Abstract
Aim:
To investigate the antiviral effects of vectors expressing specific short hairpin RNAs (shRNAs) against Hantaan virus (HTNV) infection in vitro and in vivo.
Methods:
Based on the effects of 4 shRNAs targeting different regions of HTNV genomic RNA on viral replication, the most effective RNA interference fragments of the S and M genes were constructed in pSilencer-3.0-H1 vectors, and designated pSilencer-S and pSilencer-M, respectively. The antiviral effect of pSilencer-S/M against HTNV was evaluated in both HTNV-infected Vero-E6 cells and mice.
Results:
In HTNV-infected Vero-E6 cells, pSilencer-S and pSilencer-M targeted the viral nucleocapsid proteins and envelope glycoproteins, respectively, as revealed in the immunofluorescence assay. Transfection with pSilencer-S or pSilencer-M (1, 2, 4 μg) markedly inhibited the viral antigen expression in dose- and time-dependent manners. Transfection with either plasmid (2 μg) significantly decreased HTNV-RNA level at 3 day postinfectin (dpi) and the progeny virus titer at 5 dpi. In mice infected with lethal doses of HTNV, intraperitoneal injection of pSilencer-S or pSilencer-M (30 μg) considerably increased the survival rates and mean time to death, and significantly reduced the mean virus yields and viral RNA level, and alleviated virus-induced pathological lesions in lungs, brains and kidneys.
Conclusion:
Plasmid-based shRNAs potently inhibit HTNV replication in vitro and in vivo. Our results provide a basis for development of shRNA as therapeutics for HTNV infections in humans.
Keywords: Hantaan virus, short hairpin RNA, RNA interference, antiviral therapy
Introduction
Hantaviruses represent one of the important emerging and re-emerging viral groups and have become a global threat to public health in different areas of the world1,2. Hantaviruses lead to two severe febrile diseases worldwide, ie, hemorrhagic fever with renal syndrome (HFRS) in Eurasia due to Old World hantavirus and hantavirus pulmonary syndrome (HPS) in the Americas due to New World hantavirus3. Approximately 60 000–100 000 HFRS cases are reported each year worldwide, and the mortality rate can reach 10%1,4. Hantaviruses belong to the genus Hantavirus of the Bunyaviridae family, and they are enveloped, single-strand, negative-sense, tri-segmented RNA viruses. The viral genomes consist of large (L), medium (M) and small (S) segments that encode an RNA-dependent RNA polymerase and two envelope glycoproteins (Gn and Gc) and a nucleocapsid (N) protein. Variations in the M and S segments may influence the antigenicity and virulence of hantaviruses5,6.
Extensive efforts have been made in the search for effective preventative and treatment measures for hantavirus infections1,7. Ribavirin is a broad-spectrum synthetic nucleoside analog that has been reported to effectively inhibit the replication of the Hantaan virus (HTNV, an Old World hantavirus) and Andes virus (ANDV, a New World hantavirus) in vitro and in vivo8. However, there are currently no prophylactic vaccines or therapeutic antivirals for hantavirus infection that are approved by the FDA. Therefore, the exploration of novel treatment strategies for hantavirus infection is urgent and necessary.
RNA interference (RNAi) is a short, double-strand RNA induced-process that can target the mRNA of a specific sequence for degradation9,10. Post-transcriptional gene silencing can be mediated by endogenous microRNAs (miRNAs), exogenous small interfering RNAs (siRNAs), and short hairpin RNAs (shRNAs). RNAi technology is recognized as a promising, novel nucleic acid-based tool for use against various diseases, including cancer, infectious diseases and genetic disorders. RNAi reagents have also been reported to potently and specifically inhibit the replication of different viruses, including HIV, HBV11, HSV12, SAR-CoV13, influenza virus14,15, coxsackievirus16 and others. Thus, the development of RNAis as possible antiviral agents for hantavirus infection is of considerable interest17.
In the present study, we examined the effects of shRNAs on HTNV replication using 4 shRNAs that targeted different regions of the HTNV genomic RNA. We further transfected the two most effective shRNAs into the shRNA Expression Vector, pSilencer-3.0-H1 and named the resultant vectors pSilencer-S and pSilencer-M. The antiviral effect of pSilencer-S/M against HTNV was ultimately evaluated in tissue culture and a mouse model.
