. Among the top 10 miRNAs is let-7b-5p known to influence translation of insulin receptor substrate 2 transcripts and this protein functions as an adaptor to link receptor tyrosine kinases with downstream effectors. Dynein is also regulated by this miRNA and this microtubule-activated ATPase is of uttermost importance in intracellular motility and trafficking of vesicles. A further example is the regulation of the liver receptor homolog 1 (NR5A2) by miR-1 and this nuclear receptor functions as a transcription factor and phospholipid binding protein and plays an essential role in hepatic metabolism. Repression of small heterodimer partner by NR5A2 affects VLDL synthesis and secretion and is deregulated in NAFLD as summarized in the seminal review of Bechmann and colleagues39. Lastly, the gene coding for the ER lipid raft associated protein 1 is a target of miR30a-5p and is part of the ERLIN1/ERLIN2 complex to assist ER-associated degradation of inositol 1, 4, 5-trisphosphate receptors (IP3Rs); note, alterations in ER lipid rafts have been shown to contribute to lipotoxicity40.
The chosen examples illustrate the benefit in construction molecular circuits based on co-expressed miRNA-gene targets and the knowledge gain by considering mechanistic information regarding common and exclusively regulated genes in DIS and NAFLD. In an effort to define commonalities amongst steatotic drugs four mechanistically linked genes (ELOVL2, ELOVL5, FADS1 and FADS2) were identified as consistently regulated after single and repeated treatments with N = 17 steatotic drugs with FADS1 and FADS2 catalyzing Δ5/Δ6 desaturation of fatty acids to influence arachidonic acid derived eicosanoid, prostaglandins and leukotriene production. The significant regulation of FADS1 and FADS2 will influence arachidonic acid homeostasis and associated herewith immune cell populations with a significant reduction in CD4+/CD3+ T-cell and mononuclear cells as observed in FADS1 null mice and in murine models of NASH41. The observed repression of FADS1 and 2 may therefore be consider as an adaption to drug treatment to prevent overt toxicity associated with DIS induced inflammation. While deficiency in unsaturated fatty acids impacts lipid and energy metabolism, membrane structures and signalling pathways, variations in the expression of these enzymes will help to reduce production of inflammatory lipids42. Likewise, repression of the fatty acid elongases ELOV2 and 5 denotes coordinate responses in polyunsaturated fatty acid synthesis. Note, overexpression of ELOVL2 enhanced triacylglycerol synthesis in 3T3-L1 and F442A cells and caused induction of the lipogenic genes diacylglycerol acyltransferase-2 and fatty acid-binding protein-4 to support triacylglycerol synthesis and subsequent accumulation of lipid droplets43. Modulation of the synthesis of very long-chain fatty acids by ELOV2 and 5 and subsequent desaturations by the Δ5/Δ6-desaturases FADS1 and 2 signifies the relevance of the identified DIS associated genes and assaying mechanistically relevant DIS genomic biomarkers can be easily adapted at the preclinical stage of drug development by merely comparing the hepatic gene expression signature of an experimental drug to DIS and DILI signatures along with appropriate controls.
Testimony to the findings of the present study is also a recently published comprehensive lipidomics approach for an identification of classifier lipid metabolites in urine and plasma and transcriptomic data as identified by RNA-sequencing of liver tissue from a total of N = 31 healthy controls, N = 17 steatotic and N = 20 NASH patients. The transcriptomic data correlated with the clinical stage of disease and signatures of lipids were identified that can be employed as biomarkers for disease staging44.
In an effort to define mechanistically relevant serum biomarkers research of the present study identified 24 serum miRNAs as commonly regulated in NASH (Fig. 8). These miRNAs were examined for their role in targeting N = 200 lipid droplet associated genes (supplementary Table S1); a network was constructed whereby 7 of the 24 common regulated miRNAs were involved in the translational control of 41 fatty liver disease specific gene regulations (Fig. 10b). Once again the findings highlight the possibilities by which specific miRNAs instruct pathophysiological processes in fatty liver disease with miR-7-5p targeting suppressor of cytokine signalling SOCs and calveolin-1, i.e. an essential component of membrane lipid rafts with complex functions in cell signalling while the subnetwork of let-7b-5p involves translational control of the LDL receptor, hepatic lipoprotein lipase and SNAP23 that forms a complex with other vesicle-associated membrane protein and is part of the fusion machinery in macro-vesicular steatosis. Further evidence for a coordinate responses in fatty liver disease was obtained by considering regulation of the ER lipid raft associated 2, PPARγ and insulin induced gene 1 by miR-192-5p. The coded proteins are critically involved in lipid and cholesterol homeostasis while inflammatory signalling of the Toll-like receptor 4 and STAT3 pathways was influenced by let-7i-5p and miR-125b-5p, respectively. These and other examples mentioned above clearly demonstrate the benefit of considering mechanistically related miRNA gene target associations in obtaining pathophysiological relevant information by assaying specific serum miRNAs. Indeed, ncRNAs function precisely to repress targets mRNAs and as a result influence expression of proteins and cellular phenotypes45. Although a number of miRNAs are widely expressed amongst diverse tissues, certain miRNAs appear to be organ specific. In addition, some miRNAs are conserved across species and hold promise for translational research. To date, over 1800 mature human miRNAs have been identified and recorded in the miRBase registry46.
