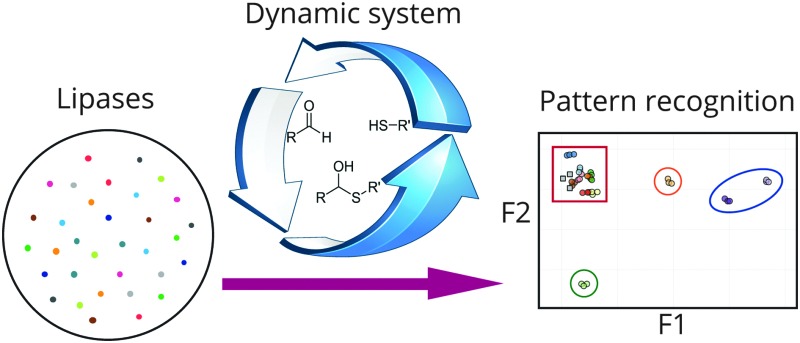
 A complex dynamic hemithioacetal system was used in combination with pattern recognition methodology to classify lipases into distinct groups.
A complex dynamic hemithioacetal system was used in combination with pattern recognition methodology to classify lipases into distinct groups.
Abstract
A complex dynamic hemithioacetal system was generated for the evaluation of lipase reactivities in organic media. In combination with pattern recognition methodology, twelve different lipases were successfully classified into four distinct groups following their reaction selectivities and reactivities. A probe lipase was further categorized using the training matrix with predicted reactivity.
Enzyme catalysis has over the last decades been increasingly explored for organic synthesis due to favorable properties such as environmental benignity, high efficiency and selectivity.1–9 A multitude of different enzymes have been applied, among which lipases have drawn special attention, in part due to their performances in organic solvents coupled to high stereoselectivities.10–13 Applications of lipases have also been recently expanded owing to new findings on enzyme modification and promiscuity, where improved or additional activities have been illustrated.14–17 However, lipases from individual sources or families may not behave the same in regard to substrates or reaction types, and in order to distinguish the specificities among different lipases, an efficient method for lipase screening and grouping would be useful, accelerating further applications in synthesis.
Classification of enzyme performances in principle requires collection and analysis of sizable data sets, involving interpretation of arrays of reaction results. This process can however be facilitated using two methodologies: dynamic chemistry and pattern recognition. Pattern recognition has been demonstrated to be a powerful technique with broad applications in classification and identification, for example in retrosynthetic analysis and protein differentiation.18–20 Methods such as principal component analysis (PCA), linear discriminant analysis (LDA), and partial least square (PLS) analysis have thus been developed for the extraction of relevant data features, generating classification patterns which can be used to predict unknown species.21–23 In conjunction to these techniques, dynamic systemic resolution (DSR), a concept based on constitutional dynamic chemistry (CDC), can be efficiently applied to generating and probing complex systems.24–28 Typical CDC systems involve reversible (covalent) reactions, making all components mutually interchange under thermodynamic control. Applied selection pressures may then exert rearrangements of the systems, causing amplification of optimal system species.29–51
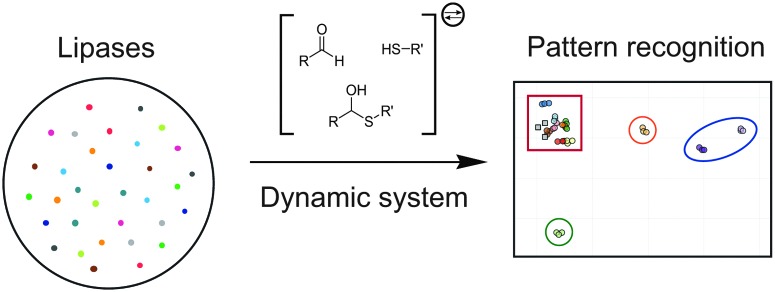
In DSR systems, kinetic steps are coupled to the reversible reactions, resulting in irreversible resolution processes.43,52–61 For example, kinetic enzymatic transformations have been used as external selection pathways, enabling rapid substrate screening. Therefore, the combination of DSR and pattern recognition would result in a powerful platform for enzyme screening and classification in reference to the enzyme reactivities and substrate selectivities. Herein, this dynamic diversity has been demonstrated and further applied to distinguish lipase selectivities. Dynamic hemithioacetal systems were thus generated and further subjected to biocatalyzed transesterification, and the resulting information subsequently analyzed by pattern recognition methodology resulting in discrete classification of lipase reactivity (Fig. 1).