Materials and methods
Cells, virus, and reagents
Vero-E6 cells were obtained from the China Center for Type Culture Collection (CCTCC) and maintained in DMEM supplemented with 10% fetal bovine serum (FBS, GIBCO, Carlsbad, CA, USA), 0.1% L-glutamine, 100 U/mL penicillin and streptomycin. DMEM containing 2% FBS was used to maintain the medium after viral infection. The stocks of Hantaan virus strain 76–118 were obtained from the Institute of Virology of the Chinese Academy of Preventive Medicine (Beijing, China) and were propagated in Vero-E6 cells. HTNV titration was performed regularly based on indirect immunofluorescence assay (IFA) as described previously18. The TCID50 was determined to be 106/0.1 mL.
shRNA design and selection
Two individual targeting sites were selected for the S (GenBank accession number AF187082) and M genes (GenBank accession number AF276987) of HTNV. The design of the shRNA sequences was performed according to methods referred to in the literature19 and involved the use of the web-based Block-iT RNAi Designer program (http://rnaidesigner.thermofisher.com/rnaiexpress/). A BLAST search (http://www.ncbi.nlm.nih.gov/BLAST) was performed to exclude the possible homologous sequences. The sequences and positions of the shRNAs are illustrated in Table 1. The designed structure contained the following sequences: 5′-AA-sense strand siRNA, CTTGCTTC (loop region); and antisense strand siRNA, TATAGTGA (T7-Oligo promoter)-3′. The target gene is presented in Table 1. A negative control was chosen to target positions 1023 to 1043 of Leishmania CRK1, the sequence of which was 5′-AATCGGGCAGTTGTTTGAGAT-3′.
Table 1. The gene sequence and position of RNA interference targeting.
| Sequence | Position | |
|---|---|---|
| shRNA-S1 | AAGGCACTGTACATGTTAACA | Position in S gene sequence: 448 |
| shRNA-S2 | AAGGATGACAGCTCATATGAG | Position in S gene sequence: 520 |
| shRNA-M1 | AAGGTGGTGGTCTAATATTTA | Position in M gene sequence: 614 |
| shRNA-M2 | AAGGACATTGACTTTGATAAC | Position in M gene sequence: 928 |
| shCRK | AATCGGGCAGTTGTTTGAGAT | 1023 to 1043 of Leishmania CRK1 |
The DNA template was synthesized by Sangon Biotech (Shanghai) Co, Ltd, and the shRNA was generated using the MessageMuter™ shRNA production kit (Qiagen) as described previously12. The Vero-E6 cells were grown in 24-well plates to 80%–90% confluence and transfected with specific or control shRNA (60 nmol/L) with RNAiFect transfection reagent (Qiagen, Germantown, MD, USA). At 24 h after transfection, the cells were infected with 100 TCID50 HTNV 76–118 (100 μL per well), and viral RNA was detected by qRT-PCR at 24 or 48 hour post-infection (hpi) to determine the interference efficiency. The viral titers were determined at 96 hpi. The shRNA S1 and shRNA M2 were found to be the most effective shRNAs in terms of the inhibition of HTNV and were selected for use in the subsequent experiments.
Construction of shRNA vectors targeting the S and M genes of HTNV
Synthesized oligonucleotides containing the S1 and M2 shRNA sequences with BamH I or Hind III sequences at the 5′ or 3′ termini were designed as illustrated in Table 2 according to the instructions of the manufacturer of RNAi Designer (Invitrogen, Carlsbad, CA, USA). The two complementary oligonucleotides were annealed and inserted into the BamH I/Hind III site of the pSilencer-3.0-H1 vector. After transformation, clone PCR and enzyme digestion, the identified shRNA expression plasmids were sent for sequence analysis for further confirmation and were subsequently designated pSilencer-S and pSilencer-M.
Table 2. The oligonucleotides inserting into pSilencer-3.0-H1 vector.
| Target | Oligos | Sequences |
|---|---|---|
| S | S Forward | 5′-GATCCGGCACTGTACATGTTAACATTCAAGAGATGTTAACATGTACAGTGCCTTTTTTGGAAA-----3′ |
| S Reverse | 3′-----GCCGTGACATGTACAATTGTAAGTTCTCTACAATTGTACATGTCACGGAAAAAACCTTTTCGA-5′ | |
| M | M Forward | 5′-GATCCGGACATTGACTTTGATAAC TTCAAGAGA GTTATCAAAGTCAATGTCCTTTTTTGGAAA-----3′ |
| M Reverse | 3′-----GCCTGTAACTGAAACTATTG AAGTTCTCT CAATAGTTTCAGTTACAGGAAAAAACCTTTTCGA-5′ |
Antiviral activity in vitro
One day before transfection, the Vero-E6 cells were seeded into 24-well plates at 5×104 cells per well. The 80%-90% confluent cells were then transfected with 0.25, 0.5, 1, 2, or 4 μg of the pSilencer-S, pSilencer-M or pSilencer 3.0 H1 (as pSilencer-blank) vectors with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. The transfection efficiencies were according to GFP expression of the co-transfected plasmid pEGFP-C1. After 24 hpi, the cells were infected with 100 TCID50/0.2 mL of HTNV76–118. At 1, 2, 3, 5, 7, and 9 day post-infection (dpi), antigen slides were prepared to detect the HTNV protein expressions by IFA.
In other parallel experiments, the cells from individual wells (2-μg plasmid transfection groups) were collected at 2 dpi for viral RNA detection. At 5 dpi, the cultures were subjected to two cycles of freezing and thawing followed by centrifugation at low speed (1000×g), and the supernatants were then serially diluted, inoculated on the cells, and finally titrated by IFA.