Lastly, the following caveats need to be considered. The study findings are based on the miRTar resource but were not obtained within the same liver tissue used for the DIS study. Nonetheless, the evidence within the miRTar repository is based on independent experimental research and for its high evolutionary conservation the results obtained can be exploited to search for transcriptional regulations of co-expressed microRNA target genes. Specifically, only miRNAs found to be expressed in liver tissue were considered and the research performed focused on the discovery of miRNA targeting genes mechanistically involved in lipid droplet formation and hepatic steatosis. Future research will involve the construction of feed-forward loops (FFLs) to decipher regulatory gene networks as we had demonstrated for the repurposing of drugs in the treatment of cystic fibrosis47.
Conclusions
New insight into co-expressed microRNA target genes in DIS and NAFLD was obtained. An identification of mechanistically linked safety biomarkers will aid drug development and contribute to an understanding of risk for drug induced steatosis and to distinguish amongst other causes of fatty liver disease. Our findings encourage independent validation of the proposed miRNA DIS and NAFLD biomarkers.
Additional Information
How to cite this article: Liu, Z. et al. Mechanistically linked serum miRNAs distinguish between drug induced and fatty liver disease of different grades. Sci. Rep. 6, 23709; doi: 10.1038/srep23709 (2016).
Supplementary Material
Acknowledgments
We thank Hong Fang for comments and discussion. We also thank Dr. Takeki Uehara/Dr. Ikuo Kato and the TGP group for helpful discussions and for expert opinion on histopathology of steatotic livers of treated rats. J Borlak receives funding from the German Federal Ministry of Education and Research as part of the Virtual Liver Network initiative (grant number 031 6154). He is also a recipient of an ORISE stipend of the FDA which is gratefully acknowledged. The views presented in this article do not necessarily reflect current or future opinion or policy of the US Food and Drug Administration. Any mention of commercial products is for clarification and not intended as endorsement.
Footnotes
Author Contributions J.B. initiated and supervised the study. Z.L. performed the computational analysis. J.B., Z.L., Y.W. and W.T. analyzed the data and discussed the findings. J.B. wrote the manuscript. All authors read and approved the final manuscript.
References
- Clark J. M. & Diehl A. Nonalcoholic fatty liver disease: An underrecognized cause of cryptogenic cirrhosis. JAMA. 289, 3000–3004 (2003). [DOI] [PubMed] [Google Scholar]
- Alisi A., Feldstein A. E., Villani A., Raponi M. & Nobili V. Pediatric nonalcoholic fatty liver disease: a multidisciplinary approach. Nat Rev Gastroenterol Hepatol. 9, 152–161 (2012). [DOI] [PubMed] [Google Scholar]
- Matteoni C. A. et al. Nonalcoholic fatty liver disease: A spectrum of clinical and pathological severity. Gastroenterol. 116, 1413–1419 (1999). [DOI] [PubMed] [Google Scholar]
- Targher G. & Arcaro G. Non-alcoholic fatty liver disease and increased risk of cardiovascular disease. Atherosclerosis. 191, 235–240 (2007). [DOI] [PubMed] [Google Scholar]
- Björnsson E. S., Bergmann O. M., Björnsson H. K., Kvaran R. B. & Olafsson S. Incidence, Presentation, and Outcomes in Patients With Drug-Induced Liver Injury in the General Population of Iceland. Gastroenterol. 144, 1419–1425 (2013). [DOI] [PubMed] [Google Scholar]
- Anstee Q. M., McPherson S. & Day C. P. How big a problem is non-alcoholic fatty liver disease? BMJ. 343, d3897 (2011). [DOI] [PubMed] [Google Scholar]
- Amacher D. E. The mechanistic basis for the induction of hepatic steatosis by xenobiotics. Expert Opin Drug Metab Toxicol. 7, 949–965 (2011). [DOI] [PubMed] [Google Scholar]
- Massart J. et al. Drug-Induced Inhibition of Mitochondrial Fatty Acid Oxidation and Steatosis. Curr Pathobiol Rep. 1, 147–157 (2013). [Google Scholar]
- Carthew R. W. & Sontheimer E. J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 136, 642–655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N. & Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- Nobuhiro Okada C.-P. L. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 28, 438–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R. et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 510, 115–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S. et al. miR-99a/100 ∼ 125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 28, 858–874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. et al. The association between HLA-DQB1 polymorphism and antituberculosis drug-induced liver injury: a Case-Control Study. J Clin Pharm Ther. 1, 208–211 (2014). [DOI] [PubMed] [Google Scholar]
- Rokavec M., Li H., Jiang L. & Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 6, 214–230 (2014). [DOI] [PubMed] [Google Scholar]
