Abstract
The translocator protein, a microglial-expressed marker of neuroinflammation, has been implicated in Alzheimer’s disease, which is characterized by alterations in vascular and inflammatory states. A TSPO variant, rs6971, determines binding affinity of exogenous radioligands in vivo; however, the effect of these altered binding characteristics on inflammatory and cerebrovascular biomarkers has not been assessed. In 2345 living subjects (Alzheimer’s Disease Neuroimaging Initiative, n = 1330) and postmortem brain samples (Religious Orders Study and Memory and Aging Project, n = 1015), we analyzed effects of rs6971 on white matter hyperintensisites, cerebral infarcts, circulating inflammatory biomarkers, amyloid angiopathy, and microglial activation. We found that rs6971 does not alter translocator protein in a way that impacts cerebrovascular and inflammatory states known to be affected in dementia.
Keywords: Microglia, Alzheimer’s, MRI, genetics, risk factors, inflammation, white matter disease
Introduction
Late-onset Alzheimer’s disease (AD) is characterized by the accumulation of Aβ plaques and neurofibrillary tangles, chronic inflammation, and progressive neurodegeneration.1–4 The inflammation hypothesis of AD posits that chronic over-activation of microglia in response to the buildup of amyloid pathology results in a positive feedback loop whereby pro-inflammatory conditions sustain the elevated expression of factors that further exacerbate amyloidogenesis.1,5–7 In addition to the damaging effects of “traditional” neuroinflammation, defined by the activation of resident microglia and infiltration of peripheral monocytes, the immune response also serves to protect against neurodegeneration; studies show that increased expression of certain inflammatory cytokines may reduce Aβ deposition8 and promote cell survival and neurovascularization.4,9
The convergence of inflammation and cerebrovascular pathology in AD is becoming increasingly clear, yet the mechanisms behind this convergence are poorly understood. It is known that cerebral ischemia is a strong activator of the immune response,10,11 and that, in parallel, vascular irregularities are themselves risk factors for AD.12,13 Aβ deposition within neurovasculature, known as cerebral amyloid angiopathy (CAA), may contribute to brain ischemia and lead to cognitive impairment.14,15 Cerebral infarcts are regions of ischemic damage associated with aging and present to a greater degree in individuals who develop AD than in those who do not.16 The presence of both macro- and micro cerebral infarcts, caused largely by cerebral small vessel disease (SVD),17 have been shown to influence risk for AD as well as global cognitive performance,18–20 and may be a consequence of accumulating cerebrovascular insults (and accompanying hypoperfusion21) that are implicated in neurodegenerative processes.22 At the neuroimaging level, white matter lesions associated with SVD can be imaged in vivo as regions of increased signal on T2-weighted MR images; these white matter hyperintensities (WMHs) have been associated with increased age, the presence of cerebral infarcts, multiple vascular disease risk factors, AD and mild cognitive impairment (MCI) diagnosis, and the progression from MCI to AD.23,24
At the molecular genetic level, the translocator protein 18 kDa (TSPO) has recently become a focus of interest in the investigation of psychiatric and neurological illnesses with potential immune mechanisms of illness. TSPO is expressed in activated microglia and used as a biomarker for neuroinflammation25,26; it is found in the outer mitochondrial membrane where it is thought to regulate cholesterol transport, steroidogenesis, and apoptosis.27 In vivo PET imaging studies using TSPO radioligands show increased binding in patients with acute brain injury,28 multiple sclerosis,29–33 MCI,34 and AD.25,35 A single genetic polymorphism located in exon 4 of the TSPO gene, rs6971 (Ala147Thr), reliably determines the binding affinity of second generation TSPO radioligands in the brain,36,37 where A/A, A/G, and G/G genotypes correspond to high, medium, and low-affinity binding phenotypes (herein referred to as HABs, MABs, and LABs, respectively). It is hypothesized that the Alanine to Threonine substitution at position 147 results in a conformational change in TSPO structure that influences its interaction with ligands.36,38,39 Differences in binding affinity may have important implications for the etiopathology of AD; TSPO ligands have been shown to ameliorate neuroinflammation in vitro,40 reverse neuropathology and behavioral decline in AD mouse models,41 reduce Aβ42-induced neurodegeneration in drosophila,42 as well as confer neuroprotective and regenerative effects in vivo and in vitro.43–46
Despite this potential involvement of TSPO in the etiopathology of neurodegenerative disorders, and the well-known links between inflammation, cerebrovascular disease, and AD, no studies to date have investigated the effect of rs6971 on MRI-based phenotypes or inflammatory biomarkers. We therefore sought to test whether TSPO rs6971 genotype was associated with in vivo structural imaging measures of cerebrovascular disease and neuroinflammation in two large clinical samples (the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the Religious Orders Study/Memory and Aging Project (ROS/MAP)). To identify potential mechanisms of action, we also analyzed the effect of genotype on levels of plasma inflammatory biomarkers in living subjects, as well as on cerebral infarcts, CAA, and densities of active microglia from postmortem brain tissue. Due to the anti-inflammatory action of TSPO ligands,47 we hypothesize that that low-affinity binding groups would have exacerbated pathology and increased levels of pro-inflammatory biomarkers vs. medium- and high-affinity binding groups, as determined by genotype.
Materials and methods
Subject characteristics and genetics
ADNI
ADNI is a large multicenter collaboration established in 2003, in which elderly subjects at various stages of cognitive impairment are assessed longitudinally for multimodal imaging, neuropsychiatric test performance, and fluid biomarkers. Details on subject recruitment and inclusion/exclusion criteria are reported elsewhere.48 A total of 699 individuals from ADNI 1 and 631 individuals from ADNI GO and 2, all self-reported Caucasian, were included in analyses (ntotal = 1330; see Table 1 for details). Data were extracted from the ADNI website (http://adni.loni.usc.edu). ADNI 1 subjects were genotyped for rs6971 using the Quad 610 BeadChip (Illumina Inc., San Diego, CA, USA), and ADNI GO and 2 subjects were genotyped using the HumanOmniExpress BeadChip (Illumina Inc., San Diego, CA, USA). APOE ɛ4 status was obtained separately by genotyping rs429358 and rs7412, according to previously published methods.49 ADNI ethics approval was obtained for each institution involved including the Research Ethics Board at the Centre for Addiction and Mental Health (Toronto, Canada). All aspects of the study were performed in accordance with the Declaration of Helsinki, US 21CFR Part 50 – Protection of Human Subjects, and Part 56 – Institutional Review Boards, and federal HIPAA regulations. All subjects provided written informed consent.
