Abstract
Hypoxic areas are a common feature of rapidly growing malignant tumors and their metastases and are typically spatially heterogeneous. Hypoxia has a strong impact on tumor cell biology and contributes to tumor progression in multiple ways. To date, only a few molecular key players in tumor hypoxia, such as hypoxia-inducible factor-1 (HIF-1), have been discovered. The distribution of biomolecules is frequently heterogeneous in the tumor volume and may be driven by hypoxia and HIF-1α. Understanding the spatially heterogeneous hypoxic response of tumors is critical. Mass spectrometric imaging (MSI) provides a unique way of imaging biomolecular distributions in tissue sections with high spectral and spatial resolution. In this paper, breast tumor xenografts grown from MDA-MB-231-HRE-tdTomato cells, with a red fluorescent tdTomato protein construct under the control of a hypoxia response element (HRE)-containing promoter driven by HIF-1α, were used to detect the spatial distribution of hypoxic regions. We elucidated the 3D spatial relationship between hypoxic regions and the localization of lipids and proteins by using principal component analysis–linear discriminant analysis (PCA-LDA) on 3D rendered MSI volume data from MDA-MB-231-HRE-tdTomato breast tumor xenografts. In this study, we identified hypoxia-regulated proteins active in several distinct pathways such as glucose metabolism, regulation of actin cytoskeleton, protein folding, translation/ribosome, splicesome, the PI3K-Akt signaling pathway, hemoglobin chaperone, protein processing in endoplasmic reticulum, detoxification of reactive oxygen species, aurora B signaling/apoptotic execution phase, the RAS signaling pathway, the FAS signaling pathway/caspase cascade in apoptosis, and telomere stress induced senescence. In parallel, we also identified colocalization of hypoxic regions and various lipid species such as PC(16:0/18:0), PC(16:0/18:1), PC(16:0/18:2), PC(16:1/18:4), PC(18:0/18:1), and PC(18:1/18:1), among others. Our findings shed light on the biomolecular composition of hypoxic tumor regions, which may be responsible for a given tumor's resistance to radiation or chemotherapy.
Hypoxia is a hallmark of cancer and triggers multiple signaling cascades that significantly impact tumor angiogenesis, increased invasion and metastasis, and selection for resistance to apoptosis as well as radiation and chemotherapy.1–3 The hypoxia-inducible factor-1 (HIF-1) pathway is one of the best-characterized signaling pathways regulated by hypoxia in cancer.4,5 HIF-1α, which is overexpressed in breast cancer, has a high impact on tumor progression by enhancing the transcription of target genes, such as P53,4,6 vascular endothelial growth factor (VEGF),7 and lysyl oxidase8 through binding to cis-acting hypoxia response elements (HREs) containing the consensus binding site 5′-RCGTG-3′ in the promoters of these genes. HIF-2α is also regulated by hypoxia and binds with HIF-1β to form the HIF-2 heterodimer. While HIF-1α and HIF-2α share a high degree of sequence similarity, HIF-2 stimulates some, but not all, of the genes activated by HIF-1, such as chemokine receptor type 4 (CXCR4),9 Ephrin-A1,10 and transmembrane glycoprotein CD44,11 among others. We have previously utilized MDA-MB-231-HRE-tdTomato breast cancer cells, which stably express tdTomato red fluorescent protein under the control of an HRE-containing promoter to detect the spatial distribution of hypoxic regions within the tumor.11 These hypoxic regions arise from inadequate oxygen supply due to spatially heterogeneous variations in vascular volume, vascular permeability, and microvessel density across the tumor volume.1,12
HIF-1α expression alone cannot be viewed as a definitive marker of tumor response to hypoxia, and recent studies have focused on determining other gene and protein signatures involved in this complex phenomenon. Chi et al.13 used human cDNA microarrays containing 42 000 elements that represent 27 291 unique genes and detected 253 genes that are up-regulated under hypoxia in human mammary epithelial cells and renal proximal tubule epithelial cells in vitro. Buffa et al.14 showed that genes involved in angiogenesis such as vascular endothelial growth factor A (VEGFA), glucose metabolism such as glucose transporter type 1 (GTR1), phosphoglycerate mutase 1 (PGAM1), enolase I (ENOA), l-lactate dehydrogenase A (LDHA), triosephosphate isomerase II (TPIS2), and fructose-bisphosphate aldolase A (ALDOA), and cell cycle regulation such as cyclin-dependent kinase inhibitor 3 (CDKN3) were among those that were likely overexpressed in hypoxic breast cancers. Using proteomics tools, Cui et al.15 identified hypoxia-regulated proteins that were overexpressed in microdissected pancreatic cancer nests. Immunohistochemistry (IHC) confirmed that these pancreatic cancer nests had significantly higher expression levels of glucose-regulated protein 78 (GRP78), macrophage migration inhibitory factor (MIF), and annexin A5 (ANXA5) than normal pancreatic tissues, suggesting that these hypoxia-regulated proteins are promising targets for pancreatic cancer diagnosis and therapy.15
Tumor hypoxia is typically spatially heterogeneous making it challenging to assess the spatial distribution of colocalizing biomolecules and investigate hypoxia-regulated pathways in solid tumors. We have previously performed a comprehensive analysis of the combined in vivo 1H MRSI and ex vivo optical imaging data obtained from the MDA-MB-231-HRE-tdTomato breast tumor model and showed a higher concentration of noninvasively detected tCho and mobile lipid droplets that colocalized with the tdTomato-fluorescing hypoxic regions, which indicated that hypoxia can up-regulate tCho and lipid CH3 levels in this breast tumor model.16–18
Mass spectrometric imaging (MSI) enables the characterization and profiling of a plethora of molecules and, at the same time, reveals their individual spatial localizations in cancer tissue sections, without the need for labeling of these molecules.19 Hence, MSI provides a unique way to make snapshots of molecular distributions in a high-throughput manner with high spectral and spatial resolution.20 We have recently identified characteristic tryptic peptides from the red fluorescent tdTomato protein by combining microscopic fluorescence imaging with matrix-assisted laser desorption ionization (MALDI)-MSI using a novel fiducial marker system,21 making it possible to detect tdTomato by MALDI-MSI. We have spatially localized a number of lipid species in two dimensions in this breast tumor xenograft model.22 It is now possible to perform 3D reconstruction and rendering of MSI tissue volumes by using block-face optical imaging methods23 or fiducial marker strategies24 to accurately align successive 2D MSI experiments of tissue sections that are cut with well-defined spacing throughout a biological sample, such as a tumor or an organ. 3D reconstruction and rendering of MSI data is useful for visualizing the characteristics of a tissue volume in 3D, and it also enables quantitative mining of 3D MSI volume data, for example, for quantifying correlations between spectral and spatial features, by using multivariate statistical analysis approaches.25,26
Differences in phospholipid signals between renal cell carcinoma and adjacent normal tissue have been studied in 2D using multivariate analysis on desorption electrospray ionization (DESI)-MSI data with a misclassification rate of about 14%.27 DESI lipid imaging was able to show a high recognition rate of 97% with cross validation for classifying subtype, grade, and concentration features of human brain tumors.28 MSI of biopsies from human colorectal cancer liver metastases revealed lipids that are significantly more1 or less abundant in the tumor region.29 Principal component analysis–linear discriminant analysis (PCA-LDA) was used for analyzing 2D MALDI-MSI data of on-tissue digested tryptic peptides from a human formaldehyde-fixed paraffin-embedded (FFPE) pancreatic tumor tissue microarray (TMA) and demonstrated that a novel tumor classification model based on direct proteome information was feasible.30 A recent study31 proposed a new computational pipeline for construction and analysis of 3D MALDI-MSI data, including unsupervised image segmentation and peak picking modules, to reveal 3D anatomic structures of the mouse kidney. Two up-to-date reviews about the challenges of statistical multivariate analysis of MALDI-MSI data, especially 3D MALDI-MSI data, have addressed that the large size of MALDI-MSI data is the main challenge for analysis.32,33
In this study, to systematically understand and evaluate the lipidomic and proteomic complexity as well as the spatial distribution of the hypoxic response of breast tumors, we performed 3D MALDI-MSI of MDA-MB-231-HRE-tdTomato breast cancer tissue sections and applied supervised statistical multivariate analyses to these high-throughput 3D MALDI-MSI data. We also generated a breast cancer protein database by performing reverse-phase liquid chromatography (RPLC)-electrospray ionization (ESI)-MS analysis of MBA-MB-231-HRE-tdTomato cells combined with accurate mass tag (AMT) strategies and identified 3D MALDI-MSI-detected tryptic peptides and proteins by searching this AMT-database.