- Cheung O. et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 48, 1810–1820 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahini N. & Borlak J. Recent insights into the molecular pathophysiology of lipid droplet formation in hepatocytes. Prog. Lipid. Res. 54, 86–112 (2014). [DOI] [PubMed] [Google Scholar]
- Igarashi Y. et al. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 43, D921–D927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-D. et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 42, D78–D85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G. & Hogue C. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 4, 2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirola C. J. et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 64, 800–812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moylan C. A. et al. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 59, 471–482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Vicario C. et al. Molecular interplay between Δ5/Δ6 desaturases and long-chain fatty acids in the pathogenesis of non-alcoholic steatohepatitis. Gut. 63, 344–355 (2014). [DOI] [PubMed] [Google Scholar]
- Murakami Y. et al. Comprehensive miRNA Expression Analysis in Peripheral Blood Can Diagnose Liver Disease. PLoS One. 7, e48366 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y. et al. Activation of the aryl hydrocarbon receptor induces hepatic steatosis via the upregulation of fatty acid transport. Arch. Biochem. Biophys. 504, 221–227 (2010). [DOI] [PubMed] [Google Scholar]
- Nakanishi N. et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem. Biophys. Res. Commun. 385, 492–496 (2009). [DOI] [PubMed] [Google Scholar]
- Kuroda S. et al. Rho-kinase inhibitor targeting the liver prevents ischemia/reperfusion injury in the steatotic liver without major systemic adversity in rats. Liver Transpl. 21, 123–131 (2015). [DOI] [PubMed] [Google Scholar]
- Chalasani N. et al. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 55, 2005–2023 (2012). [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernando C., Suarez Y., Rayner K. J. & Moore K. J. MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 22, 86–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaura Y. et al. Plasma microRNA profiles in rat models of hepatocellular injury, cholestasis, and steatosis. PLoS One. 7, e30250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. S. et al. Current and future trends in biomarker discovery and development of companion diagnostics for arthritis. Expert Rev. Mol. Diagn. 15, 219–234 (2014). [DOI] [PubMed] [Google Scholar]
- Villeneuve D. L. et al. Adverse Outcome Pathway Development II: Best Practices. Toxicol. Sci. 142, 321–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine D. J. et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 58, 777–787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlak J., Chatterji B., Londhe K. B. & Watkins P. B. Serum acute phase reactants hallmark healthy individuals at risk for acetaminophen-induced liver injury. Genome Med. 5, 86–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhill C. Motility: CO2-releasing suppository quickly and effectively relieves constipation. Nat Rev Gastroenterol Hepatol. 11, 455–455 (2014). [DOI] [PubMed] [Google Scholar]
- Lee J. H. et al. A Novel Role for the Dioxin Receptor in Fatty Acid Metabolism and Hepatic Steatosis. Gastroenterol. 139, 653–663 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palian B. M. et al. Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS genetics. 10, e1004789 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechmann L. P. et al. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 56, 952–964 (2012). [DOI] [PubMed] [Google Scholar]
- Boslem E. et al. Alteration of Endoplasmic Reticulum Lipid Rafts Contributes to Lipotoxicity in Pancreatic β-Cells. J. Biol. Chem. 288, 26569–26582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.-Y. et al. Characterization of an arachidonic acid-deficient (Fads1 knockout) mouse model. J. Lipid Res. 53, 1287–1295 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. et al. Peroxisomal and Microsomal Lipid Pathways Associated with Resistance to Hepatic Steatosis and Reduced Pro-inflammatory State. J. Biol. Chem. 285, 31011–31023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Zadravec D. & Jacobsson A. ELOVL2 overexpression enhances triacylglycerol synthesis in 3T3-L1 and F442A cells. FEBS lett. 581, 3157–3163 (2007). [DOI] [PubMed] [Google Scholar]
- Gorden D. L. et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 56, 722–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S. & Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Borlak J. & Tong W. Deciphering miRNA transcription factor feed-forward loops to identify drug repurposing candidates for cystic fibrosis. Genome Med. 6, 94–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens M. et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 18, 296–302 (2013). [DOI] [PubMed] [Google Scholar]