Table 1.
Summary statistics for ADNI samples by diagnosis.
| ADNI 1 (n = 699) | CN (n = 195) | SMC | EMCI | LMCI (n = 338) | AD (n = 166) | Diff P |
|---|---|---|---|---|---|---|
| WMHva (SD) | 0.8 (2) | – | – | 0.9 (3) | 1.3 (3) | 0.07 |
| CI (+/−)b | 18+,177− | – | – | 30+,308− | 13+,153− | 0.90 |
| Age, Y (SD) | 75 (7) | – | – | 75 (7) | 76 (7) | 0.22 |
| Sex (F/M) | 88 F, 107 M | – | – | 117 F, 221 M | 75 F, 91 M | 0.018 |
| BP (systolic) | 133 (15) | – | – | 133 (17) | 135 (18) | 0.35 |
| BP (diastolic) | 73 (10) | – | – | 74 (10) | 73 (9) | 0.61 |
| MMSE (SD) | 29 (1) | – | – | 27 (2) | 23 (2) | <0.001 |
| Education, Y (SD) | 16 (3) | – | – | 16 (3) | 15 (3) | <0.001 |
| APOE ɛ4 status (+/−) | 53+,142− | – | – | 185+,153− | 112+,54− | <0.001 |
| rs6971 genotype (LAB/MAB/HAB) | 20/88/87 | – | – | 25/135/178 | 18/66/82 | 0.33 |
| ADNI GO and 2 (n = 631) | (n = 129) | (n = 40) | (n = 242) | (n = 122) | (n = 98) | Diff P |
| WMHvc (SD) | 6.8 (13) | 8.5 (11) | 7.3 (9) | 7.7 (10) | 8.7 (10) | 0.70 |
| CI (+/−)b | 6+,113−,10 NA | 40 NA | 20+, 184−, 38 NA | 6+, 75−, 41 NA | 1+, 35−, 62 NA | 0.35 |
| Age, Y (SD) | 74 (6) | 72 (5) | 71 (7) | 72 (8) | 75 (8) | <0.001 |
| Sex (F/M) | 62 F, 67 M | 27 F, 13 M | 102 F, 140 M | 52 F, 70 M | 38 F, 60 M | 0.025 |
| BP (systolic) | 134 (16) | 131 (17) | 132 (17) | 132 (18) | 131 (17) | 0.76 |
| BP (diastolic) | 74 (10) | 72 (9) | 73 (9) | 73 (10) | 74 (10) | 0.87 |
| MMSE (SD) | 29 (1) | 29 (1) | 28 (2) | 28 (2) | 23 (2) | <0.001 |
| Education, Y (SD) | 16 (3) | 17 (3) | 16 (3) | 16 (3) | 16 (3) | 0.16 |
| APOE ɛ4 status (+/−) | 34+,95− | 12+,28− | 105+,137− | 69+,53− | 66+,32− | <0.001 |
| rs6971 genotype (LAB/MAB/HAB) | 19/51/59 | 5/20/15 | 26/108/108 | 9/56/57 | 10/42/46 | 0.75 |
Volumes are in mm3, normalized to a standard space.
Sample sizes for MRI cerebral infarcts (CI) are different than those indicated in column headers, some non-overlapping subjects were evaluated for CI: ADNI1 (nCN = 196, nLMCI = 339, nAD = 167), ADNI GO and 2 (nCN = 123, nEMCI = 207, nLMCI = 83, nAD = 36). cVolumes are in mm3. Continuous variables (age, education, MMSE, and BP) were analyzed for diagnosis group differences using ANOVA (two-tailed). Factor variables (sex, CI, APOE ɛ4 status, and rs6971 genotype) were analyzed using Fisher’s exact test (two-tailed).
LAB: low affinity binders (rs6971 A/A); MAB: medium affinity binders (rs6971 A/G); HAB: high affinity binders (rs6971 G/G); MAF: minor allele frequency; WMHv: white matter hyperintensity volume; CI: cerebral infarcts detected by MRI; MMSE: mini mental status examination; APOE: Apolipoprotein E; BP: blood pressure (mmHg); SD: standard deviation; M: male; F: female; Y: years; CN: cognitively normal; SMC: some memory concern; EMCI: early mild cognitive impairment; LMCI: late mild cognitive impairment; AD: Alzheimer’s disease.
ROS/MAP
ROS and MAP are community-based cohort studies of aging and dementia. Participants in ROS are older nuns, priests, and brothers from across the USA,50 and those in MAP are older residents of northeastern Illinois.51 Both studies were approved by the Institutional Review Board of Rush University Medical Center in compliance with the Health Insurance Portability and Accountability Act and enroll older persons without dementia who agree to annual evaluation and autopsy. All participants provided written informed consent. Follow-up rates exceed 90% and autopsy rates exceed 85%. A neuroimaging substudy was initiated in 2009.52 A total of 1015 postmortem brains were available for analysis at time of study. Full details on sample characterization and assessments have been previously published53 (supplementary methods). All subjects were genotyped for rs6971 using the Affymetrix (Santa Clara, CA, USA) Genechip 6.0 platform. APOE (rs7412 and rs429358) genotypes were imputed from MACH (version 1.0.16a) and HapMap release 22 CEU (build 36), as previously described.54
In vivo neuroimaging
ADNI
Structural MR images (including T1, T2, PD) were acquired for all subjects using a standard protocol on 1.5T scanners across multiple sites. Correction for gradient non-linearity was performed using gradwarp and standardization across sites and platforms was performed as previously published.55 The presence of cerebral infarcts was evaluated for 1151 subjects in ADNI 1, GO, and 2 using the same method, outlined by De Carli et al. on the ADNI website56 (supplementary methods). For ADNI 1, WMHv estimates were derived from T1, T2, and PD images using a method described elsewhere57,58 (supplementary methods); for ADNI GO and 2, volumes of CSF, gray matter, white matter, and WMH were calculated from fluid attenuated inversion recovery (FLAIR) and T1-weighted images using a four-tissue Bayesian segmentation method,59,60 outlined by De Carli et al. on the ADNI website61 (supplementary methods).