Fig. 1. Lipase classification through dynamic hemithioacetal systems in combination with pattern recognition methodology.
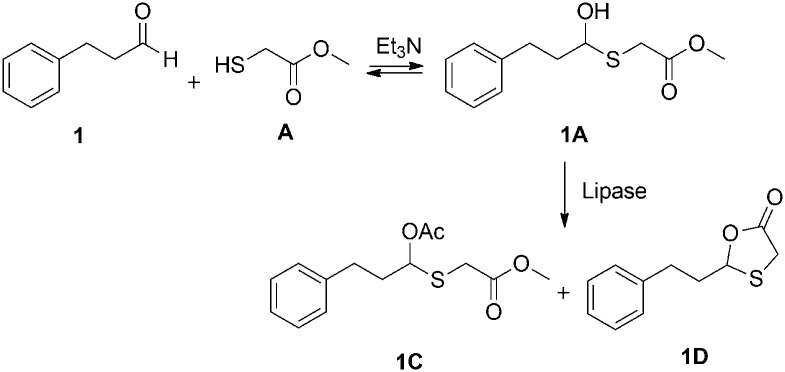
Hemithioacetal chemistry shows attractive reversibility features, and can be applied to generate dynamic systems in both aqueous and organic media.43,57,59 We have for example previously demonstrated the selective lipase-catalyzed cyclization to 1,3-oxathiolan-5-ones using sulfanylacetates.57 When additional acyl donors are added into these systems, linear (non-cyclized) acylated products can potentially be produced in addition to cyclized oxathiolanones. This leads to increased diversity of the system, thereby enabling additional information of the lipase catalysis process. In order to evaluate the potential of forming both intermolecularly acetylated and intramolecularly lactonized products, test reactions between 3-phenylpropanal and methyl 2-sulfanylacetate in the presence of phenyl acetate were first performed under the catalytic action of two different lipases: PSIm from Burkholderia (Pseudomonas) cepacia and CALB from Candida antarctica (Scheme 1). Interestingly, with triethylamine (Et3N) as base,57 both types of products were detected in the 1H-NMR spectra of the reaction mixtures at room temperature, but the ratio between these two products varied according to the lipases used. While PSIm gave predominantly the intermolecular, ‘linear’ acetylated product (1C : 1D = 6.4 : 1), CALB mostly yielded the cyclized intramolecular product (1C : 1D = 1 : 2.7). These results thereby confirmed the potential of using these dynamic systems for lipase reactivity profiling.
Scheme 1. Lipase-catalyzed acetylation/lactonization of dynamic hemithioacetal system.
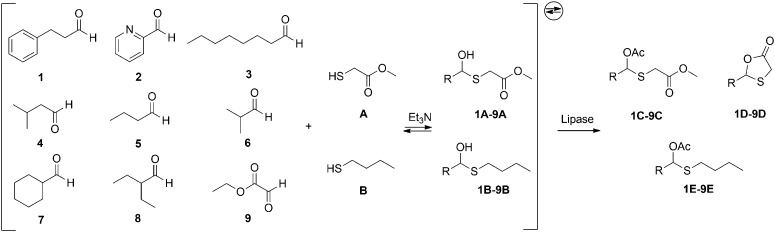
A dynamic hemithioacetal system with enhanced complexity was subsequently established. Nine aldehydes (1–9) of different structure were incorporated, including both aromatic and aliphatic compounds, structures containing short and longer chains, and linear and cyclized functional groups (Scheme 2). Besides methyl 2-sulfanylacetate, butanethiol was also included in the system, only able to form linear acetylated products. Under the same conditions as in the test reactions, twelve commonly used and commercially available lipases were chosen to challenge the system as external selectors, irreversibly transforming the intermediates 1A–9A and 1B–9B to the respective preferred products. Thus, lipases from Aspergillus niger (ANL), Candida antarctica (CALA and CALB), Candida rugosa (CRL), Mucor javanicus (MJL), Mucor miehei (MML), Pseudomonas fluorescens (PFL), porcine pancreas (PPL), Burkholderia cepacia (PSCI and PSIm), Rhizopus arrhizus (RAL), and Rhizopus niveus (RNL), were evaluated. In principle, under these conditions, 36 different intermediates of two types and 54 possible products of three different types can be generated, forming a dynamic system with the complexity of 101 different compounds.