EXPERIMENTAL MATERIALS AND METHODS
MALDI Mass Spectrometric Imaging of Breast Tumor Xenograft Models
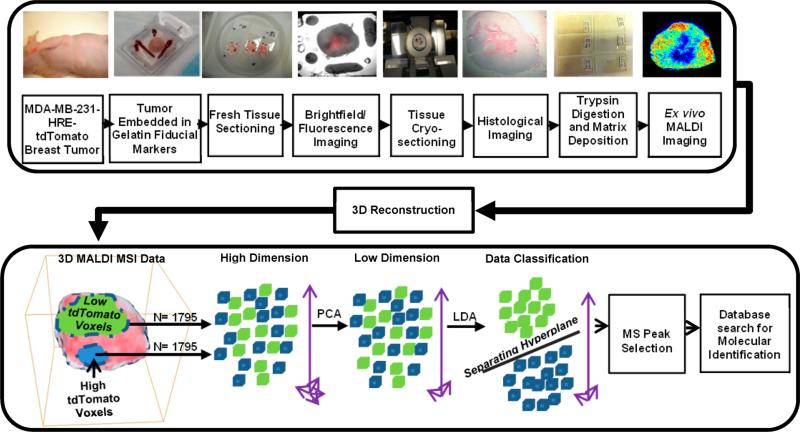
Figure 1 shows an overview of our experimental and data analysis workflow including sample preparation of breast tumor tissue, 3D MALDI-MSI, and data processing in human MDA-MB-231-HRE-tdTomato breast tumor xenografts.18,34,35 MALDI-MSI was performed using the 10 μm sections on ITO slides. Prior to MSI analysis, tissue sections were briefly washed by immersion in 70% and 90% ethanol and dried in a vacuum desiccator for 10 min. Trypsin was resuspended in water at a concentration of 0.05 μg/μL, and 5 nL per spot was deposited on the tissue in a 150 μm × 150 μm raster by CHIP-1000 Chemical Printer (Shimadzu, Japan). A solution of α-cyano-4-hydroxycinnamic acid (CHCA) matrix (Fluka, Switzerland) was prepared at a concentration of 10 mg/mL in 1:1 ACN:H2O/0.1% TFA and was applied to the tissue surface by an ImagePrep (Bruker, Germany) application system. Samples were analyzed on a MALDI-Q-TOF (Synapt HDMS, Waters, UK) instrument in time-of-flight (TOF) mode detecting the positive ions in a mass range between m/z 100 and m/z 3000. The MALDI-MS images were acquired with 150 μm × 150 μm spatial resolution. In this study, we have applied a two-step multivariate data analysis method, which consisted of consecutive PCA36 and LDA37 analyses of the hyperspectral 3D MALDI-MSI data obtained from MDA-MB-231-HRE-tdTomato breast tumor xenografts as shown in Figure 1. The identification of m/z peaks in MALDI-MSI was performed on the basis of an accurate mass tag (AMT) proteomics database,38,39 which was obtained from the MDA-MB-231-HRE-tdTomato cell line and dissected normoxic and hypoxic MDA-MB-231-HRE-tdTomato tumor tissues. Details of experimental materials and methods can be found in the Supporting Information.
Figure 1.
Overview of the experimental and data analysis workflow for 3D MALDI-MSI in MDA-MB-231-HRE-tdTomato breast tumor xenografts. The workflow includes sample preparation of breast tumor tissue, 3D MALDI-MSI, and data processing.
RESULTS
Our 3D PCA-LDA analysis of MALDI-MS images of positive ions in the lipid range from m/z 100 to m/z 1000 in four MDA-MB-231-HRE-tdTomato breast tumor xenografts resulted in 31 top-ranked principal components, which accounted for 78% variance in both PCA and LDA analyses.40 The ratio of between category variation to within category variation (B/W) of the first linear discriminant component was 2.261. Analysis of MALDI-MS images in the tryptic peptide range from m/z 1000 to m/z 3000 in four MDA-MB-231-HRE-tdTomato breast tumors resulted in 100 top-ranked principal components that were analyzed in the LDA analysis. The B/W ratio of the first linear discriminant component was 3.651.
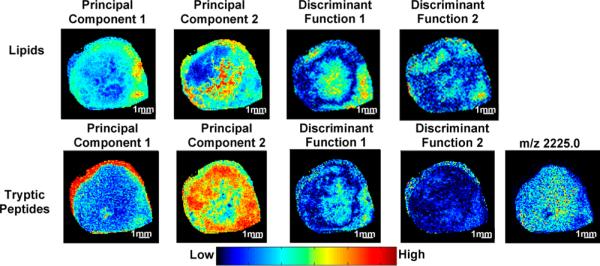
Our data analysis approach captured the overall spatial molecular heterogeneity as well as distinct regions in these breast tumor models. Figure 2 displays PCA and LDA component 1 and 2 reconstructed images for the lipid and tryptic peptide ranges, which visualize the heterogeneous distribution of these biomolecules within this representative MDA-MB-231-HRE-tdTomato breast tumor. For example, PCA component 2 and LDA component 1 reconstructed images (Figure 2, top panel) show distinct biomolecular distributions in the central area of the tumor, where hypoxic regions can be found in this representative tumor as is evident from the distribution of the tdTomato tryptic peptide at m/z 2225.0.
