Abstract
Background
Detection of type 2 diabetes (T2D) is routinely based on the presence of dysglycemia. Although disturbed lipid metabolism is a hallmark of T2D, the potential of plasma lipidomics as a biomarker of future T2D is unknown. Our objective was to develop and validate a plasma lipidomic risk score (LRS) as a biomarker of future type 2 diabetes and to evaluate its cost-effectiveness for T2D screening.
Methods
Plasma LRS, based on significantly associated lipid species from an array of 319 lipid species, was developed in a cohort of initially T2D-free individuals from the San Antonio Family Heart Study (SAFHS). The LRS derived from SAFHS as well as its recalibrated version were validated in an independent cohort from Australia – the AusDiab cohort. The participants were T2D-free at baseline and followed for 9197 person-years in the SAFHS cohort (n = 771) and 5930 person-years in the AusDiab cohort (n = 644). Statistically and clinically improved T2D prediction was evaluated with established statistical parameters in both cohorts. Modeling studies were conducted to determine whether the use of LRS would be cost-effective for T2D screening. The main outcome measures included accuracy and incremental value of the LRS over routinely used clinical predictors of T2D risk; validation of these results in an independent cohort and cost-effectiveness of including LRS in screening/intervention programs for T2D.
Results
The LRS was based on plasma concentration of dihydroceramide 18:0, lysoalkylphosphatidylcholine 22:1 and triacyglycerol 16:0/18:0/18:1. The score predicted future T2D independently of prediabetes with an accuracy of 76 %. Even in the subset of initially euglycemic individuals, the LRS improved T2D prediction. In the AusDiab cohort, the LRS continued to predict T2D significantly and independently. When combined with risk-stratification methods currently used in clinical practice, the LRS significantly improved the model fit (p < 0.001), information content (p < 0.001), discrimination (p < 0.001) and reclassification (p < 0.001) in both cohorts. Modeling studies demonstrated that LRS-based risk-stratification combined with metformin supplementation for high-risk individuals was the most cost-effective strategy for T2D prevention.
Conclusions
Considering the novelty, incremental value and cost-effectiveness of LRS it should be used for risk-stratification of future T2D.
Electronic supplementary material
The online version of this article (doi:10.1186/s12944-016-0234-3) contains supplementary material, which is available to authorized users.
Keywords: Diabetes, Endocrine disorders, Lipidomics, Diagnostic tools, Genetics
Background
Type 2 diabetes (T2D) represents a complex metabolic turmoil rather than just a derangement of blood glucose control. In the continued quest for biomarkers of T2D [1] methods that can tap the whole gamut of metabolic disruption are therefore urgently required. In this context, there is a growing realization of the existence of the “plasma lipidome” that encompasses the entire spectrum of circulating lipid molecules [2, 3]. New high-throughput and high-resolution technologies applied to plasma lipidomic profiling are now providing additional insights into disease pathophysiology and individual lipid species have been associated with existing T2D or prediabetes [4–7]. However, the potential use of plasma lipid species as biomarkers to predict future development of T2D remains unexplored.
In this study, we used rich phenotypic and lipidomic data from two large, well-characterized, population-based cohorts and investigated the potential of plasma lipid species as independent biomarkers of incident T2D. We derived a lipidomic risk score (LRS) based on significant association of plasma lipid species with time to T2D in initially T2D-free Mexican American participants (n = 771, see Additional file 1: Figures S1, S2 and Tables S1, S2) from the San Antonio Family Heart Study (SAFHS) [8–10]. After establishing the independent and additive value of this LRS in the SAFHS cohort, we validated these results in a geo-epidemiologically distinct cohort from Australia – the AusDiab cohort (n = 644, see Additional file 1: Figure S3 and Table S2) [7, 11].
Methods
Cohorts
Studies related to the development and accuracy of the LRS were conducted in the SAFHS cohort [8–10]. At baseline, this cohort consisted of 1,431 individuals from 42 large and extended pedigrees of Mexican Americans residing in San Antonio, Texas, USA. Our protocol for selecting 771 individuals without diabetes who were followed for up to three additional visits spaced approximately 5 years apart (9197.1 person-years of follow-up with a maximum follow-up of 23.53 years) is shown in (Additional file 1: Figure S1). The distribution of SAFHS participants across families, their familial relationships and clinical characteristics and are shown in, (Additional file 1: Figure S2, Table S1 and Table S2). To determine the generalizability of the LRS derived from the SAFHS data, we used an independent well-characterized, high-risk subset of predominant Europid origin individuals drawn from the AusDiab cohort [7, 11]. From this cohort, we selected 653 participants who were initially non-diabetic. Since follow-up information was not available on 9 (1.4 %) individuals we included 644 participants in association analyses who were followed up for 5929.5 person-years (maximum follow-up 12.94 years). The algorithm used for inclusion of AusDiab participants is given in Additional file 1: Figure S3 and their clinical characteristics are shown in (Additional file 1: Table S2).
Outcomes
Our primary outcome of interest was incident T2D, defined as T2D detected during follow-up visits in participants who did not have T2D at the baseline visit. At each visit, T2D was considered present when one or both of the following conditions were met: i) fasting plasma glucose ≥126 mg/dl (7.0 mmol/l) or 2-hour post-glucose load ≥200 mg/dl (11.1 mmol/l)]; ii) receipt of anti-diabetic medication. There were 122 (15.8) and 233 (36.2 %) cases of incident T2D in the SAFHS and AusDiab cohorts, respectively.
We also examined the association of the LRS with six measures of insulin resistance. These measures were [12]: fasting plasma glucose (FPG), fasting plasma insulin (FPI), homeostatic model of assessment – insulin resistance (HOMA-IR), quantitative insulin sensitivity check index (QUICKI) [13], McAuley index [14] and leptin/adiponectin ratio [15]. HOMA-IR was defined as FPG (mg/dl) x FPI (IU/L)/405; QUICKI was defined as 1/(log FPI + log FPG (mg/dl)) and McAuley’s index was calculated as exp [2.63 – 0.28 ln(FPI) – 0.31 ln(serum triglycerides in mmol/l)]. The methods used for measurement of FPG, FPI, leptin and adiponectin have been previously described [8–10, 16, 17].
Lipidomic studies
Lipidomic profiling was conducted in the Metabolomics Laboratory, Baker IDI Heart and Diabetes Institute, Australia. We quantified a total of 319 lipid species (descriptive statistics are in, Additional file 1: Table S3) in plasma using a combination of liquid chromatography and electrospray ionisation-tandem mass spectrometry.
These methods have been described extensively elsewhere [7, 18–22]. Briefly, 10 μL aliquots of plasma were combined with 200 μL CHCl3/MeOH (2:1) and 15 μL of internal standard mix. Then samples were mixed (rotary mixer, 10 min), sonicated (water bath, 30 min) and allowed to stand (20 min) at room temperature. Afterwards, the samples were centrifuged (16,000xg, 10 min) and the supernatant was dried under a stream of nitrogen at 40 °C. Extracted lipids were resuspended in 50 μL H2O saturated BuOH, followed by 50 μL of 10 mM NH4CHOO in MeOH. Extracts were centrifuged (3,350xg, 5 min) and the supernatant transferred into 0.2 mL glass inserts in vials with teflon lined caps. Lipid measurements were performed by liquid chromatography electrospray ionisation-tandem mass spectrometry using an Applied Biosystems 4000 QTRAP. Liquid chromatography was performed on a Zorbax C18, 1.8 μm, 50 × 2.1 mm column at 300 μL/min using the following gradient conditions; 0 % B to 100 % B over 8.0 min, 2.5 min at 100 % B, a return to 0 % B over 0.5 min then 3.0 min at 0 % B prior to the next injection. Diacylglycerols and triacylglycerols were separated using the same solvent system with an isocratic flow (100 μL/min) of 85 % B over six minutes. Solvent A and B consisted of tetrahydrofuran:methanol:water in the ratios (30:20:50) and (75:20:5) respectively, both containing 10 mM NH4COOH. Quantification of individual lipid species was then performed using scheduled multiple-reaction monitoring (MRM) in positive ion mode [21, 22]. Lipid concentrations were calculated by relating the peak area of each species to the peak area of the corresponding internal standard. Cholesteryl ester species were corrected for response factors determined for each species.