Scheme 2. Dynamic hemithioacetal system with enhanced complexity used for lipase evaluation.
1H-NMR was used to monitor the reaction progress in the system, which showed the typical intermediate and product peaks around 4–7 ppm. As expected, this complex dynamic system produced largely overlapping signals, difficult to resolve by conventional means. Pattern recognition methodology was then adopted to analyze the results. Among the data classification techniques, LDA is excellent at processing two or more classes of objects, and is in addition simple and robust.62 The principle of LDA analyses is based on a training matrix created by a collection of independent (explanatory) and dependent variables; where the dependent variables (lipases) are classified according to the values generated by the explanatory variables (NMR signal intensities). LDA treats the array response as the linear combination of discriminant functions or canonical scores.63,64
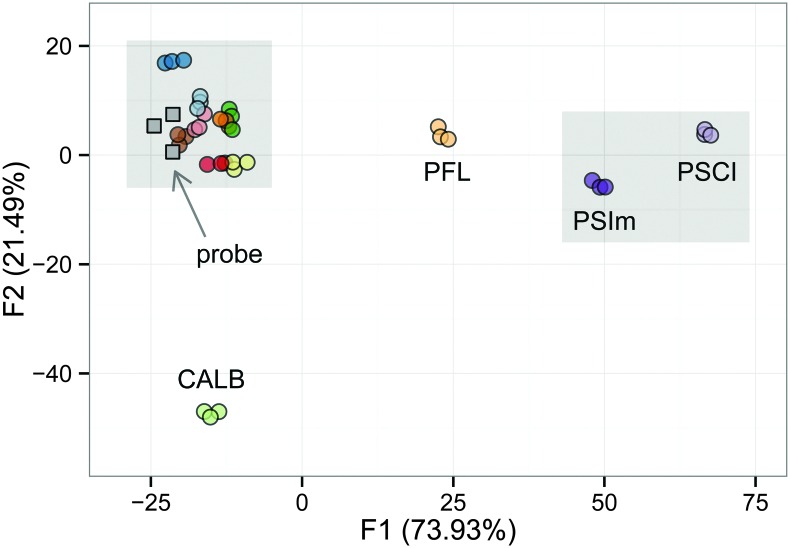
The relative signal integrations using Et3N as the internal standard were first extracted from the 1H-NMR spectra, with every 0.1 ppm in the range of 4.6–6.7 ppm, which were further employed as 21 explanatory variables in the LDA training matrix (Table S1, ESI†). The dependent variables to generate the training matrix were the twelve lipases with their repeated triplicate experimental data. For the best separation achieved in Fig. 2, the lipases were classified into groups on the factor plot where the canonical factors F1 and F2 contained variation of 73.93% and 21.49%, respectively.
Fig. 2. Factor plot obtained from the LDA analysis of lipases tested with the dynamic hemithioacetal system; ( ) ANL, (
) ANL, ( ) CALA, (
) CALA, ( ) CALB, (
) CALB, ( ) CRL, (
) CRL, ( ) MJL, (
) MJL, ( ) MML, (
) MML, ( ) PFL, (
) PFL, ( ) PPL, (
) PPL, ( ) PSCI, (
) PSCI, ( ) PSIm, (
) PSIm, ( ) RAL, (
) RAL, ( ) RNL, (
) RNL, ( ) probe.
) probe.
From the pattern generated from the training matrix, the twelve different lipases could be classified into several distinct areas in the analysis window: three discrete areas with CALB, PFL, and PSCI/PSIm, and an extended region grouping all remaining lipases (Fig. 2). Validation tests revealed some cross-correlation between PSCI and PSIm, as could be expected, and strong correlation between the lipases of the extended group. The overall cross-validation score was thus relatively low (0.39, Table S7, ESI†). However, upon grouping of the lipases in the extended region, ANL, CALA, CRL, MJL, MML, PPL, RAL, RNL, the validation result was considerably higher (0.97, Table S7, ESI†).
Furthermore, the correlation between the pattern and the enzyme activities was also studied. CALB, CRL, PFL and PSIm, were selected as representative enzymes of the different groups and evaluated in reaction tests with 3-phenylpropanal. These studies revealed clear differences between the respective enzymes. CALB, PFL and PSIm were comparatively more reactive, whereas the distributions of the formed products were opposite: CALB showed preference for the cyclized product (1D), while PSIm was selective for the linear species (1C). For PFL, the resulting ratio between the linear acetylated- and cyclized product was 29 : 1, displaying strong resemblance to the PSIm/PSCI group. On the other hand, CRL yielded only trace amount of cyclized product, which demonstrated the inactive behavior of this group of lipases toward intermolecular acetylations under these reaction conditions.