- Baker S. S., Baker R. D., Liu W., Nowak N. J. & Zhu L. Role of alcohol metabolism in non-alcoholic steatohepatitis. PLoS One. 5, e9570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Wu C.-Q., Zhang Z.-Q., Yao D.-K. & Zhu L. Loss of expression of miR-335 is implicated in hepatic stellate cell migration and activation. Exp. Cell Res. 317, 1714–1725 (2011). [DOI] [PubMed] [Google Scholar]
- Xiao Y. et al. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting IL-6/STAT3 signalling. J. Hepatol. 62, 889–896 (2015). [DOI] [PubMed] [Google Scholar]
- Lovis P., Gattesco S. & Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 389, 305–312 (2008). [DOI] [PubMed] [Google Scholar]
- Ji J. et al. MicroRNA Expression, Survival, and Response to Interferon in Liver Cancer. N. Engl. J. Med. 361, 1437–1447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X. et al. Transition from hepatic steatosis to steatohepatitis: Unique microRNA patterns and potential downstream functions and pathways. J. Gastroenterol. Hepatol. 27, 331–340 (2012). [DOI] [PubMed] [Google Scholar]
- Wang B. et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid–defined diet in C57BL/6 mice. Hepatology. 50, 1152–1161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q. et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 118, 2431–2442 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 56, 1631–1640 (2012). [DOI] [PubMed] [Google Scholar]
- Miller A. M. et al. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 8, e72324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand N. J. et al. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterol. 136, 1081–1090 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberle V. et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur. J. Cancer. 49, 3442–3449 (2013). [DOI] [PubMed] [Google Scholar]
- Tan Y.-L. et al. miR-744 is a potential prognostic marker in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 39, 359–365 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang H. et al. Serum levels of microRNAs can specifically predict liver injury of chronic hepatitis B. World J. Gastroenterol. 18, 5188–5196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey Lewis P. J. et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 54, 1767–1776 (2011). [DOI] [PubMed] [Google Scholar]
- Wang K. et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 106, 4402–4407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H. et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta. 424, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. The Autoregulatory Feedback Loop of MicroRNA-21/Programmed Cell Death Protein 4/Activation Protein-1 (MiR-21/PDCD4/AP-1) as a Driving Force for Hepatic Fibrosis Development. J. Biol. Chem. 288, 37082–37093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Upregulation of miR-15b in NAFLD models and in the serum of patients with fatty liver disease. Diabetes Res. Clin. Pract. 99, 327–334 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang N. et al. MicroRNA-101 overexpression by IL-6 and TNF-α inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Exp. Cell Res. 336, 33–42 (2015). [DOI] [PubMed] [Google Scholar]
- Xu Q., Li Y., Shang Y. F., Wang H. L. & Yao M. X. miRNA-103: molecular link between insulin resistance and nonalcoholic fatty liver disease. World J. Gastroenterol. 21, 511–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. et al. Aberrant miR199a-5p/caveolin1/PPARα axis in hepatic steatosis. J. Mol. Endocrinol. 53, 393–403 (2014). [DOI] [PubMed] [Google Scholar]
- Li Y. Y. Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease. World J. Gastroenterol. 18, 6546–6551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma H. et al. Expression of genes for microRNA-processing enzymes is altered in advanced non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 28, 1410–1415 (2013). [DOI] [PubMed] [Google Scholar]
- Sun C. et al. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int. J. Mol. Med. 35, 847–853 (2015). [DOI] [PubMed] [Google Scholar]
- Estep M. et al. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 32, 487–497 (2010). [DOI] [PubMed] [Google Scholar]