Statistical analysis
Data normalization
To ensure that the lipidomic traits are normally distributed and represented on a comparable metric, we transformed the raw plasma concentration of each lipid species by inverse-normalization that included ranking, generation of cumulative density functions and determination of z-values based on the cumulative density.
Mixed effects Cox proportional hazards modeling
In the SAFHS cohort, we used mixed effects Cox Proportional Hazards models that account for the fixed effects of the predictors as well as random effects due to kinship [23, 24]. We created a kinship matrix representing paired genetic relationships among study subjects. The elements of a kinship matrix (φ) indicate the genetic similarity (kinship coefficient) denoted by relationships between each pair of the study subjects. For example, the routinely used kinship coefficients for different relationships are as follows: identical twins, 1; parent-offspring or sibling, 0.5; and grandparent-grandchild, avuncular, half-siblings or double first cousins, 0.25. Further, the kinship coefficients for 3rd, 4th, 5th and 6th degree relatives are 0.0078, 0.0020, 0.0005 and 0.0001, respectively.
In these mixed effects Cox PH models we adjusted the association of lipid species with time to T2D for the following covariates: age, age2, sex, age x sex interaction, age2 × sex interaction, systolic and diastolic blood pressures (SBP and DBP), waist circumference, body mass index (BMI), total serum cholesterol, serum high-density lipoprotein (HDL) cholesterol, serum triglycerides and use of anti-lipid and anti-hypertensive drugs. We corrected for the potentially false positive results using Benjamini and Hochberg’s [25] method of controlling the false discovery rate (FDR). All p-values were two-tailed.
Receiver operating characteristics curves for family data
Parallel to the reasoning for using mixed effects Cox PH analyses, we also accounted for kinship structure in the receiver operating characteristics (ROC) curves using a variance components approach. Receiver operating characteristic curves plot a series of estimated sensitivity (true positive rate) and 1-specificity (false positive rate) pairs when a continuous predictor is dichotomized at various cut-offs [26, 27]. In the context of family studies, the estimates of sensitivity and specificities can be biased due to the kinships. To account for the kinships in the estimation of sensitivity and specificity we used the variance components approach and polygenic regression modeling in the following way.
For a given cut-off, we used a liability threshold model [28] for analysis of discrete traits and estimated the prevalence of T2D events separately in participants above and below the cut-off. We used polygenic regression models for this which permitted accounting for both the kinships as random effects and the abovementioned clinical predictors as fixed effects. A polygenic regression model is of the form:
where, LT is the liability threshold, μ is the overall mean LT, β is the regression coefficient vector corresponding to the covariate matrix a, gi is the polygenic effect (used to estimate the heritabilities) and ei is the measurement error. The mean (μ) represents the cumulative distribution function, the inverse of which provides probability. In the case of discrete traits this probability represents the prevalence of a condition.
Since we estimated the prevalence estimates in subset of subjects who were above or below the cut-off for a predictor, these prevalence estimates represent the post-test probability of a positive (p1) and negative (p0) result. Since the proportion of subjects above the cut-off (p) can also be estimated from the sample (through a similar polygenic regression model); we derived the Bayesian estimates of sensitivity and specificity as follows: sensitivity = p*p1/[p*p1 + (1-p)*p0] and specificity = (1-p)*(1-p0)/[(1-p)*(1-p0) + p*(1-p1)].
We repeated this procedure over the entire spectrum of observed cut-off values and plotted the ROC curve as tuples of sensitivity and 1-specificity. These estimates implicitly account for the kinship structure of the study subjects. We then used the methods described by Hanley and McNeil [26] to determine the area under the ROC curve (AUC, a measure of the predictive accuracy) and its standard error. We used the chi-square tests based on AUCs and their standard errors [29] to test for significant difference between AUCs.
Incremental value of plasma LRS
We determined the incremental value of lipidomic biomarkers to commonly used methods of risk stratification with respect to the following five aspects – model fit (assessed by likelihood ratio χ2, LRχ2), information content (Akaike information criterion, AIC), accuracy (Uno’s survival C statistic [30]), discrimination (integrated discrimination improvement, IDI) and continuous version of reclassification (net reclassification index, NRI).
Validation studies in the AusDiab cohort
In the AusDiab cohort, we used Poisson regression models to account for length time bias (using length of follow-up as an exposure variable) since the exact date of T2D diagnosis was unknown. We took three complementary approaches for validation of the LRS: i) the LRS derived from SAFHS was directly applied to the AusDiab participants; ii) the LRS was recalibrated for the AusDiab cohort; and iii) the predictive performance of the recalibrated score in AusDiab was compared to a similar set of Poisson regression models in the SAFHS cohort. To increase the generalizability of these interpretations, the confidence intervals (CI) were estimated using a bootstrap procedure on 1000 replicates. We also estimated AIC, IDI and NRI to quantify the improved prediction due to LRS in the AusDiab cohort.
Cost-effectiveness studies
We investigated if the use of LRS - alone or in combination with other screening methods – would be a cost-effective option in the setting of T2D screening. For this we considered seven potentially useful screening and intervention strategies (Fig. 4a) and compared the cost and effectiveness of these strategies.
Fig. 4.
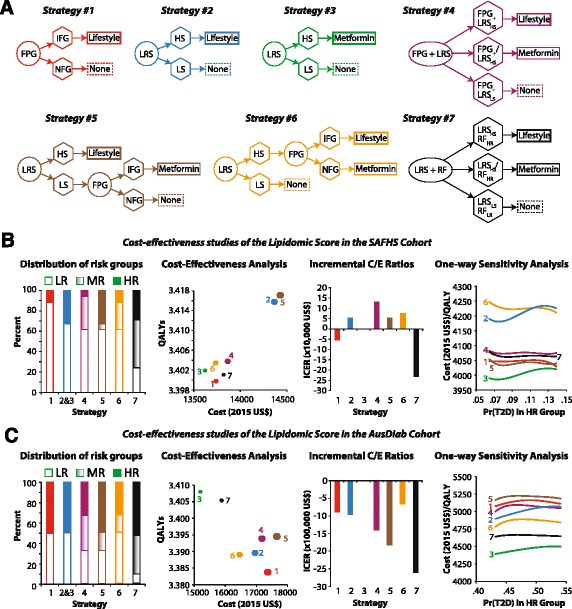
Cost effectiveness analyses of candidate screening and intervention strategies for T2D risk-stratification. a The seven strategies that were considered. The diagrams use the following convention: circles, name of the screening test; hexagons, results of the screening test; rectangles, suggested intervention; thick border for rectangles, high-risk group; thin border for rectangles, moderate-risk group; dashed border for rectangles, low-risk group. The abbreviations used in the panel are as follows: FPG, fasting plasma glucose; IFG impaired fasting glucose; NFG, normal fasting glucose; LRS, lipidomic risk score; HS, high score; LS, low score; RF, risk factor assessment. For LRS, a high score was defined on the basis of cutoffs defined by receiver-operating-characteristic curves. The strategies are color coded and the colors are consistently used in panels B and C. The full decision tree based on the seven strategies is shown in (Additional file 1: Figure S6). Details of the costs and utilities are provided in Material and Methods, (see Additional file 1: Figure S6, Tables S7 and S8). b Cost-effectiveness analyses based on the LRS in the SAFHS cohort. Leftmost plot shows the relative distribution of the risk groups based on the screening strategy used (LR, low-risk group; MR, moderate-risk group; HR, high-risk group). The plot second from left shows costs (2015 US$) and quality-adjusted life years (QALYs) for each strategy. The size of the markers is proportional to the expected risk reduction in T2D incidence. The next plot shows incremental cost-effectiveness ratios using the most cost-effective strategy (#3) as the reference strategy. The rightmost plot shows smoothed results of sensitivity analyses that used 1000 microsimulation runs. The results were smoothed using fourth-order polynomial regression lines. Sensitivity analyses shown here are for the variable (probability of T2D at 5-years in HR individuals) to which the decision was most sensitive based on the tornado diagrams shown in (Additional file 1: Figure S7). c Cost-effectiveness analyses based on the LRS in the AusDiab cohort. Key to plots is the same as those for plots in (Panel b). Full numerical results of cost effectiveness analyses for both cohorts are provided in (Additional file 1: Table S9)
All the screening strategies considered in these analyses assume:
Single payer perspective
A one-time screening with the indicated strategy;
Identification of differential risk groups (high risk, moderate risk or low risk) based on the strategy used;
Influence of the screening/interventions strategy on the 5-year observed probability of incident T2D;
A willingness-to-pay (WTP) US$ 4450.12 for a 5-year T2D prevention program. This estimate is based on the 3-year estimates of WTP reported by Johnson et al. [31], linearly extrapolated to five years and converted to 2015 US$; and
Even though the probability estimates used here are derived from both SAFHS and AusDiab, the analyses assume US target population to which the cost-effectiveness measures apply.