Information from the grouping pattern of all the lipases shown above was then applied to test a probe lipase. Thus, a lipase from Penicillium camembertii (PCL) was introduced as the testing lipase into the complex system under the same conditions as for all other lipases. As illustrated in Fig. 2, this probe lipase was positioned in the region of the extended group.
In order to examine the correlation between the found pattern and the activity of PCL, the test reaction between 3-phenylpropanal and methyl 2-sulfanylacetate with phenyl acetate as acetyl donor was challenged also by PCL catalysis. The resulting pattern demonstrated the similarity to the extended lipase region.
In conclusion, complex dynamic hemithioacetal systems were generated and coupled to enzymatic transformation, providing two types of potential products, intermolecular linear acetylated compounds and intramolecular cyclized structures. Pattern recognition methodology was further employed to evaluate the selectivities and reactivities of lipases based on the signal integrations in the 1H-NMR spectra of the reaction mixture. Thus, twelve different lipases were classified into four groups in the analysis window, which showed good correlation to their selectivities between the two types of products. Lipase PCL as a probe lipase from the training set was tested with the same dynamic system, giving major recognition into an extended lipase group, which was consistent with the enzyme reactivity.
This work was in part supported by the Swedish Research Council, the National Institutes of Health (R01GM080295 and 2R15GM066279 to MY), and the Royal Institute of Technology. YZ thanks the China Scholarship Council for a special scholarship award.
Footnotes
References
- Cuetos A., García-Ramos M., Fischereder E.-M., Díaz-Rodríguez A., Grogan G., Gotor V., Kroutil W., Lavandera I. Angew. Chem., Int. Ed. 2016;55:3144–3147. doi: 10.1002/anie.201510554. [DOI] [PubMed] [Google Scholar]
- Du Y.-L., Singh R., Alkhalaf L. M., Kuatsjah E., He H.-Y., Eltis L. D., Ryan K. S. Nat. Chem. Biol. 2016;12:194–199. doi: 10.1038/nchembio.2009. [DOI] [PubMed] [Google Scholar]
- Porter J. L., Rusli R. A., Ollis D. L. ChemBioChem. 2016;17:197–203. doi: 10.1002/cbic.201500280. [DOI] [PubMed] [Google Scholar]
- Wang J.-b., Reetz M. T. Nat. Chem. 2015;7:948–949. doi: 10.1038/nchem.2408. [DOI] [PubMed] [Google Scholar]
- Nestl B. M., Hammer S. C., Nebel B. A., Hauer B. Angew. Chem., Int. Ed. 2014;53:3070–3095. doi: 10.1002/anie.201302195. [DOI] [PubMed] [Google Scholar]
- Turner N. J., O'Reilly E. Nat. Chem. Biol. 2013;9:285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- Lumbroso A., Cooke M. L., Breit B. Angew. Chem., Int. Ed. 2013;52:1890–1932. doi: 10.1002/anie.201204579. [DOI] [PubMed] [Google Scholar]
- Hu L., Schaufelberger F., Zhang Y., Ramström O. Chem. Commun. 2013;49:10376–10378. doi: 10.1039/c3cc45551c. [DOI] [PubMed] [Google Scholar]
- Enzyme Catalysis in Organic Synthesis, ed. K. Drauz, H. Gröger and O. May, John Wiley & Sons, 2012. [Google Scholar]
- Manoel E. A., Robert J. M., Pinto M. C. C., Machado A. C. O., Besteti M. D., Coelho M. A. Z., Simas A. B. C., Fernandez-Lafuente R., Pinto J. C., Freire D. M. G. RSC Adv. 2016;6:4043–4052. [Google Scholar]
- Kitamoto Y., Kuruma Y., Suzuki K., Hattori T. J. Org. Chem. 2015;80:521–527. doi: 10.1021/jo502521e. [DOI] [PubMed] [Google Scholar]