Antiviral activity in vivo
This study was approved by the Ethics Committee of Wuhan University School of Medicine. All work with infected mice was performed in the Animal Biosafety Level 3 (ABSL-3) Laboratory of the Animal Research Center at Wuhan University and was humanely conducted in compliance with the Chinese Animal Protection Act and the National Research Council criteria.
Specific pathogen-free pregnant BALB/c mice were obtained from the Animal Research Center of Wuhan University. Suckling mice (2 d old) were intracranially inoculated with 20 μL HTNV 76–118 viral stock at 100 LD50. The mice were randomly divided into 4 groups (30 mice per group) and intraperitoneally injected with 10 μL of the plasmids (pSilencer-S, pSilencer-M or pSilencer 3.0 H1 vectors at concentrations of 3 μg/μL) or 10 μL PBS for the normal controls and viral controls at 1 and 3 dpi, respectively. The treatments were performed for the survival rate study (n=10); these mice were observed daily for 15 d following infection, and the mortality rate and mean time to death (MTD) were estimated.
Four mice from each group were sacrificed at 2, 3, 4, and 5 dpi, and brain tissues were collected for viral RNA detection by qRT-PCR. At 8 dpi, the brains, lungs and kidneys were dissected from the sacrificed animals (n=4) and used for pathological examinations with H&E staining.
At the experimental point of 5 dpi, the brains, lungs and kidneys were harvested, weighed and homogenized into a ∼10% (weight/volume) suspension in test medium. The homogenates were frozen and thawed twice and centrifuged at 3000 rounds per minute for 10 min. The virus titers were determined as described above.
At the experimental point of 2 dpi, blood samples were collected and pooled for each group. TNF-α was detected in the sera with QuantiKine quantitative sandwich enzyme immunoassays according to the manufacturer's instructions.
qRT-PCR analysis
Total RNAs were extracted from the cell or tissues using TRIzol reagents (Invitrogen, USA) following the manufacturer's instructions. First-strand cDNA was synthesized by reverse transcription of 500 ng of total RNA using random primers (Promega) and M-MLV reverse transcriptase (Promega) at 25 ºC for 10 min, 37 ºC for 60 min, 85 ºC for 5 min and 4 ºC for 4 min. The amplification was performed using the following primer sets: HTNV forward: 5′-TCTAGTTGTATCCCCATCGACTG-3′, HTNV reverse: 5′-ACATGCGGAATACAATTATGGC-3′ human GAPDH forward: 5′-GGTGGTCCTCTGACTTCAACA-3′, human GAPDH reverse: 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ and mouse GAPDH forward: 5′-ACCCAGAAGACTGTGGATGG-3′, mouse GAPDH reverse: 5′-ACACATTGGGGGTAGGAACA-3′. The plasmid pGEM-T/HTNV, pGEM-T/human GAPDH, and pGEM-T/mouse GAPDH were stored in our laboratory and used to generate the standard curves. The absolute quantification of the HTNV viral gene was performed as previously described20. The data are presented as vRNA copies/ng of GAPDH mRNA.
Immunofluorescence assay (IFA)
IFA was performed to detect the hantavirus antigens in the infected Vero-E6 cells as previously reported18 with monoclonal antibody A35, which targets the nucleocapsid protein (1:200 dilution, provided by the Institute of Virology, Chinese Academy of Preventive Medicine, Beijing, China), or rabbit polyclonal antibody targeting glycoprotein G2 (1:100 dilution, provided by Prof John W HUGGINS, Virology Division, United States Army Medical Research Institute of Infectious Disease). After incubation, FITC-conjugated goat anti-mouse or anti-rabbit IgG (Sigma Aldrich, 1:200 dilution) secondary antibodies were used. The cell cytoplasm was stained red with Evans blue. The images were collected using a fluorescence microscope (Nikon TE2000).
Statistical analysis
All of the data were analyzed with SPSS 17.0 software. The data are expressed as the mean±SD for all experiments. One-way ANOVAs were used to assess the significance of the differences between the means. P values below 0.05 were considered statistically significant.