ROS/MAP
A subset of subjects with genetic data (n = 291) underwent a multimodal neuroimaging protocol that included high-resolution T1-weighted magnetization-prepared rapid acquisition gradient-echo (MPRAGE) and T2-weighted FLAIR scans. Detailed scan acquisition parameters have been published elsewhere.62 WMHv estimates were extracted from scans using an automated pipeline, as described62,63 (supplementary methods).
In vivo plasma biomarkers
ADNI 1
Data were available from a subset of individuals from ADNI 1 (n = 520) who contributed plasma aliquots for proteomic analysis using the 190-analyte Luminex xMAP platform by Rules-Based Medicine (Myriad RBM, Austin, TX, USA) (supplementary methods). Concentrations of five inflammatory biomarkers were available that were also assayed in the ROS/MAP sample: tumor necrosis factor alpha (TNFα), interleukin-6 receptor (IL6R), C-reactive protein (CRP), vascular cell adhesion protein 1 (VCAM1, CD106), and matrix metallopeptidase 9 (MMP-9).
ROS/MAP
A subset of subjects (n = 394) had blood drawn and plasma inflammatory biomarkers quantified as previously described.62 Highly sensitive multiplexed sandwich ELISA arrays (Endogen Searchlight technologies, Billerica, MA) were used to detect plasma concentrations of five inflammatory proteins, listed above.
Postmortem neuropathology, ROS/MAP
Neuropathological evaluation
Brain autopsies were conducted at predetermined sites across the United States and neuropathological examinations were performed at Rush University Medical Centre. Full details of autopsy and pathological evaluation procedures have been previously published.18,64 Micro and Macro cerebral infarcts were quantified from fixed brain slabs using hematoxylin/eosin staining (supplementary methods) and coded as either present (one or more infarcts) or absent, as previously reported.19 Degree of cerebral amyloid angiopathy (CAA; none, mild, moderate, and severe) was assessed using immunohistochemical labeling as previously described65 (supplementary methods). A subset of brains were also analyzed for quantification of microglia at three levels of activation, across four brain regions (inferior temporal, midfrontal, posterior putamen, and ventral medial caudate), as described66 (supplementary methods). Average density (across all stages of activation), as well as the square root-transformed ratio of most active (stage 3) to least active microglia (stage 1), was calculated for each region.
Statistical analysis
All analyses were performed using R statistical software (version 3.0.2). Dichotomous outcomes (presence or absence of cerebral infarcts both in vivo and postmortem) were tested using logistic regression, with TSPO genotype (additive model), sex, age, diastolic blood pressure (hypertension y/n in ROS/MAP), education (total years), and APOE ɛ4 status as predictors. Continuous outcomes (WMHv, plasma biomarkers, microglial density) were modeled similarly using linear regression. Due to important differences in WMHv estimation methods between ADNI 1, ADNI GO and 2, and ROS/MAP, each dataset was analyzed separately. Outcomes were tested for normality using the Shapiro-Wilk test67 and corrected, if necessary (Shapiro-Wilk P < 0.01), using Box–Cox power transformations.68 Since the ADNI 2 FLAIR-derived WMHv estimates were not normalized to a standard space, models of this data also co-varied for total intracranial volume. Ordinal regression was used to evaluate the degree of amyloid angiopathy using the same modeling approach and set of covariates as above. Due to their biologically related functions in cholesterol transport69 and TSPO’s ability to up-regulate APOE,70 interactions between APOE ɛ4 status and TSPO genotype were also tested in each model. For all analyses, P-values are two-tailed and reported as Bonferroni-corrected (Pcor) or uncorrected (Praw).
Data handling
All analyses in this study were performed using de-identified data from centrally curated databases. ADNI data are stored and maintained in the Laboratory of Neuro Imaging (LONI) Image Data Archive (IDA) at University of Southern California (Los Angeles, CA, USA). Clinical, imaging, and genetic data, ROS/MAP data are stored and maintained at the Rush Alzheimer’s Disease Center (RADC) at the Rush University Medical Center (Chicago, IL, USA). For clinical evaluation in ROS/MAP, a PhD level neuropsychologist with expertise in late-life cognitive function, AD and dementia, blinded to age, gender, race and clinical data other than education, occupation, information about sensory or motor deficits and effort, reviews the results, and summarizes impairment in each of five clinical domains (orientation, attention, memory, language and perception) as probable, possible or not present and renders an opinion regarding the presence of dementia and AD. For neuropathology, JAS is one of three board-certified neuropathologists involved in reviewing and confirming postmortem assessments. Microglia counts were obtained by trained research assistants and postdoctoral fellows – inter-rater reliability is excellent. For all protocols, neuropathological examiners were blind to all clinical data including but not limited to age, gender, education, and diagnosis at time of death. In ADNI, RBM was blinded to the clinical status of subjects who contributed plasma samples. For ROS/MAP, all available data on each phenotype of interest were extracted by a trained database manager and transferred securely to CAMH for statistical analyses. For ADNI, data were downloaded directly from the LONI IDA with institutional permission.
Results
In vivo cerebral infarcts and white matter hyperintensities (ADNI and ROS/MAP)
Table 1 summarizes demographic characteristics of the ADNI 1 and ADNI GO and 2 cohorts analyzed, according to TSPO rs6971 genotype.
In the ADNI 1, GO, and 2 combined Caucasian sample (n = 1151), rs6971 genotype was not associated with the presence of cerebral infarcts (Wald χ21 = 1.98, Praw = 0.16) (Figure 1). However, there was a nominal association with infarcts in the LMCI group (Wald χ21 = 3.98, Praw = 0.047), whereby likelihood of having infarcts increased stepwise with decreasing binding affinity (O.R.LAB:HAB = 2.97, C.I.95% = (1.01,8.70)). No associations were observed in the CN (Wald χ21 = 2.67, Praw = 0.10), EMCI (Wald χ21 = 0.01, Praw = 0.94), or AD group (Wald χ21 = 1.13, Praw = 0.29), though there were no AD subjects who had both infarcts and the rs6971 LAB genotype, making additive model inference in this subgroup impossible.
Figure 1.