- de Miranda A. S., Miranda L. S., de Souza R. O. Biotechnol. Adv. 2015;33:372–393. doi: 10.1016/j.biotechadv.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Adlercreutz P. Chem. Soc. Rev. 2013;42:6406–6436. doi: 10.1039/c3cs35446f. [DOI] [PubMed] [Google Scholar]
- Stavila E., Alberda van Ekenstein G. O. R., Woortman A. J. J., Loos K. Biomacromolecules. 2014;15:234–241. doi: 10.1021/bm401514k. [DOI] [PubMed] [Google Scholar]
- Sakulsombat M., Vongvilai P., Ramström O. Chem. – Eur. J. 2014;20:11322–11325. doi: 10.1002/chem.201402615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humble M. S., Berglund P. Eur. J. Org. Chem. 2011:3391–3401. [Google Scholar]
- Vongvilai P., Linder M., Sakulsombat M., Svedendahl Humble M., Berglund P., Brinck T., Ramström O. Angew. Chem., Int. Ed. 2011;50:6592–6595. doi: 10.1002/anie.201007373. [DOI] [PubMed] [Google Scholar]
- Bi X., Li D., Liu Z. Anal. Chem. 2015;87:4442–4447. doi: 10.1021/acs.analchem.5b01034. [DOI] [PubMed] [Google Scholar]
- Jayawardena H. S. N., Wang X., Yan M. Anal. Chem. 2013;85:10277–10281. doi: 10.1021/ac402069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto S., Cativiela C., Urriolabeitia E. P. New J. Chem. 2012;36:566–569. [Google Scholar]
- Elci S. G., Moyano D. F., Rana S., Tonga G. Y., Phillips R. L., Bunz U. H. F., Rotello V. M. Chem. Sci. 2013;4:2076–2080. [Google Scholar]
- Stewart S., Ivy M. A., Anslyn E. V. Chem. Soc. Rev. 2014;43:70–84. doi: 10.1039/c3cs60183h. [DOI] [PubMed] [Google Scholar]
- Buryak A., Severin K. J. Am. Chem. Soc. 2005;127:3700–3701. doi: 10.1021/ja042363v. [DOI] [PubMed] [Google Scholar]
- Lehn J.-M. Angew. Chem., Int. Ed. 2015;54:3276–3289. doi: 10.1002/anie.201409399. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Hu L. and Ramström O., Supramolecular Systems in Biomedical Fields, The Royal Society of Chemistry, 2013, pp. 397–418. [Google Scholar]
- Constitutional Dynamic Chemistry, ed. M. Barboiu, Springer Verlag, Berlin, Heidelberg, 2012. [Google Scholar]
- Dynamic Combinatorial Chemistry: In Drug Discovery, Bioorganic Chemistry, and Materials Science, ed. B. L. Miller, John Wiley & Sons, Hoboken, NJ, 2010. [Google Scholar]
- Dynamic Combinatorial Chemistry, ed. J. N. H. Reek and S. Otto, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2010. [Google Scholar]
- Zhang Y., Barboiu M. Chem. Rev. 2016;116:809–834. doi: 10.1021/acs.chemrev.5b00168. [DOI] [PubMed] [Google Scholar]
- Kosikova T., Mackenzie H., Philp D. Chem. – Eur. J. 2016;22:1831–1839. doi: 10.1002/chem.201503740. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Ohishi T., Goseki R., Otsuka H. Polymer. 2016;82:319–326. [Google Scholar]
- Barcan G. A., Zhang X., Waymouth R. M. J. Am. Chem. Soc. 2015;137:5650–5653. doi: 10.1021/jacs.5b02161. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Barboiu M. Chem. Commun. 2015;51:15925–15927. doi: 10.1039/c5cc06805c. [DOI] [PubMed] [Google Scholar]
- Schaufelberger F., Ramström O. Chem. – Eur. J. 2015;21:12735–12740. doi: 10.1002/chem.201502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufelberger F., Hu L., Ramström O. Chem. – Eur. J. 2015;21:9776–9783. doi: 10.1002/chem.201500520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer S. A., Turner D. R. Chem. Commun. 2015;51:17375–17378. doi: 10.1039/c5cc07422c. [DOI] [PubMed] [Google Scholar]
- Li G., Zheng W., Chen Z., Zhou Y., Liu Y., Yang J., Huang Y., Li X. Chem. Sci. 2015;6:7097–7104. doi: 10.1039/c5sc02467f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Yuan Y., You L., Anslyn E. V. Chem. – Eur. J. 2015;21:8207–8213. doi: 10.1002/chem.201500105. [DOI] [PubMed] [Google Scholar]