Cost estimations
For estimating the cost of interventions, we used the 10-year cost-effectiveness data reported by the DPP Trial [32, 33]. We cumulated the first five years data from the DPP Trial in individuals undergoing Lifestyle intervention, Metformin supplementation or No intervention (placebo) group. We also considered the medical treatment costs for individuals with and without T2D outside the DPP. We then converted these costs into 2015 US$ which represent the undiscounted costs from a single-payer’s perspective. It is recommended that the costs and QALYs be discounted for future outcomes in studies of prevention strategies such as ours. However, the rate at which the costs and QALYs should be discounted (currently recommended to be 3.5 % for both [34]) is a subject of an ongoing debate [35–38]. In this study, we chose to use undiscounted costs and QALYs since our aim was to conduct proof-of-principle cost-effectiveness analyses rather than to provide a futuristic solution to screening for T2D. Details of the cost estimations are shown in (Additional file 1: Table S7). To estimate the costs of screening, we used different sources of data. For the costs associated with fasting plasma glucose (FPG), we used the costs as mentioned by Sullivan et al. [39] and converted these costs into 2015 US$. To estimate the cost of risk factor screening using a questionnaire method, we used data as reported by Zhang et al. [40] and converted these costs into 2015 US$. The cost of the LRS was established as follows: assuming a center that conducts a large number of lipidomic assays, the cost of each lipidomic species measured is ~1 US$ (total 3 US$ for three lipid species), the cost of mass spectrometry is ~4 US$ and the cost of manpower for the assay is 4 US$ totaling to US$11 per assay. For screening strategies that used only LRS, there would be an additional cost of plasma sample preparation (29 US$, based on costs reported at: http://pathology.med.wayne.edu/lipidomics/servicescosts.php) and a primary care visit (50 US$, reported in [40]). However when the LRS is combined with the fasting plasma glucose assays then these additional costs of sample preparation and primary care visit are charged only once and not duplicated for each assay separately.
Effectiveness measures
We used two measures of effectiveness – risk reduction in the incidence of T2D and quality-adjusted life years (QALYs). The estimates of QALY were derived from the DPP data which report the annual QALYs stratified by T2D status and intervention received. Risk reduction in the incidence of T2D was estimated as a function of the probability of T2D at five years and the expected probability based on efficacy of the intervention used. We used an estimate of 0.58 and 0.31 as efficacy of lifestyle intervention and metformin supplementation (500 mg once daily), respectively, based on the DPP Trial results [33, 41].
Outcomes of cost-effectiveness studies
These were: cost and effectiveness, incremental cost-effectiveness ratios (ICER) and net monetary benefits. Results were represented as cost-effectiveness plots, tornado diagrams and ICER plots.
Analytical approach for cost-effectiveness studies
We conducted cost-effectiveness studies separately for the SAHFS and AusDiab datasets. For each dataset, we used the same estimates of costs and effectiveness measures (as detailed above). The probability of being in either the high risk (HR), moderate risk (MR) or low risk (LR) was different for the two study cohorts; hence two different sets of analyses were used. For each dataset, we first studied the distribution of the risk groups and then conducted cost-effectiveness studies by i) rolling back the decision tree shown in (Additional file 1: Figure S6); and ii) plotting the cost and effectiveness. Since there were two effectiveness measures, we combined these measures into a single plot by using ordinate axis for QALYs and altering the size of marker based on the risk reduction in T2D incidence (Fig. 4b and c). We then estimated the ICERs based on estimated cost and QALYs for each strategy (see Additional file 1: Table S8). These analyses together represented the base case scenarios. Finally, we conducted sensitivity analyses by first using tornado plots (see Additional file 1: Figure S7) and then conducting a one-way sensitivity analysis for the variable to which the expected value (EV) was most sensitive. For the one-way sensitivity analyses, we used microsimulation with 1000 runs and the results were expressed as mean cost/QALY ratio for each strategy based on the 1000 runs. Further, we smoothed the results using fourth order polynomial regression analyses to capture the potential non-linearity in the association of EV with the sensitivity variable.
Statistical software
We used the following statistical programs (with the analytical purpose): Sequential Oligogenic Linkage Analysis Routines – SOLAR [42] (inverse normalization of traits, polygenic regression models, ROC); coxme in R (mixed effects Cox PH models, LRχ2, AIC); survC1 [43] in R (Uno’s survival C statistic); survIDINRI [44] in R (IDI and NRI estimation), Stata 12.0 (Stata Corp, College Station, TX; data management, FDR corrected p-values, Poisson regression analyses, improved prediction indices in the AusDiab cohort and Cuzick’s nonparametric test for linear trend), and TreeAge Pro 2015 (TreeAge Software, Inc, Williamstown, MA; cost-effectiveness analyses).
Results
Development of LRS in the SAFHS cohort
In the SAFHS cohort, there was a wide inter-individual variability in plasma concentrations of the lipid species (see Additional file 1: Table S3). Thus, we transformed these plasma concentrations using an inverse-normalization procedure for use as independent variables in mixed effects Cox regression modeling. The results demonstrated that after accounting for clinically relevant covariates, 87 of the 319 lipid species predicted T2D with a nominal p-value <0 · 05 but only 10 lipid species had a false discovery rate (FDR) corrected p-value <0 · 2 (Fig. 1a, see Additional file 1: Table S4). We then included these 10 species (see Additional file 1: Figure S4A) in backward-elimination stepwise regression models and found that the final model retained only three lipid species (Fig. 1a, see Additional file 1: Figure S4A): dihydroceramide 18:0 (Cer(d18:0/18:0)), lysoalkylphosphatidylcholine 22:1 (LPC(O-22:1)) and triacylglycerol 16:0/18:0/18:1 (TG(16:0/18:0/18:1)). LPC(O-22:1) was associated with a slower disease onset while the other two species were associated with faster progression to T2D. From these results we generated a composite LRS as shown in (Additional file 1: Figure S4B). To compare the predictive accuracy of the lipid score against each of its components, we conducted ROC analyses (see Additional file 1: Figure S4C) which showed that the predictive accuracy of the composite LRS (AUC 0 · 7566, 95 % CI 0 · 7111 – 0 · 8021) was significantly better than that of any component species.
Fig. 1.