Effects of rs6971 on cerebral infarcts in vivo and postmortem.
Odds ratios (O.R.s) for rs6971 effect on presence of cerebral infarcts in the ADNI 1, GO, and 2 combined sample (n = 1151) and ROS/MAP (n = 1015) samples. O.R.s are shown for the additive model homozygote contrast (i.e. HABs vs. LABs).
ROS/MAP: Religious Orders Study/Memory and Aging Project; ADNI: Alzheimer's Disease Neuroimaging Initiative; HAB: high-affinity binding genotype; MAB: medium-affinity binding genotype; LAB: low-affinity binding genotype.
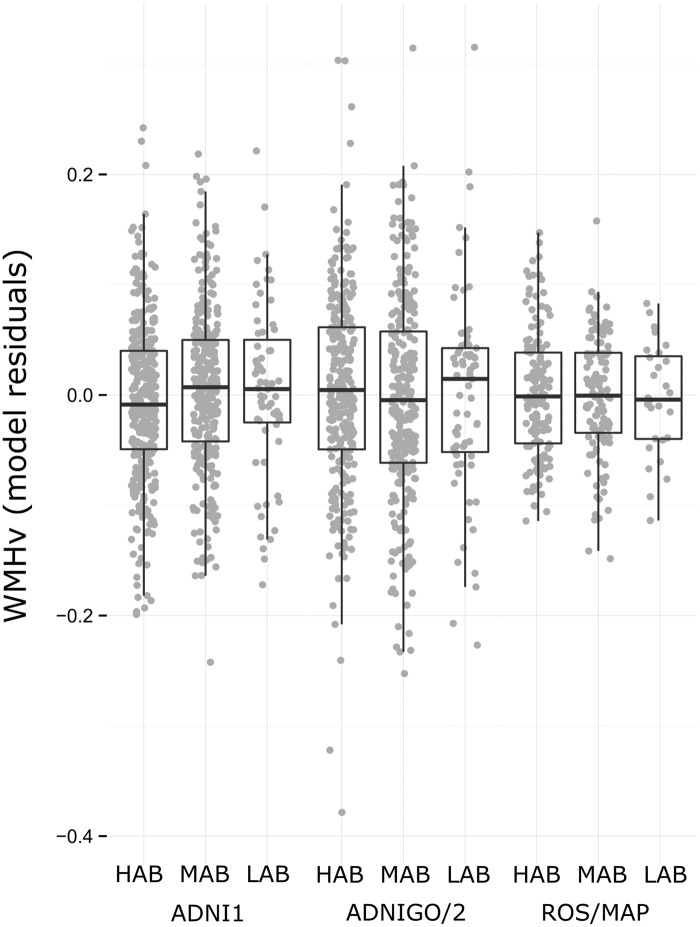
In three independent samples (ADNI 1, ADNI GO and 2, and ROS/MAP), there were no significant associations of rs6971 genotype with WMHv (Figure 2), though in ADNI 1 (n = 699), rs6971 genotype showed a trend-level association with WMHv (additive F1,692 = 2.87, Praw = 0.091). The direction of this trend in ADNI 1 was consistent with that observed in the combined sample for infarcts, with HAB subjects showing both lower risk for presence of infarcts and greater WMHv (though neither result was significant at P < 0.05). Also, in ADNI, GO, and 2 (n = 631), there was a weak association of rs6971 with WMHv in the EMCI group (n = 242, additive F1,234 = 3.55, Praw = 0.061); however, the allelic pattern of this trend was in the opposite direction as in ADNI 1, whereby LAB subjects had lower WMHv. No interactions of rs6971 genotype and APOE ɛ4 status were found (all Praw > 0.1).
Figure 2.
Effects of rs6971 on in vivo white matter hyperintensities. White matter hyperintensity volume (WMHv) plotted by rs6971 genotype group in ADNI 1 (n = 699), ADNI GO and 2 (n = 631), and ROS/MAP (n = 291) samples. WMHv is plotted as the residuals of linear models that include the following co-variates: age, sex, diastolic blood pressure (hypertension y/n for ROS/MAP), APOE ɛ4 status, education, and total intracranial volume (ADNI 2 only). No results show evidence to reject null association (all additive P > 0.05).
ROS/MAP: Religious Orders Study/Memory and Aging Project; ADNI = Alzheimer's Disease Neuroimaging Initiative; HAB: high-affinity binding genotype; MAB: medium-affinity binding genotype; LAB: low-affinity binding genotype.
Plasma inflammatory biomarkers (ADNI and ROS/MAP)
In the ADNI 1 subsample with plasma inflammatory biomarker data (n = 520), no main effect of rs6971 genotype was found for any biomarker (all Praw > 0.1) (supplementary Figure 1). Post-hoc testing revealed effects of genotype on TNFα levels in the AD subjects (n = 104, additive F1,99 = 10.92, Praw = 0.0013); however, this effect was not replicated in the ROS/MAP AD subsample (n = 85, additive F1,80 = 0.34, Praw = 0.24). Further, there were no main effects of rs6971 genotype on any biomarker in the ROS/MAP sample subset or within any diagnostic group separately, and no significant interactions of rs6971 and APOE ɛ4 status were found in either subsample (all Pcor > 0.05) (supplementary Figure 1).
Postmortem neuropathology and microglial activation (ROS/MAP)
In the whole sample (n = 1015), rs6971 was not associated with macro or micro cerebral infarcts (all Praw > 0.1, see Figure 1). No significant main effects of rs6971 genotype were found for degree of amyloid angiopathy; however, there was an uncorrected trend toward significance (additive Wald χ21 = 3.43, Praw = 0.064), whereby HAB subjects were less likely to have greater CAA severity (O.R.HAB:LAB = 0.72, C.I.95% = (0.51,1.02)) (supplementary Figure 2). This trend was seen only in the CN group (additive Wald χ21 = 3.34, Praw = 0.068); raw association P-values in the MCI and AD groups were 0.51 and 0.28, respectively. Finally, we found no associations of rs6971 with either the density of active microglia at any stage of activation nor the ratio of highly active microglia to relatively inactive microglia, in any region tested (all Praw > 0.05) (supplementary Figure 3). Interaction analyses revealed no interactions of rs6971 and APOE ɛ4 status on any postmortem pathological measure (all Praw > 0.1).