- Zarrinpar A., Gupta S., Maurya M. R., Subramaniam S. & Loomba R. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. doi: 10.1136/gutjnl-2015-309456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H. et al. MiRNA-194 Regulates Palmitic Acid-Induced Toll-Like Receptor 4 Inflammatory Responses in THP-1 Cells. Nutrients. 7, 3483–3496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave S. C. et al. A role for miR-296 in the regulation of lipoapoptosis by targeting PUMA. J. Lipid Res. 52, 1517–1525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S., Ruggieri A., Marrero J. A., Ioannou G. N. & Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 6, e23937 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leti F. et al. High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Transl Res. 166, 304–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Ge G., Pan T., Wen D. & Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 9, e105192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvai G. et al. Elevated miR-33a and miR-224 in steatotic chronic hepatitis C liver biopsies. World J. Gastroenterol. 20, 15343–15350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaaki H. et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int. 34, e302–307 (2014). [DOI] [PubMed] [Google Scholar]
- Zheng L., Lv G. C., Sheng J. & Yang Y. D. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. J. Gastroenterol. Hepatol. 25, 156–163 (2010). [DOI] [PubMed] [Google Scholar]
- Ahn J., Lee H., Jung C. H. & Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res. 56, 1665–1674 (2012). [DOI] [PubMed] [Google Scholar]
- Alisi A. et al. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab. Invest. 91, 283–293 (2011). [DOI] [PubMed] [Google Scholar]
- Tryndyak V. P. et al. Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol. Appl. Pharmacol. 262, 52–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J. Lipid Res. 50, 1756–1765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H. et al. Longitudinal study of circulating miR-122 in a rat model of non-alcoholic fatty liver disease. Clin. Chim. Acta. 446, 267–271 (2015). [DOI] [PubMed] [Google Scholar]
- Povero D. et al. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 9, e113651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. C. et al. Upregulation of miR-125b by estrogen protects against non-alcoholic fatty liver in female mice. J. Hepatol. 63, 1466–1475 (2015). [DOI] [PubMed] [Google Scholar]
- Feng Y. Y. et al. Aberrant hepatic microRNA expression in nonalcoholic fatty liver disease. Cell. Physiol. Biochem. 34, 1983–1997 (2014). [DOI] [PubMed] [Google Scholar]
- Lin X. et al. Overexpression of miR-155 in the liver of transgenic mice alters the expression profiling of hepatic genes associated with lipid metabolism. PLoS One. 10, e0118417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella S. et al. EZH2 down-regulation exacerbates lipid accumulation and inflammation in in vitro and in vivo NAFLD. Int J Mol Sci. 14, 24154–24168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M., van der Sluis R. J., Kuiper J. & Van Berkel T. J. Nonalcoholic fatty liver disease is associated with an altered hepatocyte microRNA profile in LDL receptor knockout mice. J. Nutr. Biochem. 23, 622–628 (2012). [DOI] [PubMed] [Google Scholar]
- Ahn J., Lee H., Chung C. H. & Ha T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochem. Biophys. Res. Commun. 414, 664–669 (2011). [DOI] [PubMed] [Google Scholar]
- Dattaroy D. et al. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G298–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R. E. et al. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 58, 119–125 (2013). [DOI] [PubMed] [Google Scholar]
- Takaki Y. et al. Silencing of microRNA-122 is an early event during hepatocarcinogenesis from non-alcoholic steatohepatitis. Cancer Sci. 105, 1254–1260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csak T. et al. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 35, 532–541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. D. et al. Circulating microRNA 122 in the methionine and choline-deficient mouse model of non-alcoholic steatohepatitis. J Appl Toxicol. 34, 726–732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius B. et al. Resolvin D1 primes the resolution process initiated by calorie restriction in obesity-induced steatohepatitis. FASEB J. 28, 836–848, doi: 10.1096/fj.13-235614 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang B. et al. Upregulated microRNA-199a-5p inhibits nuclear receptor corepressor 1 translation in mice with non-alcoholic steatohepatitis. Mol Med Rep. 10, 3080–3086 (2014). [DOI] [PubMed] [Google Scholar]
- Hur W. et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 64, 265–276 (2015). [DOI] [PubMed] [Google Scholar]
- Ng R. et al. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology. 60, 554–564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J. et al. Carbon monoxide protects against hepatic ischemia/reperfusion injury by modulating the miR-34a/SIRT1 pathway. Biochim. Biophys. Acta. 1852, 1550–1559 (2015). [DOI] [PubMed] [Google Scholar]
- Shpyleva S. et al. Interstrain differences in the progression of nonalcoholic steatohepatitis to fibrosis in mice are associated with altered hepatic iron metabolism. J. Nutr. Biochem. 25, 1235–1242 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.