Results
Inhibition of HTNV production in Vero-E6 cells by shRNAs
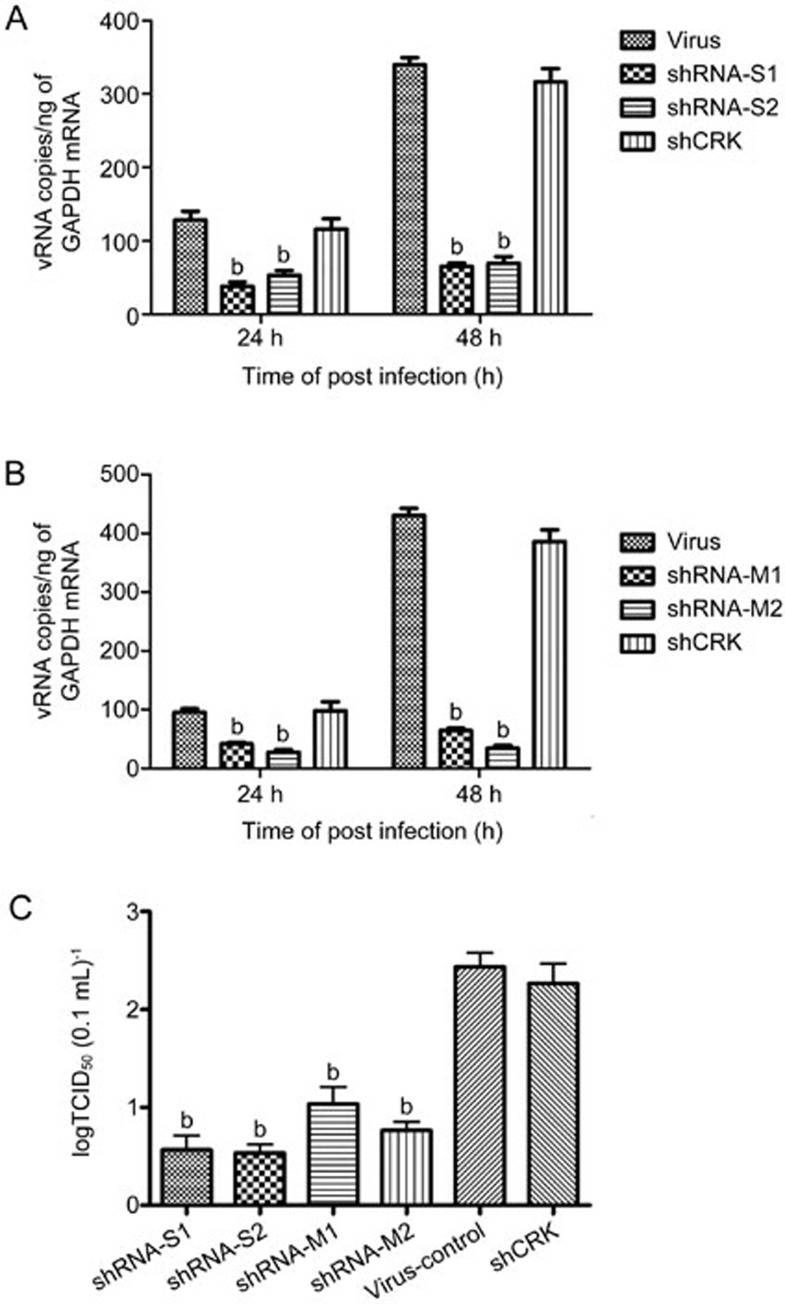
shRNAs were designed to specifically target the S and M genes of HTNV. The efficacies of the shRNAs in the inhibition of HTNV replication were evaluated by transfecting the Vero-E6 cells with 60 nmol/L of S1, S2, M1 and M2 shRNAs followed by the infection of the cells with 100 TCID50/0.2 mL of HTNV76–118. As illustrated in Figure 1A and 1B, the transfections of the 4 shRNAs resulted in the inhibitions of viral RNA transcription at 24 hpi of 65.72%±1.7% (S1), 56.8%±4.8% (S2), 55.17%±6.9% (M1) and 64.9%±5.7% (M2) (P<0.05 vs the virus group). When the cells were treated with the shRNAs at 48 hpi, the efficiencies of viral gene inhibition were 91.76%±2.29% (S1), 78.28%±6.46% (S2), 68.72%±8.8% (M1) and 95.23%±6.25% (M2) (P<0.05 vs virus group). No protective effect was observed in the Vero-E6 cells treated with shCRK.
Figure 1.
ShRNA interference with HTNV production in Vero-E6 cells. Vero-E6 cells were transfected with 60 nmol/L shRNA-S1, -S2 (A), -M1, -M2 (B), or shCRK as a control and were then were infected 24 h later with HTNV. The cells were harvested for RNA purification and real-time PCR at 24 and 48 hpi. The viral titers of the frozen-thawed culture samples collected at 96 hpi were measured (C). The data are expressed as the log10 values of the vial titers. All experiments were repeated three times, the replications produced similar results. bP<0.05.
For the yield-reduction assay, culture samples were collected at 96 hpi, and the titers were determined. The inhibitory effects of the four shRNAs against HTNV replication were evaluated. The four shRNAs reduced the HTNV yields by approximately 4 log10 (S) and 2 log10 (M) compared with the control (Figure 1C).
Based on these results and the viral antigen expression results detected by IFA (data not shown), we concluded that the shRNAs that targeted the S and M segments of the HTNV gene were able to inhibit RNA transcript and virus production in the HTNV-infected cells and that shRNA-S1 and shRNA-M2 exhibited a stronger inhibitory effect against HTNV. Thus, shRNA-S1 and shRNA-M2 were further selected for insertion into the pSilencer-3.0-H1 of the RNAi vector, and the resultant vectors were designated pSilencer-S and pSilencer-M.