Discussion
Our study is the first to analyze variation in TSPO (rs6971) with respect to structural neuroimaging and plasma biomarkers related to neuroinflammation. To our knowledge, we are also the first to analyze the effect of rs6971 on microglial activation and postmortem neuropathology. We found no significant effects of TSPO genotype with respect to postmortem or in vivo cerebral infarcts (Figure 1). While we found a marginally protective effect (0.05 < P < 0.1) of genotype on in vivo WMH burden (ADNI 1), this finding did not replicate in either of two independent samples (ADNI GO and 2, ROS/MAP) (Figure 2). Further, we found diagnosis-dependent effects of TSPO genotype on plasma levels of TNFα (ADNI 1) that did not replicate in an independent sample (ROS/MAP). After correction for multiple testing, we found no significant association of TSPO genotype with either degree of cerebral amyloid angiopathy or microglial activation in postmortem tissue. Exploratory analyses found no interactions of TSPO and APOE genotypes.
We hypothesized that genotype-driven alterations in TSPO structure may influence its downstream neuroprotective action,43–46 thereby conferring early and enduring risk across the lifespan for atherosclerotic damage, SVD, and potentially inflammatory dysregulation. The observed trend effects of TSPO genotype on vascular phenotypes (as well as amyloid angiopathy) were most prominent in the CN subgroups of both samples; this suggests that the effects of TSPO variation may be dependent on the time course of illness, with the greatest genotype differences observed in pre-symptomatic or early stage AD subjects. Interestingly, the HAB subjects appeared to be both marginally protected from micro infarcts as well as from increased CAA severity in the ROS/MAP sample, though neither result achieved statistical significance at P < 0.05. While association between micro infarcts and severe CAA has been previously shown,71 it is possible that the level of neuropathology across MCI and AD groups in the ROS/MAP sample is not high enough to observe a similarly significant pattern of TSPO effects on these measures; ROS subjects are priests, nuns, and brothers with well-documented healthy lifestyles and greater than average lifespan.72
In our ADNI 1 analysis of inflammatory biomarkers, we found a diagnosis-dependent effect of TSPO genotype on TNFα, whereby HABs had higher levels if they were diagnosed with AD. TNFα is generally regarded as a “traditional” pro-inflammatory cytokine, but recent evidence suggests that TNFα may have regulatory roles in a diverse set of processes within the central nervous system including cell viability, synaptic plasticity, and learning and memory.73,74 This finding, however, was not replicated in ROS/MAP, suggesting that the association is likely null. This null association is further supported by our negative observations for the other four biomarkers tested. Despite evidence for the cross-talk of peripheral and central immune systems,75 it is possible that blood-based inflammatory biomarkers were unable to capture brain-specific molecular effects of TSPO variation.
In line with our primarily null findings, the evolving literature on TSPO function is somewhat equivocal, and genome-wide association studies of AD and AD-related phenotypes have not demonstrated significant associations with TSPO variants. It is possible that the relationship between TSPO genotype and AD is modulated by additional factors that have not yet been identified. Our main effect results align with a very recent investigation of TSPO genotype effects on in vivo amyloid and cognition in the ADNI cohort,76 which found no differences between HAB, MAB, and LAB subjects across or within diagnostic subgroups. At the molecular level, the role of TSPO in steroidogenesis has recently been called into question77 following two studies: one showing that testicular production of testosterone was unaltered in a conditional TSPO knockout mouse,78 and the other showing that the global knockout mouse was viable79 (contrary to previous reports80). These studies, however, do not preclude the possibility that, when present, different forms of TSPO may function in potentially divergent ways. To test this directly in the context of immune system activation, we examined the effect of genotype on microglial activation in human postmortem brain tissue, finding no genotypic group differences. Importantly, these results suggest that the mechanism via which TSPO genotype may influence downstream pathology and disease risk does not involve the modulation of microglial activation.
There are several limitations to this study. First, while efforts were made to ensure genotype groups were matched for socio-demographic factors, potential confounding effects of unaccounted-for subclinical pathology or other environmental factors cannot be ruled out. Second, population stratification due to ethnic diversity is a concern in any genetic analysis and must be considered as a potential confounder. The observed minor allele frequencies (MAFs) for TSPO rs6971 in the ROS/MAP and ADNI samples were 0.32 and 0.31, respectively, consistent with observations in other Caucasian populations (HapMap-CEU MAF = 0.29 (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/snp_details_phase3?name=rs6971&source=hapmap28_B36&tmpl=snp_details_phase3)). Third, we recognize that the literature shows discordance between patterns of inflammatory biomarkers measured in serum vs. CSF,81 and thus our plasma biomarker results may not apply to brain-specific inflammation. Fourth, M1- and M2-like microglial phenotypes82,83 have been shown to have distinct and often opposing roles in disease.84,85 Given that we did not quantify the relative densities of M1- and M2-like microglia, it is possible that a genotype effect on either specific cellular population may have been missed. Fifth, AD is being increasingly recognized as a pathologically heterogeneous disorder,86 and great variability exists even at the level of our well-defined endophenotypes (i.e. location and type of infarcts and hyperintensities); therefore, heterogeneity between subjects and within diagnostic groups likely contributed to our negative findings. Finally, study design differences between ADNI and ROS/MAP should be acknowledged when considering our findings. While inter-study variability in image processing pipelines (for WMHv) and plasma protein quantification methods could potentially drive differences between sample results, the fact that we found no significant effects of genotype in either sample (other than for plasma TNFα in ADNI AD subjects only) should alleviate concerns regarding false negatives.
In conclusion, our findings do not support genetic variation in TSPO at rs6971 as a cerebrovascular and inflammatory risk factor related to neurodegeneration. In particular, the observed null effects of rs6971 on levels of microglial density across brain regions suggest that TSPO structural changes due to genotype likely do not alter interactions with endogenous ligands in a manner that influences microglial activation. While we provide the first insight into the potential effects of rs6971 variation on structural imaging and postmortem brain pathology in humans, continued in vitro and animal model experiments of differential TSPO binding would be required to verify the pathway-level impact of this variation with respect to AD etiopathology.