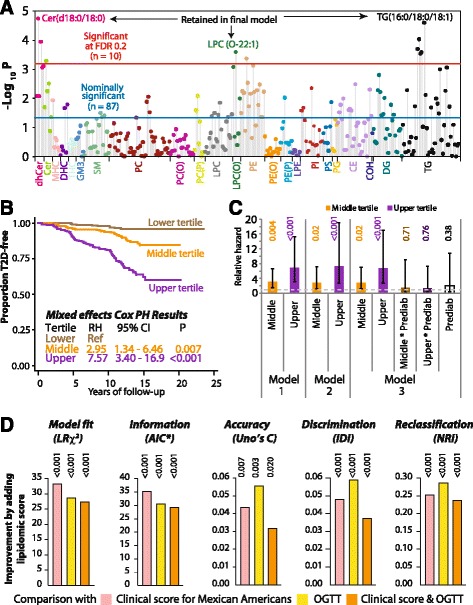
Development and assessment of the LRS in the SAFHS cohort. a Manhattan plot showing the association of the entire lipidome with incident T2D. All results are from mixed effects Cox proportional hazards models that account for degree of relationship among participants. All models are adjusted for age, age2, sex, age x sex interaction, age2 × sex interaction, systolic and diastolic blood pressures, waist circumference, body mass index, total serum cholesterol, serum high-density lipoprotein cholesterol, serum triglycerides and use of anti-lipid and anti-hypertensive drugs. The dots are color-coded for different lipid classes. The blue horizontal line represents a nominal type I error rate of 0.05 while the red horizontal line represents a global, FDR-corrected type I error rate of 0.2. All lipid species above the red line were simultaneously included in stepwise regression models and the species that were retained in the final multivariate model are indicated at the top of the plot. b Association of the LRS with incident T2D. Based on the regression coefficients from this final model, we derived a LRS as 0 · 4176*i(Cer(d18:0/18:0))-0 · 3443*i(LPC(O-22:1)) + 0 · 5361*i(TG(16:0_18:0_18:1)) where i(L) represents the inverse-normalized plasma concentration of lipid species L. The individuals were classified into three groups based on tertiles of the LRS. Shown is a Kaplan-Meier plot of progression to T2D based on the LRS. The results of mixed effects Cox proportional hazards models are after adjusting for the same covariates as those listed in Panel A. RH, relative hazard. c Association of the LRS with incident T2D independent of prediabetes. The plot shows relative hazard (colored bars), 95 % confidence intervals (error bars) and p-values (rotated numbers above bars). Model 1 – stratified mixed effects Cox PH model with presence of prediabetes as the stratifying variable; Model 2 – mixed effects Cox PH model restricted to NGT participants only; Model 3 – Interactive model that included interaction terms between presence of prediabetes and the LRS tertiles. All models use participants in the lower tertile of LRS as the reference category. NGT, normal glucose tolerance; Prediab, presence of prediabetes. d Clinical value of the LRS as a biomarker of incident T2D. Plots show bars representing improvement in the indicated characteristic achieved by adding LRS to the color coded models. Three models were used: clinical score for Mexican Americans (pink bars), oral glucose tolerance test (yellow bars) and clinical score combined with oral glucose tolerance test (orange bars). Statistical significance of the improvement is shown by the rotated numbers above the bars. *, the plot shows decrease in this parameter as a measure of improvement; LRχ2, likelihood ratio χ 2 statistic; AIC, Akaike information criterion; Uno’s C, Uno’s C statistic for survival data; IDI, integrated discrimination improvement; NRI, continuous net reclassification index
Prediction of future T2D based on LRS in the SAFHS cohort
We stratified the SAFHS participants into tertiles of the LRS. Participants in the middle and upper tertile groups of the LRS progressed ~3 and ~8 times faster to T2D, respectively, as compared to those in the lower tertile group (Fig. 1b) even after accounting for all the relevant clinical and biochemical confounders. Corroborating these observations we found that by 15 years of follow up 3 · 5, 12 · 5 and 30 · 7 % of the participants in the lower, middle and upper tertile groups had progressed to T2D, respectively (see Additional file 1: Figure S4D). Since prediabetes [defined as the presence of impaired fasting glucose (IFG) and/or impaired glucose tolerance (IGT)] was a strong determinant of incident T2D risk in the SAFHS participants (see Additional file 1: Figure S5), we investigated if our results of lipidomic associations were confounded by prediabetes. For this, we ran three complementary regression models – a stratified model (prediabetes as the stratifying variable), a model restricted to participants with normal glucose tolerance (NGT) and an interactive model that included interaction between the LRS tertiles and prediabetes. Our results (Fig. 1c) showed that irrespective of the modeling approach used, the participants in the middle and upper tertile of the LRS progressed significantly faster to T2D as compared to the participants in the lower tertile.
Corroborating these results, we observed that the median LRS scores showed a clear gradient in association with the T2D status (see Additional file 1: Table S5). Individuals, who were NGT at baseline and remained NGT throughout follow-up had the lowest median score, while those were prediabetic at baseline but did not develop T2D during follow-up had a higher median LRS (0.38 versus −0.24). The highest median LRS, however, was observed for individuals who developed T2D during follow-up (0.78). Cuzick’s test for linear trend showed that these median LRS values were linearly related to the T2D status and highly significant (p < 1.0x10−22).
LRS-based improvement in clinical prediction of T2D in the SAFHS cohort
We then examined if the addition of the LRS to commonly used clinical predictors of T2D risk can significantly augment the predictive performance. We considered two sets of commonly used predictors: a clinical score tailored for Mexican Americans [45] and the oral glucose tolerance test (OGTT). The clinical score is based on age, sex, fasting glucose, systolic blood pressure, HDL cholesterol, BMI and family history of T2D [45]. Figure 1d shows that addition of the LRS to the clinical score and to the OGTT, singly or in combination, resulted in an improved model fit (higher likelihood χ2), information content (lower Akaike Informatin Criterion, AIC) and predictive accuracy (increased c-statistic of Uno). Integrated discrimination improvement (IDI) showed a 3 · 7 % – 5 · 9 % improvement in discrimination and the continuous net reclassification index (NRI) indicated an improved reclassification by addition of the LRS. The observed improvement in all indexes was statistically significant (Fig. 1d).
Association of LRS with insulin resistance
Mechanistically, insulin resistance (IR) provides an initial trigger to the pathogenesis of T2D [46–48]. Given the fact that the LRS predicted progression to T2D even in the NGT individuals, we considered whether the LRS was associated with IR in the SAFHS individuals. In the absence of data on the diagnostic euglycemic clamp, we used six previously established measures of IR [12]: fasting plasma glucose (FPG), fasting plasma insulin (FPI), homeostatic model of assessment – insulin resistance (HOMA-IR), quantitative insulin sensitivity check index (QUICKI) [13], McAuley index [14] and leptin/adiponectin ratio [15]. All the six indices of IR showed a consistently linear association with the LRS tertiles in all as well as in NGT individuals (Fig. 2). These findings afford a strong support to the hypothesis that the LRS may be detecting subclinical insulin resistance.
Fig. 2.