Antiviral effects of pSilencer-S and pSilencer-M in HTNV-infected Vero-E6 cells
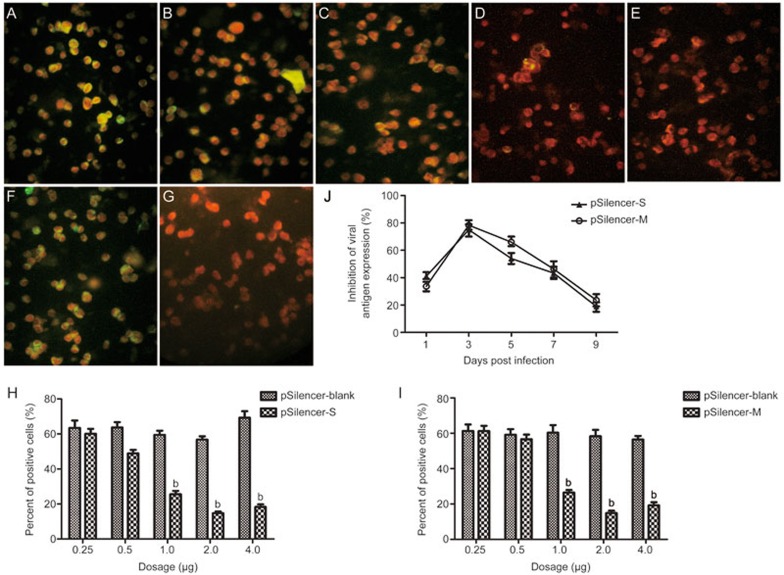
The RNAi pSilencer-S and pSilencer-M plasmids were constructed, and their antiviral effects were further evaluated by detecting the viral protein synthesis and RNA transcript and progeny virus titers in the HTNV-infected cells. The transfection efficiencies were evaluated according to the GFP expression of the co-transfected plasmid pEGFP-C1 and reached approximately 70%–80%. Treatment with pSilencer-S or pSilencer-M at concentrations of 1, 2, and 4 μg significantly decreased viral the antigen expression in the HTNV-infected Vero-E6 cells at 3 dpi (Figure 2). The percentages of positive cells declined with increases in the dose of transfected RNAi plasmids (Figure 2H and 2I). Only 14.69%±1.78% of the cells were positive for nucleocapsid protein staining in the 2-μg pSilencer-S treatment group compared to 56.8%±3.21% in the pSilencer-blank treatment group (Figure 2H). Similarly, the corresponding glycoprotein G2 positive staining rates were 14.8%±2.41% and 58.33%±6.24% in the 2-μg pSilencer-M treatment group (Figure 2I). These results indicate that the 2-μg plasmid transfection achieved the most effective inhibition of viral protein synthesis.
Figure 2.
Viral antigen detection in the shRNA expressing plasmid-treated Vero-E6 cells with HTNV infections. The Vero-E6 cells were transfected with 0.25, 0.5, 1.0, 2.0 and 4.0 μg of pSilencer-S (A–E), pSilence-M (data not shown) or pSilence-blank (F) followed by infection with HTNV 76–118. The samples were fixed at 72 hpi and stained with the monoclonal antibody A35 targeting the nucleocapsid (A–G, green color). Evans blue was applied to indicate the cytoplasm (red). Normal cells are shown in (G). The percentages of positive cells were assessed by counting the green-labeled cells and red-labeled cells in 4 quadrants of each slide, and the results are plotted in (H) (pSilencer-S) and (I) (pSilence-M). Representative fluorescence images are shown. The experiments were repeated three times, and the replications produced similar results. bP<0.05. (J) presents the dynamic inhibitory curves of pSilencer-S and pSilencer-M in terms of the viral antigen expressions in the HTNV-infected Vero-E6 cells. The cells were transfected with 2.0 μg shRNA expression plasmid followed by infection with HTNV 76–118. Slides were prepared at 1, 3, 5, 7, and 9 dpi to detect the viral antigens as described above. The inhibitory rates were calculated using the following formula: inhibitory rate (%)=(the percentage of positive cellsviral control group–the percentage of positive cellstreatment group)/the percentage of positive cellsviral control group×100%. All presented data are the results of experiments performed in triplicate.
We further detected reductions of viral antigen expression in the HTNV-infected Vero-E6 cells treated with 2 μg plasmid throughout the course of a 9-d infection. The greatest inhibitory rates were noted at 3 dpi (76.1%±1.3% for pSilencer-S and 78.2%±2.4% for pSilencer-M, Figure 2J). Moreover, the antiviral effect lasted at least 7 d (43.5%±3.4% for pSilencer-S and 46.7%±3.2% for pSilencer-M at 7 dpi).