Supplementary Material
Acknowledgement
We would like to thank all of the study participants and acknowledge the essential contributions of Chaya Gopin and Kimberly Cameron to the recruitment and clinical assessments of those participants. We are indebted to the participants in the Religious Orders Study and the Rush Memory and Aging Project. We thank the staff of the Rush Alzheimer's Disease Center.
DF is supported by a Vanier Canada Graduate Scholarship (CIHR) as well as the Peterborough K.M. Hunter Graduate Scholarship. ANV is supported by the CAMH Foundation, Ontario Ministry of Research and Innovation, Canadian Institutes of Health Research, Brain and Behavior Research Foundation, Canada Foundation for Innovation, and National Institute of Mental Health R01MH099167 and R01MH102324.
Authors’ note
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found online (http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf).
Funding
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. This study was supported by the National Institutes of Health (NIH) grants U01AG024904, P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01NS084965, R01MH099167 and R01MH102324, the Department of Defense (DOD) award number W81XWH-12-2-0012, the Illinois Department of Public Health, and the Translational Genomics Research Institute.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. ANV had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Authors’ contribution
Dr. Felsky reports no relevant disclosures and was responsible for analytical study design, data compilation and organization, final data analyses, and manuscript writing and editing. Dr. De Jager reports no relevant disclosures and was responsible for study design of the genetic component of ROS/MAP and manuscript editing. Dr. Schneider reports no relevant disclosures and was responsible for neuropathological data collection and quality control for the ROS/MAP dataset, data processing, and manuscript editing. Dr. Arfanakis reports no relevant disclosures and was responsible for data collection of the ROS/MAP imaging sub-study, data processing, and manuscript editing. Dr. Fleischman reports no relevant disclosures and was responsible for ROS/MAP study design and data collection, including imaging and inflammatory biomarker sub-studies, and manuscript editing. Dr. Arvanitakis reports no relevant disclosures and was responsible for study design of ROS/MAP inflammatory biomarker sub-study and manuscript editing. Dr. Honer reports no relevant disclosures and was responsible for study design in the MAP sample and manuscript editing. Ms. Pouget reports no relevant disclosures and was responsible for collaborative input into overall study design and manuscript editing. Dr. Mizrahi reports no relevant disclosures and was responsible for collaborative input into overall study design and manuscript editing. Dr. Pollock reports no relevant disclosures and was responsible for collaborative input into overall study design, facilitating data transfer, and manuscript editing. Dr. Kennedy reports no relevant disclosures and was responsible for collaborative input into overall study design and manuscript editing. Dr. Bennett reports no relevant disclosures and was responsible for overall ROS/MAP study design, clinical data collection, facilitation of data transfer, and manuscript editing. Dr. Voineskos reports no relevant disclosures and was responsible for assisting in the development of the overall study design, facilitating collaboration, assisting in data analysis and interpretation, and manuscript editing.
Supplementary material
Supplementary material for this paper can be found at http://jcbfm.sagepub.com/content/by/supplemental-data
References
- 1.Akiyama H. Inflammatory response in Alzheimer’s disease. Tohoku J Exp Med 1994; 174: 295–303. [DOI] [PubMed] [Google Scholar]
- 2.Heneka MT, O’Banion MK, Terwel D, et al. Neuroinflammatory processes in Alzheimer’s disease. J Neural Transm (Vienna) 2010; 117: 919–947. [DOI] [PubMed] [Google Scholar]
- 3.Hommet C, Mondon K, Camus V, et al. Neuroinflammation and β amyloid deposition in Alzheimer’s disease: in vivo quantification with molecular imaging. Dement Geriatr Cogn Disord 2014; 37: 1–18. [DOI] [PubMed] [Google Scholar]
- 4.Rubio-Perez JM, Morillas-Ruiz JM. A review: inflammatory process in Alzheimer’s disease, role of cytokines. Sci World J 2012; 2012: e756357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kummer MP, Vogl T, Axt D, et al. Mrp14 deficiency ameliorates amyloid β burden by increasing microglial phagocytosis and modulation of amyloid precursor protein processing. J Neurosci 2012; 32: 17824–17829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kummer MP, Hermes M, Delekarte A, et al. Nitration of tyrosine 10 critically enhances amyloid β aggregation and plaque formation. Neuron 2011; 71: 833–844. [DOI] [PubMed] [Google Scholar]
- 7.Bagasra O, Michaels FH, Zheng YM, et al. Activation of the inducible form of nitric oxide synthase in the brains of patients with multiple sclerosis. Proc Natl Acad Sci U S A 1995; 92: 12041–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarty P, Herring A, Ceballos-Diaz C, et al. Hippocampal expression of murine TNFα results in attenuation of amyloid deposition in vivo. Mol Neurodegener 2011; 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass CK, Saijo K, Winner B, et al. Mechanisms underlying inflammation in neurodegeneration. Cell 2010; 140: 918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Zoppo G, Ginis I, Hallenbeck JM, et al. Inflammation and stroke: putative role for cytokines, adhesion molecules and iNOS in brain response to ischemia. Brain Pathol 2000; 10: 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iadecola C, Alexander M. Cerebral ischemia and inflammation. Curr Opin Neurol 2001; 14: 89–94. [DOI] [PubMed] [Google Scholar]
- 12.de la Torre JC. Alzheimer disease as a vascular disorder: nosological evidence. Stroke J Cereb Circ 2002; 33: 1152–1162. [DOI] [PubMed] [Google Scholar]
- 13.Casserly I, Topol EJ. Convergence of atherosclerosis and Alzheimer’s disease: inflammation, cholesterol, and misfolded proteins. Lancet 2004; 363: 1139–1146. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg SM, Gurol ME, Rosand J, et al. Amyloid angiopathy-related vascular cognitive impairment. Stroke J Cereb Circ 2004; 35: 2616–2619. [DOI] [PubMed] [Google Scholar]