Association of LRS with measures of insulin resistance (IR) in SAFHS participants. Each plot shows the median (colored bars) and inter-quartile range (error bars) for the indicated measure of IR across tertiles of LRS. L, M and U represent lower, middle and upper tertiles of the LRS, respectively. K-W P, Kruskall-Wallis test significance value; R2, variability in the measure of IR explained by tertiles of LRS estimated using quantile regression; FPG, fasting plasma glucose; FPI, fasting plasma insulin; HOMA-IR, homeostatic model of assessment – insulin resistance; QUICKI, quantitative insulin sensitivity check index; L/A Ratio, leptin/adiponectin ratio; NGT, normal glucose tolerance
Validation of LRS in the AusDiab cohort
Next, we validated the LRS in another independent cohort, the AusDiab cohort. The participants in the AusDiab cohort were on an average ~20 years older, were equally distributed across sex, had lower BMI but higher blood pressures, had higher total serum cholesterol and triglycerides and a higher prevalence of existing prediabetes at baseline as compared to the SAFHS participants (see Additional file 1: Table S2). Despite these differences, a direct validation of the LRS derived from the SAFHS showed that the score was associated with an increased risk of incident T2D in AusDiab participants even after accounting for clinical covariates or prediabetes at baseline (Fig. 3a, left panel). Since practical application of the LRS can include population-specific refinement, we recalibrated the LRS for the AusDiab cohort and again observed an increased risk of incident T2D (Fig. 3b, left panel). To examine the robustness of these findings we ran the analyses in 1000 bootstrap samples and found that the LRS continued to be a significant predictor of T2D in the AusDiab cohort. Similar to the observations in the SAFHS cohort, the AusDiab cohort also demonstrated a statistically significant linear trend in median LRS based on T2D status (see Additional file 1: Table S5). Moreover, to permit a direct comparison of this recalibrated score in the AusDiab cohort, we also ran corresponding mixed effects Poisson regression models in the SAFHS participants (see Additional file 1: Table S6) and observed that the LRS indeed provided excellent corroborative results in both cohorts. Further, we found that in all the modeling approaches taken in the AusDiab cohort, addition of the LRS (whether derived from SAFHS or recalibrated for the AusDiab cohort) was always associated with a statistically significant improvement in the information content, discrimination and reclassification (Fig. 3a and b, Right panels). Together, these results demonstrate that the LRS derived from the SAFHS participants was generalizable to an independent and distinct cohort.
Fig. 3.
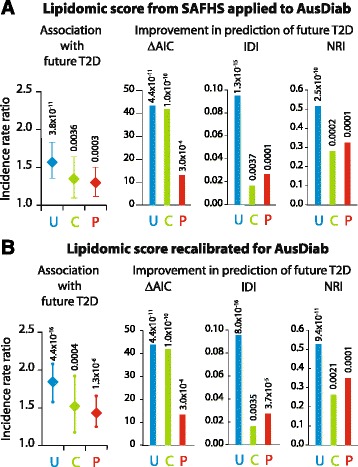
Validation of the LRS in the AusDiab cohort. a Direct application of the LRS derived from SAFHS cohort to the participants from AusDiab cohort. Left panel shows incidence rate ratio for future T2D associated with one standard deviation change in the LRS. The results are from three Poisson regression models that used duration of follow-up as an exposure variable: U – unadjusted; C – adjusted for clinical covariates; P – adjusted for presence of prediabetes. The clinical covariates used for adjustment in the multivariate models were: age, sex, systolic and diastolic blood pressure, body mass index, total cholesterol, HDL cholesterol, serum triglycerides, family history of diabetes, and use of anti-hypertensive and lipid-lowering drugs. The Right Panel shows three bar charts each of which depicts improvement in the indicated parameter upon addition of the LRS to the indicated regression model. Rotated numbers at the top of the bars are p-values. ΔAIC, improvement in Akaike Information Criterion; IDI, integrated discrimination improvement; NRI, continuous net reclassification index. b LRS recalibrated for AusDiab. Based on the results of Poisson regression analyses in the AusDiab, a recalibrated LRS was calculated as follows: 0.2259*i(Cer(d18:0/18:0))-0.2397*i(LPC 22:1) + 0.3267*i(TG(16:0_18:0_18:1)) where i(L) represents the inverse-normalized plasma concentration of lipid species L. Key to the plots in (Panel b) is the same for those in (Panel a)
Cost-effectiveness modeling of LRS based screening and intervention strategies
Considering these results in totality, we next investigated whether T2D risk-stratification based on the LRS would be an economically viable alternative to existing risk stratification strategies. It is being recognized that screening for T2D with FPG combined with lifestyle intervention in high-risk individuals may be cost-effective compared to no screening at al. [39, 49, 50] We therefore considered whether the LRS – alone or in combination with FPG or clinical risk factors – would be cost-effective as compared to FPG alone for outcomes measured at the end of five years. We used the probability estimates from SAFHS and AusDiab cohorts, cost and utility estimates from the DPP trial [33, 41] and screening costs from other published studies [39, 50–52]. Details of the probabilities, costs and utilities are provided in Materials and Methods, (see Additional file 1: Tables S7 and S8). We compared seven potentially useful screening-and-intervention strategies (Fig. 4a, Additional file 1: Figure S6) and found (Fig. 4b, Additional file 1: Table S9) that the strategy of risk stratification using the LRS followed by metformin treatment for the high-risk individuals would be the most cost-effective screening strategy. In both SAFHS and AusDiab, this strategy was associated with least costs (see Additional file 1: Table S9) and was more cost-effective than the strategy of FPG followed by lifestyle intervention (negative incremental QALYs in both cohorts, see Additional file 1: Table S9). Similarly, the strategy of risk factor stratification combined with the LRS was also associated with negative incremental effectiveness in both cohorts. On the other hand, the proportion of potential T2D cases prevented by the LRS/metformin strategy was the least as compared to other candidate strategies considered. Therefore, practical use of the screening-and-intervention policy will likely be a trade-off between attempting to increase QALYs versus targeting aggressive reduction in T2D incidence. Lastly, the decision in favor of LRS/metformin strategy was most sensitive to the probability of T2D at five years in the high risk individuals (see Additional file 1: Figure S7). However, over the entire range of sensitivity analyses, the strategy of LRS/metformin for high-risk individuals remained the most cost-effective solution in both cohorts (Fig. 4b and c).
Discussion
Our study reports five novel findings: i) the plasma LRS independently, accurately and additively predicted incident T2D; ii) given the LRS could accurately risk-stratify even the NGT participants indicates that the plasma lipidomic changes may precede those in glucose metabolism; iii) these changes are associated with insulin resistance; iv) the LRS is generalizable as validated in a distinctly different cohort; and v) the LRS combined with metformin supplementation for high-risk individuals is the most cost-effective screening strategy. Of note, supportive evidence for the biological plausibility of the three significant lipid species as T2D biomarkers is plentiful. We have previously shown that the dihydroceramide/ceramide, triacyglycerol and phosphatidylcholine axes are important targets in T2D pathophysiology [7]. The enzyme ceramide synthase 1 (specifically involved in the biosynthesis of Cer(d18:0/18:0), [53]) is inversely associated with alterations in murine models of insulin resistance [54, 55]. The negative association between lysoalkylphosphatidylcholine species and T2D risk is in agreement with the beneficial associations of LPC(O) observed in studies of overfeeding [56] and insulin resistance [57]. Lastly, the TG(16:0/18:0/18:1) has been reported as a prominent triacylglycerol species that discriminated between cases and controls of diabetes in the Framingham Offspring Study participants [58].
Two important findings from our study should be highlighted. First, it is instructive that the majority of the significant lipid species enlisted in (Additional file 1: Figure S4A) contained palmitate or stearate moieties both of which are preferred substrates for stearoyl-CoA desaturase [59] (SCD) – an enzyme involved in converting saturated to unsaturated fatty acids. This observation points towards a possibility of the derangement of SCD pathway as a forerunner of T2D. Interestingly, the EPIC-InterAct study [60] also found that the group of saturated fatty acids comprising myristic acid, palmitic acid and stearic acid is associated with a significantly increased risk of T2D. Second, since there is a strong genetic basis to T2D it is conceivable that the LRS may share some genetic influences with T2D. Genetic studies in future need to establish putative overlaps that can partly explain the genetics of T2D. Such studies need to carefully dissect out the potential associations of genetic variants, epigenetic infrastructure and gene expression on T2D susceptibility through an altered lipidomic signature.