Figure 2.
Display of PC1, PC2, DA1, and DA2 score images of a representative MDA-MB-231-HRE-tdTomato breast tumor xenograft. The top row shows examples from the lipid range of m/z 100 to m/z 1000, and the bottom row shows examples from the tryptic peptide range of m/z 1000 to m/z 3000, as well as the corresponding tdTomato tryptic peptide image at m/z 2225.0.
By combining the analysis results from all four MDA-MB-231-HRE-tdTomato tumors, we obtained 237 common m/z peaks from tryptic peptides that were increased in hypoxic regions of the 3D MALDI-MSI data. The LDA loading ranks from Table 1 represent the loading abundance in the LDA loading spectra from all four tumors. These m/z peaks were identified using our AMT-database with the requirement that 2 or more tryptic peptides per protein needed to be found in the 3D MALDI-MSI data sets and AMT-database, which resulted in identification of 110 proteins as listed in Table S1, Supporting Information. The top 10 of these hypoxia-regulated proteins are listed in Table 1, including plectin (PLEC) involved in the organization of extracellular matrix, deoxyuridine 5′-triphosphate nucleotidohydrolase (DUT) an enzyme involved in nucleotide metabolism, and glucose metabolism related proteins such as triosephosphate isomerase (TPIS) and LDHA, as well as Von Hippel-Lindau-binding protein 1 (PFD3), which in complex with Von Hippel-Lindau (VHL) protein can translocate from the cytoplasm to the nucleus. The VHL protein acts as target recruitment subunit in the E3 ubiquitin ligase complex which recruits hydroxylated HIF-1α under normoxic conditions.41 Some of these proteins, such as ENOA and hypoxia up-regulated protein 1 (HYOU1), were previously reported to be regulated by tumor hypoxia (Table S1, Supporting Information).42,43 ENOA is a multifunctional enzyme that, in addition to its role in glycolysis, participates in growth control, hypoxia tolerance, and allergic responses.44 HYOU1 has a pivotal role in cytoprotective cellular mechanisms triggered by oxygen deprivation and is highly expressed in macrophages within aortic atherosclerotic plaques and in breast cancers.42 Twelve hypoxia-regulated proteins that we identified are associated with glycolysis and glucose metabolism, such as ENOA and fructose-bisphosphate aldolase A (ALDOA), glyceraldehyde-3-phosphate dehydrogenase (G3P), malate dehydrogenase, mitochondrial (MDHM), l-lactate dehydrogenase A (LDHA), and phosphoglycerate mutase 1 (PGAM1), as well as the subunit of the cytochrome c oxidase (COX41) and cytochrome b-c1 complex subunit 1 (QCR1) and protein disulfide-isomerase (PDIA1) involved in cell redox homeostasis (Table S1, Supporting Information). We have also detected extracellular matrix modifying proteins such as cathepsin (CATD) and collagen-binding protein (SERPH) to be up-regulated by hypoxia in breast tumors (Table S1, Supporting Information). Some hypoxia-regulated proteins that we identified are associated with the PI3K/Akt/mTOR signaling pathway, such as peptidyl-prolyl cis–trans isomerase A (PPIA) (Table S1, Supporting Information), 14-3-3 protein beta/alpha (1433B). We also identified ATP dependent RNA helicase (DDX3X) to be up-regulated by hypoxia, which was reported to be directly modulated by HIF-1α in breast epithelial cells.45 In addition, we identified several proteins in our study that have not previously been reported to be up-regulated by hypoxia, such as coronin (COR1B), ATP dependent RNA helicase (DDX17), von Hippel-Lindau-binding protein 1 (VBP1), membrane-organizing extension spike protein (MOES), radixin (RADI), and T-complex protein 1 subunit beta (TCPB) (Table S1, Supporting Information).
Table 1.
List of the Top 10 Proteins That Have Been Identified from the 3D MALDI-MSI Data by 3D PCA-LDA, Ranging from m/z 1000 to m/z 3000 in Four MDA-MB-231-HRE-tdTomato Breast Tumor Xenografts
| loading rank |
description | m/z | peptide squence | ion abundance in high tdTomato regions (mean ± standard errors) |
ion abundance in low tdTomato regions (mean ± standard errors) |
ratio of ion abundance in high tdTomato regions to low tdTomato regions |
|---|---|---|---|---|---|---|
| 1 | Plectin (PLEC_HUMAN) | 1066.56393 | R.QLAEAHAQAKA | 1006.26 ± 273.78 | 112.05 ± 1.97 | 8.98 |
| 38 | 1100.56941 | R.GGAEGELQALR.A | 749.96 ± 220.47 | 97.66 ± 5.02 | 7.68 | |
| 44 | 1046.51123 | K.SLAAEEEAAR.Q. | 750.13 ± 216.48 | 103.03 ± 3.24 | 7.28 | |
| 49 | 1101.56465 | R.QLAEGTAQ.QR.L | 740.79 ± 223.35 | 94.85 ± 5.81 | 7.81 | |
| 61 | 1062.52477 | R.LQLEACETR.T | 732.77 ± 207.58 | 101.08 ± 4.07 | 7.25 | |
| 125 | 1160.55415 | R.SDEGQLSPATR.G | 706.56 ± 204.69 | 98.21 ± 5.51 | 7.19 | |
| 142 | 1286.6851 | R.WQAVLAQTDVR.Q | 721.06 ± 193.92 | 98.70 ± 3.38 | 7.31 | |
| 154 | 1015.55303 | R.LSVAAQEAAR.L | 779.47 ± 199.95 | 112.79 ± 4.50 | 6.91 | |
| 180 | 1287.69024 | KAQVEQELTTLR.L | 713.51 ± 194.42 | 96.51 ± 4.49 | 7.39 | |
| 3 | deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial (DUT_HUMAN) | 1082.55884 | R.LSEHATAPTR.G | 893.43 ± 237.87 | 103.21 ± 2.03 | 8.66 |