- 15.Viswanathan A, Greenberg SM. Cerebral amyloid angiopathy in the elderly. Ann Neurol 2011; 70: 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeer SE, Prins ND, den Heijer T, et al. Silent brain infarcts and the risk of dementia and cognitive decline. N Engl J Med 2003; 348: 1215–1222. [DOI] [PubMed] [Google Scholar]
- 17.Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol 2010; 9: 689–701. [DOI] [PubMed] [Google Scholar]
- 18.Arvanitakis Z, Leurgans SE, Barnes LL, et al. Microinfarct pathology, dementia, and cognitive systems. Stroke J Cereb Circ 2011; 42: 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider JA, Wilson RS, Cochran EJ, et al. Relation of cerebral infarctions to dementia and cognitive function in older persons. Neurology 2003; 60: 1082–1088. [DOI] [PubMed] [Google Scholar]
- 20.van Rooden S, Goos JDC, van Opstal AM, et al. Increased number of microinfarcts in Alzheimer disease at 7-T MR imaging. Radiology 2014; 270: 205–211. [DOI] [PubMed] [Google Scholar]
- 21.Suter O-C, Sunthorn T, Kraftsik R, et al. Cerebral hypoperfusion generates cortical watershed microinfarcts in Alzheimer disease. Stroke J Cereb Circ 2002; 33: 1986–1992. [DOI] [PubMed] [Google Scholar]
- 22.Farkas E, Luiten PG. Cerebral microvascular pathology in aging and Alzheimer’s disease. Prog Neurobiol 2001; 64: 575–611. [DOI] [PubMed] [Google Scholar]
- 23.Yoshita M, Fletcher E, Harvey D, et al. Extent and distribution of white matter hyperintensities in normal aging, MCI, and AD. Neurology 2006; 67: 2192–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brickman AM. Contemplating Alzheimer’s disease and the contribution of white matter hyperintensities. Curr Neurol Neurosci Rep 2013; 13: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cagnin A, Brooks DJ, Kennedy AM, et al. In-vivo measurement of activated microglia in dementia. Lancet 2001; 358: 461–467. [DOI] [PubMed] [Google Scholar]
- 26.Venneti S, Lopresti BJ, Wiley CA. The peripheral benzodiazepine receptor (translocator protein 18kDa) in microglia: from pathology to imaging. Prog Neurobiol 2006; 80: 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veenman L, Papadopoulos V, Gavish M. Channel-like functions of the 18-kDa translocator protein (TSPO): regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr Pharm Des 2007; 13: 2385–2405. [DOI] [PubMed] [Google Scholar]
- 28.Chen M-K, Guilarte TR. Translocator protein 18kDA (TSPO): molecular sensor of brain injury & repair. Pharmacol Ther 2008; 118: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harberts E, Datta D, Chen S, et al. Translocator protein 18 kDa (TSPO) expression in multiple sclerosis patients. J Neuroimmune Pharmacol 2013; 8: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh U, Fujita M, Ikonomidou VN, et al. Translocator protein PET imaging for glial activation in multiple sclerosis. J Neuroimmune Pharmacol 2011; 6: 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banati RB, Newcombe J, Gunn RN, et al. The peripheral benzodiazepine binding site in the brain in multiple sclerosis: quantitative in vivo imaging of microglia as a measure of disease activity. Brain J Neurol 2000; 123(Pt 11): 2321–2337. [DOI] [PubMed] [Google Scholar]
- 32.Colasanti A, Guo Q, Muhlert N, et al. In vivo assessment of brain white matter inflammation in multiple sclerosis with 18F-PBR111 PET. J Nucl Med 2014; 55: 1112–1118. [DOI] [PubMed] [Google Scholar]
- 33.Takano A, Piehl F, Hillert J, et al. In vivo TSPO imaging in patients with multiple sclerosis: a brain PET study with [18F]FEDAA1106. EJNMMI Res 2013; 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuno F, Kosaka J, Ota M, et al. Increased binding of peripheral benzodiazepine receptor in mild cognitive impairment-dementia converters measured by positron emission tomography with [11C]DAA1106. Psychiatry Res 2012; 203: 67–74. [DOI] [PubMed] [Google Scholar]
- 35.Kreisl WC, Lyoo CH, McGwier M, et al. In vivo radioligand binding to translocator protein correlates with severity of Alzheimer’s disease. Brain J Neurol 2013; 136: 2228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen DR, Yeo AJ, Gunn RN, et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab 2012; 32: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizrahi R, Rusjan PM, Kennedy J, et al. Translocator protein (18 kDa) polymorphism (rs6971) explains in-vivo brain binding affinity of the PET radioligand [(18)F]-FEPPA. J Cereb Blood Flow Metab 2012; 32: 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murail S, Robert J-C, Coïc Y-M, et al. Secondary and tertiary structures of the transmembrane domains of the translocator protein TSPO determined by NMR. Stabilization of the TSPO tertiary fold upon ligand binding. Biochim Biophys Acta 2008; 1778: 1375–1381. [DOI] [PubMed] [Google Scholar]
- 39.Korkhov VM, Sachse C, Short JM, et al. Three-dimensional structure of TSPO by electron cryomicroscopy of helical crystals. Structure 2010; 18: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlstetter M, Nothdurfter C, Aslanidis A, et al. Translocator protein (18 kDa) (TSPO) is expressed in reactive retinal microglia and modulates microglial inflammation and phagocytosis. J Neuroinflammation 2014; 11: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barron AM, Garcia-Segura LM, Caruso D, et al. Ligand for translocator protein reverses pathology in a mouse model of Alzheimer’s disease. J Neurosci 2013; 33: 8891–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin R, Angelin A, Da Settimo F, et al. Genetic analysis of dTSPO, an outer mitochondrial membrane protein, reveals its functions in apoptosis, longevity, and Ab42-induced neurodegeneration. Aging Cell 2014; 13: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferzaz B, Brault E, Bourliaud G, et al. SSR180575 (7-chloro-N,N,5-trimethyl-4-oxo-3-phenyl-3,5-dihydro-4H-pyridazino[4,5-b]indole-1-acetamide), a peripheral benzodiazepine receptor ligand, promotes neuronal survival and repair. J Pharmacol Exp Ther 2002; 301: 1067–1078. [DOI] [PubMed] [Google Scholar]
- 44.Girard C, Liu S, Cadepond F, et al. Etifoxine improves peripheral nerve regeneration and functional recovery. Proc Natl Acad Sci U S A 2008; 105: 20505–20510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu JK, Choi HB, McLarnon JG. Peripheral benzodiazepine receptor ligand PK11195 reduces microglial activation and neuronal death in quinolinic acid-injected rat striatum. Neurobiol Dis 2005; 20: 550–561. [DOI] [PubMed] [Google Scholar]