Conclusions
The lipidomic risk score based on plasma concentration of three, non-redundant plasma lipid species is an independent and additive biomarker of incident T2D. In the context of biomarkers for cardiovascular risk, Hlatky et al. [61] have suggested that “a novel risk marker should be evaluated in several phases, including initial proof of concept, prospective validation in independent populations, documentation of incremental information when added to standard risk markers, assessment of effects on patient management and outcomes, and ultimately, cost-effectiveness.” Our study extends this concept to T2D risk-stratification and demonstrates strong evidence for plasma LRS as a biomarker for future T2D.The strengths of our study include the scale of lipidomic profiling, prospective evaluation, two well-characterized cohorts, replicability of the accurate and independent value of LRS, its clinical value as assessed by IDI and NRI, and the cost-effectiveness analyses. The global epidemic of T2D beckons a combination of preventive strategies aimed at its control [62]. Considering the immense costs of diabetes management, it is incumbent upon us to evaluate effective preventive strategies which will hinge heavily on accurate and early detection [1, 63]. In that vein and to that end, our study suggests that while dysglycemia remains a direct measure of T2D, other subtle metabolic changes may precede it and methods aimed at detecting these early signals can play a significant role in the prevention and control of T2D.
Ethics approval and consent to participate
The Institutional Review Board of the University of Texas Health Science Center at San Antonio approved the SAFHS study and informed consent was obtained from all the study participants. The lipidomic analysis of the AusDiab study was approved by the Alfred Hospital, Ethics Committee, Project No: 104/10.
Availability of data and materials
The data are available with JEC, JB and PJM and will be shared upon request.
Acknowledgements
The authors would like to acknowledge the late Dr. Jeremy Jowett. Dr. Jowett was a very close collaborator and colleague for many years and was a key member of the research team at Baker IDI in Melbourne Australia. He helped foster the initial lipidomic collaboration between scientists in the Metabolomics group at Baker IDI and scientists at UTRGV, and worked closely as part of this collaboration until his death in September of 2012.
We gratefully acknowledge the expert help with health economic analyses provided by Professor Paul Scuffham, Professor of Health Economics, School of Medicine, Griffith University, Australia.
We are also very grateful to the participants of the San Antonio Family Heart Study for their continued involvement in our research programs. The AusDiab study co-coordinated by the Baker IDI Heart and Diabetes Institute, gratefully acknowledges the support and assistance given by: K Anstey, B Atkins, B Balkau, E Barr, A Cameron, S Chadban, M de Courten, D Dunstan, A Kavanagh, S Murray, N Owen, K Polkinghorne, T Welborn, and all the study participants. Also, for funding or logistical support, the AusDiab investigators are grateful to: Australian Government Department of Health and Ageing, Abbott Australasia Pty Ltd, Alphapharm Pty Ltd, Amgen Australia, AstraZeneca, Bristol-Myers Squibb, City Health Centre-Diabetes Service-Canberra, Department of Health and Community Services - Northern Territory, Department of Health and Human Services – Tasmania, Department of Health – New South Wales, Department of Health – Western Australia, Department of Health – South Australia, Department of Human Services – Victoria, Diabetes Australia, Diabetes Australia Northern Territory, Eli Lilly Australia, Estate of the Late Edward Wilson, GlaxoSmithKline, Jack Brockhoff Foundation, Janssen-Cilag, Kidney Health Australia, Marian & FH Flack Trust, Menzies Research Institute, Merck Sharp & Dohme, Novartis Pharmaceuticals, Novo Nordisk Pharmaceuticals, Pfizer Pty Ltd, Pratt Foundation, Queensland Health, Roche Diagnostics Australia, Royal Prince Alfred Hospital, Sydney, Sanofi Aventis, sanofi-synthelabo, and the Victorian Government’s OIS Program.
Funding
This work was supported in part by National Institutes of Health (NIH) grants R01 DK082610 and R01 DK079169; by NIH grants R01 HL045522, R01 MH078143, R01 MH078111 and R01 MH083824 (SAFHS data collection) and by NIH grant R37 MH059490 (analytical methods and software used). The AT&T Genomics Computing Center supercomputing facilities used for this work were supported in part by a gift from the AT&T Foundation with support from the National Center for Research Resources Grant Number S10 RR029392. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program grants C06 RR013556 and C06 RR017515 from the National Center for Research Resources of the National Institutes of Health. The AusDiab cohort was supported by funding from the Dairy Health and Nutrition Consortium, Australia, The National Health and Medical Research Council of Australia #233200 and #1007544, the OIS Program of the Victorian Government, Australia and by Award Number 1R01DK088972-01 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, USA.
Abbreviations
- AIC
Akaike information criterion
- AUC
area under curve
- EV
expected value
- FDR
false discovery rate
- FPG
fasting plasma glucose
- FPI
fasting plasma insulin
- HOMA-IR
homeostatic model of assessment – insulin resistance
- HR
high-risk
- ICER
incremental cost-effectiveness ratio
- IDI
integrated discrimination improvement
- IFG
impaired fasting glucose
- IGT
impaired glucose tolerance
- LR
low risk
- LRS
lipidomic risk score
- MR
moderate risk
- NRI
net reclassification index
- OGTT
oral glucose tolerance test
- QALY
quality-adjusted life year
- QUICKI
quantitative insulin sensitivity check index
- ROC
receiver operating characteristic curve
- SAFHS
San Antonio Family Heart Study
- T2D
Type 2 diabetes
- WTP
willingness-to-pay
Additional files
This file contains Supplementary Figures S1-S7 and Supplementary Tables S1-S9. (DOCX 427 kb)
Footnotes
Peter J. Meikle and Joanne E. Curran jointly coordinated this research study.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JB, JC and PJM designed and conceptualized the study. MM and HK conducted analyses and wrote the manuscript. SAFHS coauthors: CB (conducted experiments); TDD, LA, MCM, AGC, SW-B. (technical support and conceptual advice); and RD (data collection). AusDiab coauthors: JMW and CKB (conducted experiments and analyses); GW (analyses); DJM (conceptual advice); PZ and JS (study design, data collection and conceptual advice). DCG contributed to SAFHS data collection. PJM and JEC jointly coordinated the study. All authors reviewed and approved the manuscript. JEC and JB are the guarantors for this study.
Contributor Information
Manju Mamtani, Phone: (956) 882 7511, Email: manju.mamtani@utrgv.edu.
Hemant Kulkarni, Email: hemant.kulkarni@utrgv.edu.
Gerard Wong, Email: gerard_wong@sics.a-star.edu.sg.
Jacquelyn M. Weir, Email: jacqui.weir@bakeridi.edu.au
Christopher K. Barlow, Email: chris.barlow@monash.edu
Thomas D. Dyer, Email: thomas.dyer@utrgv.edu
Laura Almasy, Email: laura.almasy@utrgv.edu.
Michael C. Mahaney, Email: michael.mahaney@utrgv.edu
Anthony G. Comuzzie, Email: tony@txbiomedgenetics.org
David C. Glahn, Email: david.glahn@yale.edu
Dianna J. Magliano, Email: dianna.magliano@bakeridi.edu.au
Paul Zimmet, Email: paul.zimmet@bakeridi.edu.au.
Jonathan Shaw, Email: jonathan.shaw@bakeridi.edu.au.
Sarah Williams-Blangero, Email: sarah.williams-blangero@utrgv.edu.
Ravindranath Duggirala, Email: ravindranath.duggirala@utrgv.edu.
John Blangero, Email: john.blangero@utrgv.edu.