| 54 | 1284.68405 | R.ARPAEVGGMQLR.F | 726.66 ± 193.50 | 102.99 ± 1.39 | 7.06 | |
| 3 | 40S ribosomal protein S2 (RS2_HUMAN) | 1082.52985 | R.GCTATLGNFAK.A | 893.43 ± 237.87 | 103.21 ± 2.03 | 8.66 |
| 141 | 1025.59892 | R.GTGIVSAPVPK.K | 746.72 ± 218.44 | 105.12 ± 3.78 | 7.10 | |
| 3 | triosephosphate isomerase (TPIS_HUMAN) | 1082.57813 | R.KFFVGGNWK.M | 893.43 ± 237.87 | 103.21 ± 2.03 | 8.66 |
| 84 | 1294.63854 | A.TPQ.QAQEVHEK.LK.VAHALAEGLGVIACIGEKLDERE | 739.23 ± 202.20 | 98.18 ± 3.25 | 7.53 | |
| 182 | 2992.57705 | AGITEK.V | 293.67 ± 79.22 | 48.92 ± 5.45 | 6.00 | |
| 200 | 1269.65069 | R.IIYGGSVTGATCK.E | 721.40 ± 201.32 | 96.34 ± 3.68 | 7.49 | |
| 5 | plasminogen activator inhibitor 1 RNA- binding protein (PAIRB_HUMAN) | 1255.63887 | R.RPDQQLQGEGK.I | 755.45 ± 191.67 | 104.81 ± 1.51 | 7.21 |
| 237 | 1460.70873 | K.SAAQAAAQTNSNAAGK.Q | 781.81 ± 200.12 | 123.57 ± 14.24 | 6.33 | |
| 6 | protein S100-A11 (S10AB_HUMAN) | 1019.50033 | K.ISSPTETER.C | 726.01 ± 200.71 | 107.21 ± 2.45 | 6.77 |
| 80 | 1141.59595 | K.NQKDPGVLDR.M | 790.57 ± 208.05 | 99.82 ± 3.43 | 7.92 | |
| 7 | ATP-dependent RNA helicase (DDX17 HUMAN,DDX18_HUMAN,DDX3X_HUMAN,DDX3Y_HUMAN,DDX5_HUM) | 1283.64118 | R.MLDMGFEPQIR.K | 760.57 ± 193.62 | 107.23 ± 1.84 | 7.09 |
| 86 | 1336.63874 | R.QTMLFSATQTR.K | 702.61 ± 195.82 | 97.25 ± 3.30 | 7.22 | |
| 164 | 1252.62798 | R.TAQEVETYRR.S | 716.36 ± 195.50 | 100.47 ± 3.73 | 7.13 | |
| 7 | L-lactate dehydrogenase A chain (LDHA_HUMAN) | 1283.62256 | V.SGKDYNVTANSK.L | 760.57 ± 193.62 | 107.23 ± 1.84 | 7.09 |
| 65 | 1071.54036 | R.FRYLMGER.L | 748.54 ± 232.27 | 95.23 ± 5.14 | 7.86 | |
| 212 | 1495.77502 | K.IVSGKDYNVTANSK.L | 754.60 ± 202.60 | 101.29 ± 0.99 | 7.45 | |
| 8 | Von Hippel-Lindau-binding protein 1 (PFD3_HUMAN) | 1051.5418 | K.KLDEQYQK.Y | 814.67 ± 249.20 | 102.53 ± 3.17 | 7.95 |
| 15 | 1310.63682 | K.KKESTNSMETR.F | 725.23 ± 190.66 | 100.76 ± 0.59 | 7.20 | |
| 9 | Keratin, type II cytoskeletal 8 (K2C8_HUMAN) | 1067.52279 | S.RSYTSGPGSR.I | 924.64 ± 265.22 | 108.79 ± 1.07 | 8.50 |
| 52 | 1081.56359 | K.SYKVSTSGPR.A | 803.44 ± 232.35 | 98.58 ± 6.10 | 8.15 | |
| 66 | 1060.56326 | R.KLLEGEESR.I | 775.70 ± 206.40 | 97.91 ± 4.92 | 7.92 |
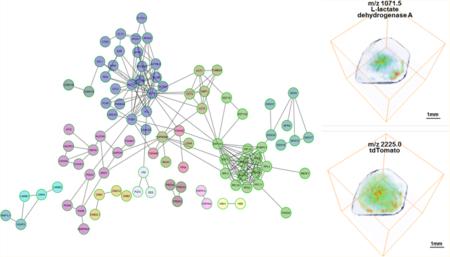
The discovered hypoxia-regulated proteins from Table 1 were analyzed with the protein–protein interaction database Reactome (http://www.reactome.org/) to generate a functional protein interaction network, which clusters the discovered proteins into distinct biological pathways as displayed in Figure 3. Hypoxia-regulated proteins clustered in several distinct pathways such as glucose metabolism, regulation of actin cytoskeleton, protein folding, translation/ribosome, spliceosome, the PI3K-Akt signaling pathway, hemoglobin chaperone, protein processing in endoplasmic reticulum, detoxification of reactive oxygen species, aurora B signaling/apoptotic execution phase, the RAS signaling pathway, the FAS signaling pathway/caspase cascade in apoptosis, and telomere stress induced senescence.
Figure 3.
Functional protein–protein interaction network of the hypoxia-up-regulated proteins that were identified by our 3D MALDI-MSI analysis of the MDA-MB-231-HRE-tdTomato breast tumor xenograft model. The hypoxia-up-regulated proteins are also listed in Table S1, Supporting Information. The Reactome Database was used to generate this network display (http://www.reactome.org/) and clusters the discovered proteins into distinct biological pathways that were up-regulated in hypoxic regions in these breast tumors.
Similarly, we generated a lipid list for the range of m/z 100 to m/z 1000 from all four MDA-MB-231-HRE-tdTomato tumors. The LDA loading ranks from Table 2 represent the loading abundance in the LDA loading spectra from all four tumors. Some m/z candidates were structurally identified by MS/MS analysis. Since most of the metabolites and small peptides were not structurally verified by MS/MS analysis, we did not list them in the table. However, the distribution of several m/z values in the metabolite and small peptide range such as phosphocholine (PCho) at m/z 184.1 significantly differentiated hypoxic and normoxic regions as is evident from Figure S2, Supporting Information. A total of 34 hypoxia-regulated lipid-related ions were identified from four tumors as listed in Table S2, Supporting Information. Table 2 lists the top 20 of these final hypoxia-regulated lipids, which include distinct phosphatidylcholine (PC) species such as PC(16:0/18:1) at m/z 760.5 [M + H]+, PC(18:1/18:1) at m/z 786.5 [M + H]+, and PC(16:1/18:4) at m/z 790.4 [M+K]+, as well as sphingomyelin (SM) SM(d18:1/24:0) at m/z 837.5 [M + Na]+, and other lipids species (Table 2). The average number of fatty acid double bonds per PC molecule was about 1.7 in both hypoxic and normoxic regions (Table 2). Hence, there was no difference in the degree of saturation in the PC species and all other lipids when comparing hypoxic with normoxic regions in MDA-MB-231-HRE-tdTomato tumors.