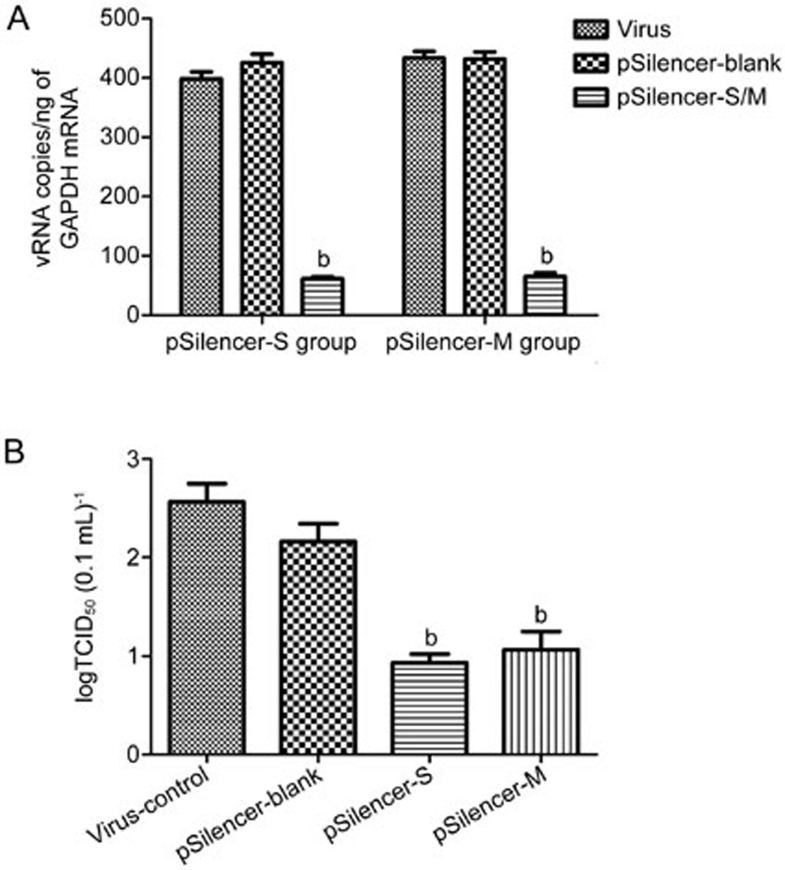
The efficacies of the inhibitions of HTNV replication by pSilencer-S and pSilencer-M were also evaluated by detecting viral RNA transcription at 48 hpi following 2-μg plasmid treatment. As illustrated in Figure 3A, when the cells were treated with 2 μg of the RNAi plasmid, the efficiencies of S and M gene inhibition reached 89.2%±3.4% and 92.7%±2.1% at 48 hpi, respectively. At 96 hpi, the culture samples were harvested, serially diluted and assayed to determine the viral titers. As illustrated in Figure 3B, the two shRNA expression plasmids reduced the HTNV yields by approximately 2 log10 compared with the control (Figure 3B).
Figure 3.
ShRNA expression plasmid interference with HTNV production in Vero-E6 cells. Vero-E6 cells were transfected with 2.0 μg of pSilence-S, pSilence-M or pSilence-blank and then infected 24 h later with HTNV. The cells were harvested for RNA purification and real-time PCR at 48 hpi (A). The viral titers of the frozen-thawed culture samples were measured at 5 dpi and are shown in (B). All experiments were repeated three times, and the replicates produced similar results. bP<0.05.
Taken together, these results indicate that pSilencer-S and pSilencer-M significantly reduced viral RNA transcripts, viral antigen synthesis and progeny virus production in the HTNV-infected Vero-E6 cells.
Protective efficacies of pSilencer-S and pSilencer-M in mice
Suckling mice were infected with HTNV as described above and treated with pSilencer-S or pSilencer-M at 1 and 3 dpi, respectively. Compared with the pSilencer-blank group, the pSilencer-S- and pSilencer-M-treated animals died later and exhibited reduced clinical signs including weight loss, ruffled fur, huddling tendencies, paralysis of the hind legs and spasms. All placebo-treated and pSilencer-blank-treated mice died with an MTD of 9 d, whereas the treatments with the RNAi plasmids increased the survival rates and MTDs (Figure 4A). The RNAi plasmid treatment increased the survival rate to 27.3% and the MTD to 12–13 d (P<0.05).
Figure 4.
Effects of the pSilence-S and -M treatments in the lethal HTNV challenge model. Suckling mice were infected with lethal doses of HTNV 76–118 and treated with pSilence-S or pSilence-M at 1 and 3 dpi, respectively. For each group, 8 mice were monitored for survival (A), 4 mice per group were sacrificed on the indicated days, and samples were collected for virological analyses (B–D). On 8 dpi, the viral loads in the lungs, kidneys and brains were determined by viral titration (B). The HTNV RNA profiles in the brains were determined with qRT-PCR on 2, 3, 4, and 5 dpi. bP<0.05.
The RNAi plasmid therapy resulted in significant reductions in the viral titers from the brain, lung, and kidney samples collected on 8 dpi (Figure 4B), which were homogenized, serially diluted and assayed to determine the viral titers by IFA. No infectious viral particles were detected in the lungs, kidneys, or brains of the survivors at 15 dpi. The dynamic viral loads in the brains were analyzed by qRT-PCR, which indicated that pSilencer-S and pSilencer-M significantly decreased HTNV gene expression on 2, 3 and 4 dpi (Figure 4C and 4D).