Peter J. Meikle, Email: peter.meikle@bakeridi.edu.au
Joanne E. Curran, Email: joanne.curran@utrgv.edu
References
- 1.Herder C, Kowall B, Tabak AG, Rathmann W. The potential of novel biomarkers to improve risk prediction of type 2 diabetes. Diabetologia. 2014;57:16–29. doi: 10.1007/s00125-013-3061-3. [DOI] [PubMed] [Google Scholar]
- 2.Oresic M, Hanninen VA, Vidal-Puig A. Lipidomics: a new window to biomedical frontiers. Trends Biotechnol. 2008;26:647–652. doi: 10.1016/j.tibtech.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Meikle PJ, Wong G, Barlow CK, Kingwell BA. Lipidomics: potential role in risk prediction and therapeutic monitoring for diabetes and cardiovascular disease. Pharmacol Ther. 2014;143:12–23. doi: 10.1016/j.pharmthera.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quehenberger O, Dennis EA. The human plasma lipidome. N Engl J Med. 2011;365:1812–1823. doi: 10.1056/NEJMra1104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotronen A, Velagapudi VR, Yetukuri L, Westerbacka J, Bergholm R, Ekroos K, Makkonen J, Taskinen MR, Oresic M, Yki-Jarvinen H. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia. 2009;52:684–690. doi: 10.1007/s00125-009-1282-2. [DOI] [PubMed] [Google Scholar]
- 7.Meikle PJ, Wong G, Barlow CK, Weir JM, Greeve MA, Macintosh GL, Almasy L, Comuzzie AG, Mahaney MC, Kowalczyk A, et al. Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS One. 2013;8:e74341. doi: 10.1371/journal.pone.0074341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacCluer JW, Stern MP, Almasy L, Atwood LA, Blangero J, Comuzzie AG, Dyke B, Haffner SM, Henkel RD, Hixson JE, et al. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr Rev. 1999;57:S59–65. doi: 10.1111/j.1753-4887.1999.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 9.Voruganti VS, Lopez-Alvarenga JC, Nath SD, Rainwater DL, Bauer R, Cole SA, Maccluer JW, Blangero J, Comuzzie AG. Genetics of variation in HOMA-IR and cardiovascular risk factors in Mexican-Americans. J Mol Med (Berl) 2008;86:303–311. doi: 10.1007/s00109-007-0273-3. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.CIR.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 11.Dunstan DW, Zimmet PZ, Welborn TA, Cameron AJ, Shaw J, de Courten M, Jolley D, McCarty DJ. The Australian Diabetes, Obesity and Lifestyle Study (AusDiab)--methods and response rates. Diabetes Res Clin Pract. 2002;57:119–129. doi: 10.1016/S0168-8227(02)00025-6. [DOI] [PubMed] [Google Scholar]
- 12.Ascaso JF, Pardo S, Real JT, Lorente RI, Priego A, Carmena R. Diagnosing insulin resistance by simple quantitative methods in subjects with normal glucose metabolism. Diabetes Care. 2003;26:3320–3325. doi: 10.2337/diacare.26.12.3320. [DOI] [PubMed] [Google Scholar]
- 13.Katz A, Nambi SS, Mather K, Baron AD, Follmann DA, Sullivan G, Quon MJ. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab. 2000;85:2402–2410. doi: 10.1210/jcem.85.7.6661. [DOI] [PubMed] [Google Scholar]
- 14.McAuley KA, Williams SM, Mann JI, Walker RJ, Lewis-Barned NJ, Temple LA, Duncan AW. Diagnosing insulin resistance in the general population. Diabetes Care. 2001;24:460–464. doi: 10.2337/diacare.24.3.460. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Jaramillo P, Gomez-Arbelaez D, Lopez-Lopez J, Lopez-Lopez C, Martinez-Ortega J, Gomez-Rodriguez A, Triana-Cubillos S. The role of leptin/adiponectin ratio in metabolic syndrome and diabetes. Horm Mol Biol Clin Investig. 2014;18:37–45. doi: 10.1515/hmbci-2013-0053. [DOI] [PubMed] [Google Scholar]
- 16.Kent JW, Jr, Comuzzie AG, Mahaney MC, Almasy L, Rainwater DL, VandeBerg JL, MacCluer JW, Blangero J. Intercellular adhesion molecule-1 concentration is genetically correlated with insulin resistance, obesity, and HDL concentration in Mexican Americans. Diabetes. 2004;53:2691–2695. doi: 10.2337/diabetes.53.10.2691. [DOI] [PubMed] [Google Scholar]
- 17.Comuzzie AG, Mitchell BD, Cole S, Martin LJ, Hsueh WC, Rainwater DL, Almasy L, Stern MP, Hixson J, MacCluer JW, Blangero J. The genetics of obesity in Mexican Americans: the evidence from genome scanning efforts in the San Antonio family heart study. Hum Biol. 2003;75:635–646. doi: 10.1353/hub.2003.0073. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB, Bellis C, Dyer TD, Johnson MP, Rainwater DL, et al. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension. 2013;62:621–626. doi: 10.1161/HYPERTENSIONAHA.113.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB, Bellis C, Dyer TD, Johnson MP, Rainwater DL, et al. Variability in associations of phosphatidylcholine molecular species with metabolic syndrome in Mexican-American families. Lipids. 2013;48:497–503. doi: 10.1007/s11745-013-3781-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamtani M, Meikle PJ, Kulkarni H, Weir JM, Barlow CK, Jowett JB, Bellis C, Dyer TD, Almasy L, Mahaney MC, et al. Plasma dihydroceramide species associate with waist cicrumference in Mexican American families. Obesity. 2014;22:950–6. doi: 10.1002/oby.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir JM, Wong G, Barlow CK, Greeve MA, Kowalczyk A, Almasy L, Comuzzie AG, Mahaney MC, Jowett JB, Shaw J, et al. Plasma lipid profiling in a large population-based cohort. J Lipid Res. 2013;54:2898–2908. doi: 10.1194/jlr.P035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meikle PJ, Wong G, Tsorotes D, Barlow CK, Weir JM, Christopher MJ, MacIntosh GL, Goudey B, Stern L, Kowalczyk A, et al. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler Thromb Vasc Biol. 2011;31:2723–2732. doi: 10.1161/ATVBAHA.111.234096. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J. Mixed-effects Cox models of alcohol dependence in extended families. BMC Genet. 2005;6(Suppl 1):S127. doi: 10.1186/1471-2156-6-S1-S127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Therneau TM. coxme: Mixed Effects Cox Models., R package 2.2-3 edition; 2012. https://cran.r-project.org/web/packages/coxme/index.html.