Table 2.
List of the Top 20 Lipids That Have Been Identified from 3D MALDI MSI Data by 3D PCA-LDA, Ranging from m/z 100 to m/z 1000 in Four MDA-MB-231-HRE-tdTomato Breast Tumor Xenografts
| loading rank | m/z | ID | ion | ion abundance in high tdTomato regions (mean ± standard error) | ion abundance in low tdTomato regions (mean ± standard error) | ratio of ion abundance in high tdTomato regions to low tdTomatos regions |
|---|---|---|---|---|---|---|
| 1 | 786.5 | PC 18:1/18:1 | [M + H]+ | 1689.7 ± 261.71 | 577.19 ± 249.28 | 2.93 |
| 2 | 760.5 | PC 16:0/18:1 | [M + H]+ | 1888.14 ± 249.59 | 630.23 ± 273.25 | 3.00 |
| 3 | 798.5 | PC 16:0/18:1 | [M + K]+ | 1131.06 ± 283.07 | 406.36 ± 223.62 | 2.78 |
| 4 | 788.5 | PC 18:0/18:1 | [M + H]+ | 1536.64 ± 202.72 | 514.16 ± 223.42 | 2.99 |
| 5 | 810.5 | PC 18:0/18:1 | [M + Na]+ | 1306.4 ± 274.33 | 477.4 ± 242.67 | 2.74 |
| 6 | 824.5 | PC 18:1/18:1 | [M + K]+ | 1052.65 ± 302.21 | 402.23 ± 230.59 | 2.62 |
| 7 | 782.5 | PC 16:0/18:1 | [M + Na]+ | 1276.73 ± 312.53 | 496.08 ± 288.39 | 2.57 |
| 8 | 826.5 | PC 18:0/18:1 | [M + K]+ | 1046.28 ± 336.34 | 475.48 ± 316.11 | 2.20 |
| 9 | 761.4 | isotope of m/z 760.5 | - | 1313.26 ± 232.81 | 454.42 ± 224.22 | 2.89 |
| 10 | 796.5 | PC 16:0/18:2 | [M + K]+ | 1014.73 ± 259.08 | 365.78 ± 203.87 | 2.77 |
| 11 | 787.4 | isotope of m/z 786.5 | - | 1297.53 ± 232.96 | 450.43 ± 217.51 | 2.88 |
| 12 | 789.4 | isotope of m/z 788.5 | - | 1161.54 ± 235.28 | 403.91 ± 202.59 | 2.88 |
| 13 | 808.5 | PC 18:1/18:1 | [M + Na]+ | 1099.06 ± 308.28 | 423.54 ± 246.14 | 2.59 |
| 14 | 811.5 | PI 10:0/22:0 | [M + H]+ | 1168.07 ± 255.77 | 419.46 ± 231.27 | 2.78 |
| 15 | 767.4 | lipid | unknown | 845.76 ± 274.35 | 311.68 ± 188.17 | 2.71 |
| 16 | 809.5 | lipid | unknown | 997.64 ± 293.27 | 377.82 ± 229.82 | 2.64 |
| 17 | 763.4 | isotope of m/z 808.5 | [M + Na]+ | 1100.46 ± 235.03 | 388.01 ± 200.49 | 2.84 |
| 18 | 790.4 | PC 16:1/18:4 | [M + K]+ | 980.94 ± 256.92 | 355.47 ± 197.5 | 2.76 |
| 19 | 801.4 | PC 16:0/18:0 | [M + K]+ | 888.25 ± 310.67 | 359.87 ± 235.78 | 2.47 |
| 20 | 837.5 | SM d18:1/24:0 | [M + Na]+ | 1029.89 ± 301.97 | 410.84 ± 267.38 | 2.51 |
Further validation of spatial colocalization between lipid and tryptic peptide m/z peaks and tdTomato at m/z 2225.0 was demonstrated in the Supporting Information in Figures S2, S3, and S4. The identified hypoxia-up-regulated lipids and proteins can be displayed in 2D and 3D to visualize their colocalization with hypoxic tumor regions, which were identified by increased tdTomato expression in this MDA-MB-231-HRE-tdTomato breast tumor xenograft model. To this end, representative biomolecular MALDI-MS images are displayed in both 2D (Figure 4A) and 3D (Figure 4B). Figure 4 clearly demonstrates that PS(14:0/22:6) at m/z 818.4 and a tryptic peptide of hypoxia up-regulated protein (HYOU1) at m/z 1047.4, as well as LDHA at m/z 1071.5, colocalized with hypoxic regions, in which high levels of a tdTomato tryptic peptide at m/z 2225.0 were detected in this breast tumor xenograft model. The m/z values of all identified hypoxia-up-regulated lipids and proteins, including the lipids and tryptic peptides presented as biomolecular images, are listed in Tables S1 and S2, Supporting Information.
Figure 4.
Display of representative biomolecular MALDI-MS images in (A) 2D and (B) 3D. PS(14:0/22:6) at m/z 818.4 and a tryptic peptide of hypoxia up-regulated protein (HYOU1) at m/z 1047.4, as well as LDHA at m/z 1071.5, colocalized with hypoxic regions, in which high levels of a tdTomato tryptic peptide at m/z 2225.0 were detected in MDA-MB-231-HRE-tdTomato breast tumor xenografts. This was evident in the 2D as well as the 3D display. The entire list of m/z values of hypoxia-up-regulated lipids and proteins, including all detected tryptic peptides per protein, is given in Tables S1 and S2, Supporting Information, respectively.
DISCUSSION
In this study, we have performed multivariate analysis of MALDI-MSI voxels in 3D that were pooled from an entire breast tumor volume to ensure that the lipids, peptides, and proteins that we have identified through this analysis in hypoxic and normoxic tumor regions are statistically and biologically meaningful. Such an approach is demanded by the vast heterogeneity of the biomolecular distribution inside breast tumors, which makes more extensive imaging technology, especially the extension from 2D to 3D, necessary in cancer research. MALDI-MSI technology provides a comprehensive way to discover and localize biomarkers in breast tumors. We have extended our previous studies and have applied our recently developed methods to mine for hypoxia-related lipids, peptides, and proteins in 3D MALDI-MSI volume data from MDA-MB-231-HRE-tdTomato breast tumor xenografts.