Four animals from each group were sacrificed at 8 dpi, and the brains, lungs and kidneys were collected for pathological examination. As illustrated in Figure 5, in the viral control group, the lung sections displayed thickened alveolar walls, interstitial lymphocyte and macrophage infiltrations, and hemorrhages. The brain tissues exhibited scattered hemorrhages, congestion, edema, and focal necrosis. The kidney exhibited focal renal interstitial hemorrhages and congestion. Both the pSilencer-S and pSilencer-M treatments decreased the severities of the viral lesions in the brain, lung and kidney tissues.
Figure 5.
Effects of the pSilence-S and -M treatments on the organ histopathologies in the suckling mice infected with HTNV. The suckling mice were infected with lethal doses of HTNV 76–118 and treated with pSilence-S or pSilence-M at 1 and 3 dpi, respectively. Hematoxylin and eosin (H&E)-stained sections of the lungs, kidneys and brains collected from each experimental group at 8 dpi are shown.
Discussion
HTNV infection has emerged as one of the severest infectious diseases threatening global health and has a significant worldwide economic influence2. In the present study, we demonstrated that vector-based shRNAs targeting specific sequences in the S and M segments of HTNV inhibited the expression and replication of HTNV in vitro and in vivo. Our results provide a new therapeutic candidate for the treatment of HTNV infections.
Since the discovery of RNAi, synthetic siRNA duplexes that generally consist of 21–23 nucleotides with 2-nt 3′ overhangs have been used to target viral RNAs in a sequence-specific manner following transfection21,22. However, the application of siRNAs has been limited by their short half-lives, the high costs of synthetic siRNAs and instability due to degradation by nucleases. Another method for inducing RNAi is via the transfection of plasmids that express antiviral short-hairpin RNAs (shRNAs), and this method is also an efficient means of eliciting RNAi in vivo15,21. Our results indicated that the shRNA expression plasmids pSilencer-S and pSilencer-M potently inhibited HTNV production in the cell lines at 72 hpi and even at 96 hpi in the newborn mice. These results provide a basis for the further development of siRNA expression plasmids for HTNV infection prophylaxis and therapy.
With the exception of ribavirin, no antiviral drugs for the treatment of hantavirus infection have been identified. Several members of the Bunyaviridae family, particularly HTNV, are sensitive to ribavirin8. We have reported that ribavirin can induce an up to 3.6-fold decrease in the vRNA level in HTNV infection at 4 dpi20, which is equivalent to the in vitro effects of pSilencer-S and -M observed in our experiments. With regard to the in vivo administration, the RNAi plasmid treatments increased the survival rate to 27.3% in a lethal HTNV-infected suckling mouse model. Zhou et al reported that the NP-specific siRNA expression plasmid pBabe-NP protected two of the eight mice (2/8) challenged with the lethal dose of avian influenza virus (H5N1) that killed all of the control mice15; this result is similar to the antiviral effects of the other siRNA expression plasmids against HTNV observed in our in vivo experiments. However, we noticed that ribavirin has been reported to be capably of affording 100% protection against lethal Andes virus infections in hamsters8 and also increases the survival rate to 81.8% in SEOV-infected suckling ICR mice23. The explanation of these phenomena may be related to the delivery of siRNA. The shRNA expression plasmid was distributed in the brain because the blood-brain barrier (BBB) of newborn mice is immature24. However, as a nonviral vector, the pSilencer shRNA expression vector does not readily cross the cellular membrane and is not stably introduced into the cells. Further studies are required to solve this problem, which is frequently considered a hurdle for the development of siRNA-based therapeutics25.
To our knowledge, this is the first report of the inhibition of hantavirus infection with an shRNA; thus, this report enriches the antiviral spectrum of RNAi therapy. In recent years, known and emerging viruses have posed increasingly serious threats to public health. Effective vaccines and antiviral drugs are not available for the majority of these viruses. The transfection of shRNA-encoding plasmids is probably best-suited for the treatment of acute viral infections, particularly among people infected with virus strains that are resistant to conventional antivirals and in cases of severe or re-emergent disease. However, a number of barriers to medical application remain to be solved, eg, improvements in delivery strategies and the safety and stability of siRNA and other issues.
Author contribution
Yuan-yuan LIU, Zhan-qiu YANG and Hai-rong XIONG designed the research plan; Yuan-yuan LIU, Liang-jun CHEN, Yan ZHONG, Meng-xin SHEN, Nian MA, Bing-yu LIU and Fan LUO performed the research; Yuan-yuan LIU and Wei HOU analyzed the data; and Yuan-yuan LIU, Zhan-qiu YANG and Hai-rong XIONG wrote the paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC Project No 81371865, 81101258 and 81000734) and the Natural Science Foundation of Hubei Province (No 2014CFB185).