- Mondal M., Hirsch A. K. Chem. Soc. Rev. 2015;44:2455–2488. doi: 10.1039/c4cs00493k. [DOI] [PubMed] [Google Scholar]
- Brachvogel R.-C., von Delius M. Chem. Sci. 2015;6:1399–1403. doi: 10.1039/c4sc03528c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasale D. B., Biswas S., Konda M., Das A. K. RSC Adv. 2015;5:1529–1537. [Google Scholar]
- Hsu C. W., Miljanic O. S. Angew. Chem., Int. Ed. 2015;54:2219–2222. doi: 10.1002/anie.201409741. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ramström O. Chem. – Eur. J. 2014;20:3288–3291. doi: 10.1002/chem.201304690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda E., Rademann J. Nat. Commun. 2014;5:5170. doi: 10.1038/ncomms6170. [DOI] [PubMed] [Google Scholar]
- Wilson A., Gasparini G., Matile S. Chem. Soc. Rev. 2014;43:1948–1962. doi: 10.1039/c3cs60342c. [DOI] [PubMed] [Google Scholar]
- Herrmann A. Chem. Soc. Rev. 2014;43:1899–1933. doi: 10.1039/c3cs60336a. [DOI] [PubMed] [Google Scholar]
- Misuraca M. C., Moulin E., Ruff Y., Giuseppone N. New J. Chem. 2014;38:3336–3349. [Google Scholar]
- Ulrich S., Dumy P. Chem. Commun. 2014;50:5810–5825. doi: 10.1039/c4cc00263f. [DOI] [PubMed] [Google Scholar]
- Santana A. G., Jimenez-Moreno E., Gomez A. M., Corzana F., Gonzalez C., Jimenez-Oses G., Jimenez-Barbero J., Asensio J. L. J. Am. Chem. Soc. 2013;135:3347–3350. doi: 10.1021/ja3120218. [DOI] [PubMed] [Google Scholar]
- Chung M.-K., Severin K., Lee S. J., Waters M. L., Gagné M. R. Chem. Sci. 2011;2:744. [Google Scholar]
- Capela M., Mosey N. J., Xing L., Wang R., Petitjean A. Chemistry. 2011;17:4598–4612. doi: 10.1002/chem.201002389. [DOI] [PubMed] [Google Scholar]
- Hu L., Zhang Y., Ramström O. Sci. Rep. 2015;5:11065. doi: 10.1038/srep11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Vongvilai P., Sakulsombat M., Fischer A., Ramström O. Adv. Synth. Catal. 2014;356:987–992. doi: 10.1002/adsc.201301033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Zhang Y., Ramström O. Org. Biomol. Chem. 2014;12:3572–3575. doi: 10.1039/c4ob00365a. [DOI] [PubMed] [Google Scholar]
- Hu L., Ramstrom O. Chem. Commun. 2014;50:3792–3794. doi: 10.1039/c4cc00944d. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Hu L., Ramström O. Chem. Commun. 2013;49:1805–1807. doi: 10.1039/c3cc38203f. [DOI] [PubMed] [Google Scholar]
- Sakulsombat M., Zhang Y., Ramström O. Chem. – Eur. J. 2012;18:6129–6132. doi: 10.1002/chem.201102139. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Angelin M., Larsson R., Albers A., Simons A., Ramström O. Chem. Commun. 2010;46:8457–8459. doi: 10.1039/c0cc02479a. [DOI] [PubMed] [Google Scholar]
- Caraballo R., Dong H., Ribeiro J. P., Jiménez-Barbero J., Ramström O. Angew. Chem., Int. Ed. 2010;49:589–593. doi: 10.1002/anie.200903920. [DOI] [PubMed] [Google Scholar]
- Vongvilai P., Ramström O. J. Am. Chem. Soc. 2009;131:14419–14425. doi: 10.1021/ja9052015. [DOI] [PubMed] [Google Scholar]
- Vongvilai P., Angelin M., Larsson R., Ramström O. Angew. Chem., Int. Ed. 2007;46:948–950. doi: 10.1002/anie.200603740. [DOI] [PubMed] [Google Scholar]
- Jurs P. C., Bakken G. A., McClelland H. E. Chem. Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- McFarland J. W., Gans D. J. J. Med. Chem. 1987;30:46–49. doi: 10.1021/jm00384a008. [DOI] [PubMed] [Google Scholar]
- Coomans D., Massart D. L., Kaufman L. Anal. Chim. Acta. 1979;112:97–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.