The MALDI-MSI detection of hypoxic regions in the HRE-tdTomato tumor model11,17,22 was possible as a result of the on-tissue digestion and MALDI-MSI detection of the fluorescent tdTomato protein, which gives rise to an abundant tryptic peptide at m/z 2225.0 in MALDI-MSI spectra.21 We have prelabeled all voxels as either low-tdTomato or high-tdTomato voxels based on their tdTomato tryptic peptide abundance. Instead of using the tdTomato tryptic peptide peak at m/z 2225.0 as a prelabeling peak to discriminate between hypoxic and normoxic voxels, any other detected mass peak that is able to discriminate between hypoxic and normoxic tumor regions could be used. Of course, it is also possible to select mass spectral peaks that discriminate other tumor microenvironmental regions such as acidic, stromal, or necrotic regions among many others depending on the focus of study. Supervised labeling and annotating MALDI-MSI images is time-consuming and expensive. As a result, it is desirable to reduce the number of required labels without compromising classification accuracy. Semisupervised learning techniques46 and active learning (AL) strategies47 have been studied for annotation and classification of hyperspectral MSI data and image segmentation.48 A nonlinear multivariate discriminant analysis method, such as the kernel method,49 may also be applied to the feature extraction and classification of 3D MALDI-MSI data, which we will test in future studies. One of the assumptions of PCA-LDA analysis is that the distribution of the data follows a normal distribution. However, our MALDI-MSI data is skewed and not normally distributed, so that in this study we performed a preprocessing step using logarithmic transformation to eliminate skewed characteristics of the data in order to approximate the data's distribution to a normal distribution. Furthermore, other generalized linear regression models with loose assumptions of data distribution may be attempted to improve the data analysis accuracy in future studies employing 3D MALDI-MSI.
We identified 12 hypoxia-regulated proteins that are associated with glycolysis and glucose metabolism. In gluconeogenesis, phosphoenolpyruvate (PEP) is reduced to fructose 1,6-bisphosphate with ALDOA catalyzing the last reaction.50 In glycolysis, fructose 1,6-bisphosphate is oxidized to PEP with ALDOA catalyzing the first reaction.50 LDHA catalyzes the conversion of l-lactate and NAD+ to pyruvate and NADH in the final step of anaerobic glycolysis.51 PGAM1 is a glycolytic enzyme that catalyzes the internal transfer of a phosphate group from C-3 to C-2, which results in the conversion of 3-phosphoglycerate (3PG) to 2-phosphoglycerate (2PG) through a 2,3-bisphosphoglycerate intermediate.52 Several of the hypoxia-regulated glycolytic proteins that we identified are regulated by HIF-1, such as ALDOA, LDHA, and G3P.5 G3P is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting d-glyceraldehyde 3-phosphate into 3-phospho-d-glyceroyl phosphate.53 With our 3D MALDI-MSI approach, we have also detected extracellular matrix modifying proteins such as CATD and SERPH to be up-regulated by hypoxia in breast tumors. CATD was detected as an extracellular protein loosely bound to the extracellular-matrix in breast cancer.54 SERPH, localized to the endoplasmic reticulum, binds specifically to collagen and could be involved as a chaperone in the biosynthetic pathway of collagen in breast cancer.55 Some hypoxia-regulated proteins that we identified belong to PPIA, which is associated with the PI3K/Akt/mTOR signaling pathway. A recent finding demonstrated that hypoxia can suppress mammalian TORC1 (mTORC1) activity by releasing TSC2 from its growth factor-induced association with inhibitory 14-3-3 proteins.56 Hypoxia is a common finding in advanced human tumors and is often associated with metastatic dissemination and poor prognosis.1–3 Cancer cells adapt to hypoxia by utilizing physiological adaptation pathways that promote a switch from oxidative to glycolytic metabolism and an activation of different signaling pathways, such as PI3K/Akt/mTOR signaling pathway, RAS signaling pathway, and the FAS signaling pathway.57–59
Not only breast cancer cells but also stromal cells such as fibroblasts, endothelial cells, and macrophages exist in the hypoxic regions of the MDA-MB-231 breast tumor model.60 This heterogeneous mixture of different cell types within hypoxic tumor regions makes our study difficult to compare with homogeneous cell culture experiments in vitro. Our study is in good agreement with a recent study that identified three common hypoxia-regulated proteins using MALDI-MSI combined with quantitative proteomics in breast cancer, including galectin-1 (LEG1), cytoplasmic 1 actin (ACTB), and one hypoxia down-regulated protein histone H2B type 1-M (HIST1H2BM).61 Our findings presented in this study revealed several hypoxia up-regulated proteins in solid tumors that were also identified in cell culture studies of hypoxia followed by quantitative proteomics, such as 1433B, ALDOA, PGAM1, LDHA, G3P, ENOA, ANXA5, cofilin-1 (COF1), clathrin heavy chain 1 (CLH1), elongation factor 1-alpha 1 (EF1A1), prelamin-A/C (LMNA), lamin-B1 (LMNB1), mitochondrial malate dehydrogenase (MDHM), and triose-phosphate isomerase (TPIS).61
Small metabolites are also detected by MALDI-MSI. Here, we revealed several metabolite peaks that have a high LDA loading rank in the analytical results, for example, PCho at m/z 184.1. In our previous studies,17,62 we illustrated the spatial colocalization between hypoxia and PCho at m/z 184.1 by multimodal molecular imaging of MDA-MB-231-HRE-tdTomato breast tumor xenografts. This multimodal molecular imaging comprises ex vivo secondary ion mass spectrometry (SIMS) imaging and fluorescence optical imaging technology. Additional studies of all other ions detected below m/z 1000 may further elucidate the molecular mechanism underlying breast cancer hypoxia.
We identified strong positive correlations between PC species, such as PC(16:0/18:1), PC(18:1/18:1), and PC-(18:0/18:1), as well as SM(d18:1/24:0),63 and the tdTomato tryptic peptide at m/z 2225.0 in hypoxic regions, suggesting that the PC species listed in Table 2 are increased in the hypoxic regions of MDA-MB-231-HRE-tdTomato breast tumor xenografts. However, no difference was observed in the degree of fatty acid saturation in these PC species when comparing hypoxic with normoxic regions. In one of our previous studies, which was performed in 2D on the central tumor sections only, we analyzed the lipid composition in hypoxic breast tumor regions compared with that in normoxic tumor regions and found that the concentrations of PC(16:0/18:1), PC(18:1/18:1), and PC(18:0/18:1) were higher in hypoxic breast tumor regions compared to normoxic regions.22 These same PC species were also identified in the hypoxic breast tumor regions of all four tumors in the presented 3D MALDI-MSI analysis. Other to date unidentified lipid species were also increased in hypoxic breast tumor regions compared to normoxic regions (see Table S2, Supporting Information). In this study, we have for the first time utilized multivariate methods applied to the analysis of the entire 3D breast tumor volume to systematically analyze the lipid distributions in hypoxic tumor regions compared with normoxic tumor regions. The effect of hypoxia in isolated hamster hearts and cardiac myocytes in culture decreased overall phospholipid biosynthesis,64,65 which was mainly caused by a decrease in high-energy nucleotide levels due to hypoxia,64 and also attributed to phospholipid degradation by phospholipase C.65 Since cancer cells are well-known to be able to maintain unaltered high-energy nucleotide levels under hypoxic conditions due to their strong reliance on glycolysis,66 it is not surprising that we detected an overall increase in several phospholipid species in hypoxic breast tumor regions.