References
- Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev 2010; 23: 412–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger DH, Figueiredo LT, Song JW, Klempa B. Hantaviruses--globally emerging pathogens. J Clin Virol 2015; 64: 128–36. [DOI] [PubMed] [Google Scholar]
- Maes P, Clement J, Gavrilovskaya I, Van Ranst M. Hantaviruses: immunology, treatment, and prevention. Viral Immunol 2004; 17: 481–97. [DOI] [PubMed] [Google Scholar]
- Kariwa H, Yoshimatsu K, Arikawa J. Hantavirus infection in East Asia. Comp Immunol Microbiol Infect Dis 2007; 30: 341–56. [DOI] [PubMed] [Google Scholar]
- Schonrich G, Rang A, Lutteke N, Raftery MJ, Charbonnel N, Ulrich RG. Hantavirus-induced immunity in rodent reservoirs and humans. Immunol Rev 2008; 225: 163–89. [DOI] [PubMed] [Google Scholar]
- Zeier M, Handermann M, Bahr U, Rensch B, Muller S, Kehm R, et al. New ecological aspects of hantavirus infection: a change of a paradigm and a challenge of prevention-a review. Virus Genes 2005; 30: 157–80. [DOI] [PubMed] [Google Scholar]
- Torre D, Pugliese A. Drug targets in infections with other emerging viruses: influenza viruses, metapneumovirus and hantaviruses. Infect Disord Drug Targets 2009; 9: 148–58. [DOI] [PubMed] [Google Scholar]
- Safronetz D, Haddock E, Feldmann F, Ebihara H, Feldmann H. In vitro and in vivo activity of ribavirin against Andes virus infection. PLoS One 2011; 6: e23560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev 2007; 59: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistella M, Marsden PA. Advances, nuances, and potential pitfalls when exploiting the therapeutic potential of RNA interference. Clin Pharmacol Ther 2015; 97: 79–87. [DOI] [PubMed] [Google Scholar]
- Ren GL, Bai XF, Zhang Y, Chen HM, Huang CX, Wang PZ, et al. Stable inhibition of hepatitis B virus expression and replication by expressed siRNA. Biochem Biophys Res Commun 2005; 335: 1051–9. [DOI] [PubMed] [Google Scholar]
- Liu YY, Deng HY, Yang G, Jiang WL, Grossin L, Yang ZQ. Short hairpin RNA-mediated inhibition of HSV-1 gene expression and function during HSV-1 infection in Vero cells. Acta Pharmacol Sin 2008; 29: 975–82. [DOI] [PubMed] [Google Scholar]
- Li T, Zhang Y, Fu L, Yu C, Li X, Li Y, et al. siRNA targeting the leader sequence of SARS-CoV inhibits virus replication. Gene Ther 2005; 12: 751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, et al. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci U S A 2003; 100: 2718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, He H, Wu Y, Duan M. RNA interference of avian influenza virus H5N1 by inhibiting viral mRNA with siRNA expression plasmids. J Biotechnol 2008; 135: 140–4. [DOI] [PubMed] [Google Scholar]
- Yuan J, Cheung PK, Zhang HM, Chau D, Yang D. Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. J Virol 2005; 79: 2151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. Progress in RNAi-based antiviral therapeutics. Methods Mol Biol 2011; 721: 67–75. [DOI] [PubMed] [Google Scholar]
- Deng HY, Luo F, Shi LQ, Zhong Q, Liu YJ, Yang ZQ. Efficacy of arbidol on lethal hantaan virus infections in suckling mice and in vitro. Acta Pharmacol Sin 2009; 30: 1015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol 2004; 22: 326–30. [DOI] [PubMed] [Google Scholar]
- Wei F, Li JL, Ling JX, Chen LJ, Li N, Liu YY, et al. Establishment of SYBR green-based qPCR assay for rapid evaluation and quantification for anti-Hantaan virus compounds in vitro and in suckling mice. Virus Genes 2013; 46: 54–62. [DOI] [PubMed] [Google Scholar]
- Deng Y, Wang CC, Choy KW, Du Q, Chen J, Wang Q, et al. Therapeutic potentials of gene silencing by RNA interference: principles, challenges, and new strategies. Gene 2014; 538: 217–27. [DOI] [PubMed] [Google Scholar]
- Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol 2007; 25: 1435–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ME, Kariwa H, Mizutani T, Tanabe H, Yoshimatsu K, Arikawa J, et al. Characterization of in vitro and in vivo antiviral activity of lactoferrin and ribavirin upon hantavirus. J Vet Med Sci 2001; 63: 637–45. [DOI] [PubMed] [Google Scholar]
- Saunders NR, Dreifuss JJ, Dziegielewska KM, Johansson PA, Habgood MD, Mollgard K, et al. The rights and wrongs of blood-brain barrier permeability studies: a walk through 100 years of history. Front Neurosci 2014; 8: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MJ, Martins S, Sarmento B. siRNA as a tool to improve the treatment of brain diseases: Mechanism, targets and delivery. Ageing Res Rev 2015; 21: 43–54. [DOI] [PubMed] [Google Scholar]