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 26.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 27.Hanley JA, McNeil BJ. Maximum attainable discrimination and the utilization of radiologic examinations. J Chronic Dis. 1982;35:601–611. doi: 10.1016/0021-9681(82)90012-1. [DOI] [PubMed] [Google Scholar]
- 28.Hunt KJ, Duggirala R, Goring HH, Williams JT, Almasy L, Blangero J, O’Leary DH, Stern MP. Genetic basis of variation in carotid artery plaque in the San Antonio Family Heart Study. Stroke. 2002;33:2775–2780. doi: 10.1161/01.STR.0000043827.03966.EF. [DOI] [PubMed] [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 30.Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson FR, Manjunath R, Mansfield CA, Clayton LJ, Hoerger TJ, Zhang P. High-risk individuals’ willingness to pay for diabetes risk-reduction programs. Diabetes Care. 2006;29:1351–1356. doi: 10.2337/dc05-2221. [DOI] [PubMed] [Google Scholar]
- 32.Herman WH, Edelstein SL, Ratner RE, Montez MG, Ackermann RT, Orchard TJ, Foulkes MA, Zhang P, Saudek CD, Brown MB, Diabetes Prevention Program Research G Effectiveness and cost-effectiveness of diabetes prevention among adherent participants. Am J Manag Care. 2013;19:194–202. [PMC free article] [PubMed] [Google Scholar]
- 33.Group DPPR. The 10-year cost-effectiveness of lifestyle intervention or metformin for diabetes prevention: an intent-to-treat analysis of the DPP/DPPOS. Diabetes Care. 2012;35:723–730. doi: 10.2337/dc11-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.NICE . Guide to the methods of technology appraisal. London: NICE; 2004. [Google Scholar]
- 35.Brouwer WB, Niessen LW, Postma MJ, Rutten FF. Need for differential discounting of costs and health effects in cost effectiveness analyses. BMJ. 2005;331:446–448. doi: 10.1136/bmj.331.7514.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliver A. A normative perspective on discounting health outcomes. J Health Serv Res Policy. 2013;18:186–189. doi: 10.1177/1355819613485671. [DOI] [PubMed] [Google Scholar]
- 37.Claxton K, Paulden M, Gravelle H, Brouwer W, Culyer AJ. Discounting and decision making in the economic evaluation of health-care technologies. Health Econ. 2011;20:2–15. doi: 10.1002/hec.1612. [DOI] [PubMed] [Google Scholar]
- 38.Claxton K, Sculpher M, Culyer A, McCabe C, Briggs A, Akehurst R, Buxton M, Brazier J. Discounting and cost-effectiveness in NICE - stepping back to sort out a confusion. Health Econ. 2006;15:1–4. doi: 10.1002/hec.1081. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan SD, Garrison LP, Jr, Rinde H, Kolberg J, Moler EJ. Cost-effectiveness of risk stratification for preventing type 2 diabetes using a multi-marker diabetes risk score. J Med Econ. 2011;14:609–616. doi: 10.3111/13696998.2011.602160. [DOI] [PubMed] [Google Scholar]
- 40.Zhang P, Engelgau MM, Valdez R, Benjamin SM, Cadwell B, Narayan KM. Costs of screening for pre-diabetes among US adults: a comparison of different screening strategies. Diabetes Care. 2003;26:2536–2542. doi: 10.2337/diacare.26.9.2536. [DOI] [PubMed] [Google Scholar]
- 41.Group DPPR. Within-trial cost-effectiveness of lifestyle intervention or metformin for the primary prevention of type 2 diabetes. Diabetes Care. 2003;26:2518–2523. doi: 10.2337/diacare.26.9.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uno H. survC1: C-statistics for risk prediction models with censored survival data., R package version 1.0-2 edition; 2013. https://mran.revolutionanalytics.com/package/survC1/.
- 44.Uno H, Cai T. survIDINRI: IDI and NRI for comparing competing risk prediction models with censored survival data. 2013. https://mran.revolutionanalytics.com/package/survIDINRI/.
- 45.Stern MP, Williams K, Haffner SM. Identification of persons at high risk for type 2 diabetes mellitus: do we need the oral glucose tolerance test? Ann Intern Med. 2002;136:575–581. doi: 10.7326/0003-4819-136-8-200204160-00006. [DOI] [PubMed] [Google Scholar]
- 46.Batsis JA, Nieto-Martinez RE, Lopez-Jimenez F. Metabolic syndrome: from global epidemiology to individualized medicine. Clin Pharmacol Ther. 2007;82:509–524. doi: 10.1038/sj.clpt.6100355. [DOI] [PubMed] [Google Scholar]
- 47.Haffner SM, Miettinen H. Insulin resistance implications for type II diabetes mellitus and coronary heart disease. Am J Med. 1997;103:152–162. doi: 10.1016/S0002-9343(97)00027-2. [DOI] [PubMed] [Google Scholar]
- 48.Zimmet P, Dowse G, Bennett P. Hyperinsulinaemia is a predictor of non-insulin-dependent diabetes mellitus. Diabete Metab. 1991;17:101–108. [PubMed] [Google Scholar]
- 49.Echouffo-Tcheugui JB, Ali MK, Griffin SJ, Narayan KM. Screening for type 2 diabetes and dysglycemia. Epidemiol Rev. 2011;33:63–87. doi: 10.1093/epirev/mxq020. [DOI] [PubMed] [Google Scholar]
- 50.Hoerger TJ, Hicks KA, Sorensen SW, Herman WH, Ratner RE, Ackermann RT, Zhang P, Engelgau MM. Cost-effectiveness of screening for pre-diabetes among overweight and obese U.S. adults. Diabetes Care. 2007;30:2874–2879. doi: 10.2337/dc07-0885. [DOI] [PubMed] [Google Scholar]
- 51.Ackermann RT, Edelstein SL, Narayan KM, Zhang P, Engelgau MM, Herman WH, Marrero DG, Diabetes Prevention Program Research G Changes in health state utilities with changes in body mass in the Diabetes Prevention Program. Obesity (Silver Spring) 2009;17:2176–2181. doi: 10.1038/oby.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asche CV, Hippler SE, Eurich DT. Review of models used in economic analyses of new oral treatments for type 2 diabetes mellitus. Pharmacoeconomics. 2014;32:15–27. doi: 10.1007/s40273-013-0117-7. [DOI] [PubMed] [Google Scholar]
- 53.Lahiri S, Futerman AH. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J Biol Chem. 2005;280:33735–33738. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 54.Frangioudakis G, Diakanastasis B, Liao BQ, Saville JT, Hoffman NJ, Mitchell TW, Schmitz-Peiffer C. Ceramide accumulation in L6 skeletal muscle cells due to increased activity of ceramide synthase isoforms has opposing effects on insulin action to those caused by palmitate treatment. Diabetologia. 2013;56:2697–2701. doi: 10.1007/s00125-013-3035-5. [DOI] [PubMed] [Google Scholar]
- 55.Frangioudakis G, Garrard J, Raddatz K, Nadler JL, Mitchell TW, Schmitz-Peiffer C. Saturated- and n-6 polyunsaturated-fat diets each induce ceramide accumulation in mouse skeletal muscle: reversal and improvement of glucose tolerance by lipid metabolism inhibitors. Endocrinology. 2010;151:4187–4196. doi: 10.1210/en.2010-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heilbronn LK, Coster AC, Campbell LV, Greenfield JR, Lange K, Christopher MJ, Meikle PJ, Samocha-Bonet D. The effect of short-term overfeeding on serum lipids in healthy humans. Obesity (Silver Spring) 2013;21:E649–659. doi: 10.1002/oby.20508. [DOI] [PubMed] [Google Scholar]
- 57.Nestel PJ, Straznicky N, Mellett NA, Wong G, De Souza DP, Tull DL, Barlow CK, Grima MT, Meikle PJ. Specific plasma lipid classes and phospholipid fatty acids indicative of dairy food consumption associate with insulin sensitivity. Am J Clin Nutr. 2014;99:46–53. doi: 10.3945/ajcn.113.071712. [DOI] [PubMed] [Google Scholar]
- 58.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sampath H, Ntambi JM. The fate and intermediary metabolism of stearic acid. Lipids. 2005;40:1187–1191. doi: 10.1007/s11745-005-1484-z. [DOI] [PubMed] [Google Scholar]
- 60.Forouhi NG, Koulman A, Sharp SJ, Imamura F, Kroger J, Schulze MB, Crowe FL, Huerta JM, Guevara M, Beulens JW, et al. Differences in the prospective association between individual plasma phospholipid saturated fatty acids and incident type 2 diabetes: the EPIC-InterAct case-cohort study. Lancet Diabetes Endocrinol. 2014;2:810–818. doi: 10.1016/S2213-8587(14)70146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hlatky MA, Greenland P, Arnett DK, Ballantyne CM, Criqui MH, Elkind MS, Go AS, Harrell FE, Jr, Hong Y, Howard BV, et al. Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119:2408–2416. doi: 10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmet PZ, Magliano DJ, Herman WH, Shaw JE. Diabetes: a 21st century challenge. The Lancet Diabetes and Endocrinology. 2014;2:56–64. doi: 10.1016/S2213-8587(13)70112-8. [DOI] [PubMed] [Google Scholar]
- 63.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O’Sullivan J, Cheng S, Rhee EP, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123:4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available with JEC, JB and PJM and will be shared upon request.