Using 3D MALDI-MSI analysis based on tdTomato-voxel classification of hypoxic regions, we identified specific PC and SM species that are either broken down in normoxic regions or experience increased biosynthesis in hypoxic regions of this breast tumor model. We also identified specific proteins that are up-regulated in hypoxic regions of breast tumors. Our findings provide the breast cancer research community with a comprehensive analysis of hypoxia triggered lipidomic and proteomic changes in solid breast tumors. The obtained lists of lipids and proteins may be translated into the clinic for further validation as potential biomarkers of hypoxia, similar to recent clinical studies,67,68 and they may provide a potential therapeutic target for the treatment of hypoxic breast cancers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Paul T. Winnard Jr. for help with generating the MDA-MB-231-HRE-tdTomato cell line. We thank Ms. Menglin Cheng for technical laboratory support. This work was supported by the National Institutes of Health (NIH) grants R01 CA134695 and R01 CA154725 and by The Netherlands Organization for Scientific Research (NWO) program of the “Foundation for Fundamental Research on Matter (FOM)”. A portion of this work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at the Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
Additional information as noted in the text. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/ac504503x.
REFERENCES
- 1.Hockel M, Vaupel PJ. Natl. Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 2.Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KS, Chapman JD, Eckelman WC, Fyles AW, Giaccia AJ, Hill RP, Koch CJ, Krishna MC, Krohn KA, Lewis JS, Mason RP, Melillo G, Padhani AR, Powis G, Rajendran JG, Reba R, Robinson SP, Semenza GL, Swartz HM, Vaupel P, Yang D, Croft B, Hoffman J, Liu G, Stone H, Sullivan D. Int. J. Radiat. Biol. 2006;82:699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]
- 3.Williams KJ, Cowen RL, Stratford IJ. Breast Cancer Res. 2001;3:328–331. doi: 10.1186/bcr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng QW, Dillehay LE, Madan A, Semenza GL, Bedi A. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 5.Semenza GL. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 7.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Mol. Cell. Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JTA, Jeffrey SS, Giaccia AJ. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 9.Liang ZX, Yoon YH, Votaw J, Goodman MM, Williams L, Shim H. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita T, Ohneda K, Nagano M, Miyoshi C, Kaneko N, Miwa Y, Yamamoto M, Ohneda O, Fujii-Kuriyama YJ. Biol. Chem. 2008;283:18926–18936. doi: 10.1074/jbc.M709133200. [DOI] [PubMed] [Google Scholar]
- 11.Krishnamachary B, Penet MF, Nimmagadda S, Mironchik Y, Raman V, Solaiyappan M, Semenza GL, Pomper MG, Bhujwalla ZM. PLoS One. 2012;7:e44078. doi: 10.1371/journal.pone.0044078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iakovlev VV, Pintilie M, Morrison A, Fyles AW, Hill RP, Hedley DW. Lab. Invest. 2007;87:1206–1217. doi: 10.1038/labinvest.3700680. [DOI] [PubMed] [Google Scholar]
- 13.Chi JT, Wang Z, Nuyten DSA, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Borresen-Dale AL, Giaccia A, Longaker MT, Hastie T, Yang GP, van de Vijver MJ, Brown PO. PLoS Med. 2006;3:395–409. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buffa FM, Harris AL, West CM, Miller CJ. Br. J. Cancer. 2010;103:1136–1136. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui YZ, Zhang DL, Jia Q, Li TL, Zhang WD, Han JX. Cancer Invest. 2009;27:747–755. doi: 10.1080/07357900802672746. [DOI] [PubMed] [Google Scholar]
- 16.Glunde K, Shah T, Winnard PT, Jr., Raman V, Takagi T, Vesuna F, Artemov D, Bhujwalla ZM. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, Greenwood TR, Artemov D, Raman V, Winnard PT, Jr., Heeren RM, Bhujwalla ZM, Glunde K. Neoplasia. 2012;14:732–741. doi: 10.1593/neo.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L, Greenwood TR, van Hove ERA, Chughtai K, Raman V, Winnard PT, Heeren RMA, Artemov D, Glunde K. NMR Biomed. 2013;26:285–298. doi: 10.1002/nbm.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chughtai K, Heeren RM. Chem. Rev. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calligaris D, Caragacianu D, Liu XH, Norton I, Thompson CJ, Richardson AL, Golshan M, Easterling ML, Santagata S, Dillon DA, Jolesz FA, Agar NY. R. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chughtai K, Jiang L, Post H, Winnard PT, Jr., Greenwood TR, Raman V, Bhujwalla ZM, Heeren RM, Glunde K. J. Am. Soc. Mass Spectrom. 2013;24:711–717. doi: 10.1007/s13361-012-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chughtai K, Jiang L, Greenwood TR, Glunde K, Heeren RM. J. Lipid Res. 2012;54:333–344. doi: 10.1194/jlr.M027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Nat. Methods. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 24.Chughtai K, Jiang L, Greenwood TR, Klinkert I, Amstalden van Hove ER, Heeren RM, Glunde K. Anal. Chem. 2012;84:1817–1823. doi: 10.1021/ac203373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klerk LA, Broersen A, Fletcher IW, van Liere R, Heeren RMA. Int. J. Mass Spectrom. 2007;260:222–236. [Google Scholar]
- 26.Eijkel GB, Kaletas BK, van der Wiel IM, Kros JM, Luider TM, Heeren RMA. Surf. Interface Anal. 2009;41:675–685. [Google Scholar]
- 27.Dill AL, Eberlin LS, Zheng C, Costa AB, Ifa DR, Cheng L, Masterson TA, Koch MO, Vitek O, Cooks RG. Anal. Bioanal. Chem. 2010;398:2969–2978. doi: 10.1007/s00216-010-4259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, Cooks RG, Agar NY. Cancer Res. 2012;72:645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas A, Patterson NH, Marcinkiewicz MM, Lazaris A, Metrakos P, Chaurand P. Anal. Chem. 2013;85:2860–2866. doi: 10.1021/ac3034294. [DOI] [PubMed] [Google Scholar]
- 30.Djidja MC, Claude E, Snel MF, Francese S, Scriven P, Carolan V, Clench MR. Anal. Bioanal. Chem. 2010;397:587–601. doi: 10.1007/s00216-010-3554-6. [DOI] [PubMed] [Google Scholar]
- 31.Trede D, Schiffler S, Becker M, Wirtz S, Steinhorst K, Strehlow J, Aichler M, Kobarg JH, Oetjen J, Dyatlov A, Heldmann S, Walch A, Thiele H, Maass P, Alexandrov T. Anal. Chem. 2012;84:6079–6087. doi: 10.1021/ac300673y. [DOI] [PubMed] [Google Scholar]
- 32.Alexandrov T. BMC Bioinf. 2012;13(Suppl 16):S11. doi: 10.1186/1471-2105-13-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. J. Proteomics. 2012;75:4962–4989. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Cailleau R, Young R, Olive M, Reeves WJ., Jr. J. Natl. Cancer Inst. 1974;53:661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glunde K, Jie C, Bhujwalla ZM. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 36.Wold S, Esbensen K, Geladi P. Chemom. Intell. Lab. Syst. 1987;2:37–52. [Google Scholar]
- 37.Yu H, Yang H. Pattern Recognit. 2001;34:2067–2070. [Google Scholar]
- 38.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. J. Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 39.Schober Y, Schramm T, Spengler B, Rompp A. Rapid Commun. Mass Spectrom. 2011;25:2475–2483. doi: 10.1002/rcm.5135. [DOI] [PubMed] [Google Scholar]
- 40.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd ed. Wiley; New York: 2001. [Google Scholar]
- 41.Tanimoto K, Makino Y, Pereira T, Poellinger L. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozawa K, Kuwabara K, Tamatani M, Takatsuji K, Tsukamoto Y, Kaneda S, Yanagi H, Stern DM, Eguchi Y, Tsujimoto Y, Ogawa S, Tohyama M. J. Biol. Chem. 1999;274:6397–6404. doi: 10.1074/jbc.274.10.6397. [DOI] [PubMed] [Google Scholar]
- 43.Sedoris KC, Thomas SD, Miller DM. BMC Cancer. 2010;10:157. doi: 10.1186/1471-2407-10-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray R, Miller DM. Mol. Cell. Biol. 1991;11:2154–2161. doi: 10.1128/mcb.11.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Botlagunta M, Krishnamachary B, Vesuna F, Winnard PT, Jr., Bol GM, Patel AH, Raman V. PLoS One. 2011;6:e17563. doi: 10.1371/journal.pone.0017563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruand J, Alexandrov T, Sistla S, Wisztorski M, Meriaux C, Becker M, Salzet M, Fournier I, Macagno E, Bafna V. J. Proteome Res. 2011;10:4734–4743. doi: 10.1021/pr2005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanselmann M, Roder J, Kothe U, Renard BY, Heeren RM, Hamprecht FA. Anal. Chem. 2013;85:147–155. doi: 10.1021/ac3023313. [DOI] [PubMed] [Google Scholar]
- 48.Hanselmann M, Kothe U, Kirchner M, Renard BY, Amstalden ER, Glunde K, Heeren RMA, Hamprecht FA. J. Proteome Res. 2009;8:3558–3567. doi: 10.1021/pr900253y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shawe-Taylor J, Cristianini N. Kernel Methods for Pattern Analysis. Cambridge University Press; New York: 2004. [Google Scholar]
- 50.Marsh JJ, Lebherz HG. Trends Biochem. Sci. 1992;17:110–113. doi: 10.1016/0968-0004(92)90247-7. [DOI] [PubMed] [Google Scholar]
- 51.Chung FZ, Tsujibo H, Bhattacharyya U, Sharief FS, Li SSL. Biochem. J. 1985;231:537–541. doi: 10.1042/bj2310537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Dworkowski FSN, Cook PF. J. Biol. Chem. 2006;281:25568–25576. doi: 10.1074/jbc.M601154200. [DOI] [PubMed] [Google Scholar]
- 53.Semenza GL. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 54.Tandon AK, Clark GM, Chamness GC, Chirgwin JM, Mcguire WL. N. Engl. J. Med. 1990;322:297–302. doi: 10.1056/NEJM199002013220504. [DOI] [PubMed] [Google Scholar]
- 55.Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA, Nebert DW, Vasiliou V. Hum. Genomics. 2013;7:22. doi: 10.1186/1479-7364-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma V, Dixit D, Koul N, Mehta VS, Sen E. J. Mol. Med. 2011;89:123–136. doi: 10.1007/s00109-010-0683-5. [DOI] [PubMed] [Google Scholar]
- 58.Menendez JA, Lupu R. Arch. Immunol. Ther. Exp. 2004;52:414–426. [PubMed] [Google Scholar]
- 59.Agani F, Jiang BH. Curr. Cancer Drug Targets. 2013;13:245–251. doi: 10.2174/1568009611313030003. [DOI] [PubMed] [Google Scholar]
- 60.Radisky D. Cold Spring Harbor Perspect. Biol. 2012 doi: 10.1101/cshperspect.a013458. DOI: 10.1101/cshperspect.a013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Djidja MC, Chang J, Hadjiprocopis A, Schmich F, Sinclair J, Mrsnik M, Schoof EM, Barker HE, Linding R, Jorgensen C, Erler JT. J. Proteome Res. 2014;13:2297–2313. doi: 10.1021/pr401056c. [DOI] [PubMed] [Google Scholar]
- 62.van Hove ERA, Blackwell TR, Klinkert I, Eijkel GB, Heeren RMA, Glunde K. Cancer Res. 2010;70:9012–9021. doi: 10.1158/0008-5472.CAN-10-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fahy E, Cotter D, Sud M, Subramaniam S. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2011;1811:637–647. doi: 10.1016/j.bbalip.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong JT, Man RYK, Choy PC. Lipids. 1996;31:1059–1067. doi: 10.1007/BF02522463. [DOI] [PubMed] [Google Scholar]
- 65.Nachas N, Pinson A. FEBS Lett. 1992;298:301–305. doi: 10.1016/0014-5793(92)80081-q. [DOI] [PubMed] [Google Scholar]
- 66.Robey IF, Lien AD, Welsh SJ, Baggett BK, Gillies RJ. Neoplasia. 2005;7:324–330. doi: 10.1593/neo.04430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steurer S, Borkowski C, Odinga S, Buchholz M, Koop C, Huland H, Becker M, Witt M, Trede D, Omidi M, Kraus O, Bahar AS, Seddiqi AS, Singer JM, Kwiatkowski M, Trusch M, Simon R, Wurlitzer M, Minner S, Schlomm T, Sauter G, Schluter H. Int. J. Cancer. 2013;133:920–928. doi: 10.1002/ijc.28080. [DOI] [PubMed] [Google Scholar]
- 68.Steurer S, Seddiqi AS, Singer JM, Bahar AS, Eichelberg C, Rink M, Dahlem R, Huland H, Sauter G, Simon R, Minner S, Burandt E, Stahl PR, Schlomm T, Wurlitzer M, Schluter H. Anticancer Res. 2014;34:2255–2261. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.