- 46.Veiga S, Azcoitia I, Garcia-Segura LM. Ro5-4864, a peripheral benzodiazepine receptor ligand, reduces reactive gliosis and protects hippocampal hilar neurons from kainic acid excitotoxicity. J Neurosci Res 2005; 80: 129–137. [DOI] [PubMed] [Google Scholar]
- 47.Leaver KR, Reynolds A, Bodard S, et al. Effects of translocator protein (18 kDa) ligands on microglial activation and neuronal death in the quinolinic-acid-injected rat striatum. ACS Chem Neurosci 2011; 3: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s disease neuroimaging initiative (ADNI) clinical characterization. Neurology 2010; 74: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saykin AJ, Shen L, Foroud TM, et al. Alzheimer’s disease neuroimaging initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement 2010; 6: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett DA, Schneider JA, Arvanitakis Z, et al. Overview and findings from the religious orders study. Curr Alzheimer Res 2012; 9: 628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett DA, Schneider JA, Buchman AS, et al. Overview and findings from the rush memory and aging project. Curr Alzheimer Res 2012; 9: 646–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleischman DA, Leurgans S, Arfanakis K, et al. Gray-matter macrostructure in cognitively healthy older persons: associations with age and cognition. Brain Struct Funct 2014; 219: 2029–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett DA, Schneider JA, Aggarwal NT, et al. Decision rules guiding the clinical diagnosis of Alzheimer’s disease in two community-based cohort studies compared to standard practice in a clinic-based cohort study. Neuroepidemiology 2006; 27: 169–176. [DOI] [PubMed] [Google Scholar]
- 54.Chibnik LB, Shulman JM, Leurgans SE, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol 2011; 69: 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jack CR, Bernstein MA, Fox NC, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27: 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeCarli C, Carmichael O and He J. MRI infarct assessment in ADNI, http://adni.bitbucket.org/docs/MRI_INFARCTS/UCD%20ADNI%20MRI%20Infarct%20Assessment%20Method.pdf (2013, accessed 26 December 2015).
- 57.Schwarz C, Fletcher E, DeCarli C, et al. Fully-automated white matter hyperintensity detection with anatomical prior knowledge and without FLAIR. In: Prince JL, Pham DL, Myers KJ. (eds). Information processing in medical imaging, Berlin Heidelberg: Springer, 2009, pp. 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshita M, Fletcher E, DeCarli C. Current concepts of analysis of cerebral white matter hyperintensities on magnetic resonance imaging. Top Magn Reson Imaging 2005; 16: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeCarli C, Murphy DG, Teichberg D, et al. Local histogram correction of MRI spatially dependent image pixel intensity nonuniformity. J Magn Reson Imaging 1996; 6: 519–528. [DOI] [PubMed] [Google Scholar]
- 60.DeCarli C, Miller BL, Swan GE, et al. Predictors of brain morphology for the men of the NHLBI twin study. Stroke J Cereb Circ 1999; 30: 529–536. [DOI] [PubMed] [Google Scholar]
- 61.DeCarli C, Maillard P and Fletcher E. Four tissue segmentation in ADNI II, http://adni.bitbucket.org/docs/UCD_ADNI2_WMH/UCD%20ADNI%20II%204%20tissue%20segmentation%20Method.pdf (2013, accessed 26 December 2015).
- 62.Arfanakis K, Fleischman DA, Grisot G, et al. Systemic inflammation in non-demented elderly human subjects: brain microstructure and cognition. PLoS One 2013; 8: e73107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zacharaki EI, Kanterakis S, Bryan RN, et al. Measuring brain lesion progression with a supervised tissue classification system. Med Image Comput Comput-Assist Interv 2008; 11: 620–627. [DOI] [PubMed] [Google Scholar]
- 64.Bennett DA, Schneider JA, Buchman AS, et al. The rush memory and aging project: study design and baseline characteristics of the study cohort. Neuroepidemiology 2005; 25: 163–175. [DOI] [PubMed] [Google Scholar]
- 65.Arvanitakis Z, Leurgans SE, Wang Z, et al. Cerebral amyloid angiopathy pathology and cognitive domains in older persons. Ann Neurol 2011; 69: 320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradshaw EM, Chibnik LB, Keenan BT, et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci 2013; 16: 848–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Royston P. Remark AS R94: a remark on algorithm AS 181: the W-test for normality. Appl Stat 1995; 44: 547. [Google Scholar]
- 68.Box GEP, Cox DR. An analysis of transformations. J R Stat Soc Ser B Methodol 1964; 26: 211–252. [Google Scholar]
- 69.Dimitrova-Shumkovska J, Veenman L, Roim I, et al. The 18 kDa translocator protein and atherosclerosis in mice lacking apolipoprotein E. In: Valenzuela Baez R (ed.) Lipid metabolism. InTech, 2013, www.intechopen.com/books/lipid-metabolism/the-18-kda-translocator-protein-and-atherosclerosis-in-mice-lacking-apolipoprotein-e (accessed 23 May 2014).
- 70.Taylor J, Allen A, Graham A. Targeting mitochondrial 18kDa translocator protein (TSPO) regulates macrophage cholesterol efflux and lipid phenotype. Clin Sci 2014; 127: 603–613. doi:10.1042/CS20140047. [DOI] [PubMed] [Google Scholar]
- 71.Soontornniyomkij V, Lynch MD, Mermash S, et al. Cerebral microinfarcts associated with severe cerebral β-amyloid angiopathy. Brain Pathol 2010; 20: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Negash S, Bennett DA, Wilson RS, et al. Cognition and neuropathology in aging: multidimensional perspectives from the rush religious orders study and rush memory and aging project. Curr Alzheimer Res 2011; 8: 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olmos G, Llado J. Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediators Inflamm 2014; 2014: 861231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frankola KA, Greig NH, Luo W, et al. Targeting TNF-α to elucidate and ameliorate neuroinflammation in neurodegenerative diseases. CNS Neurol Disord Drug Targets 2011; 10: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perry VH, Cunningham C, Holmes C. Systemic infections and inflammation affect chronic neurodegeneration. Nat Rev Immunol 2007; 7: 161–167. [DOI] [PubMed] [Google Scholar]
- 76.Fan Z, Harold D, Pasqualetti G, et al. Can studies of neuroinflammation in a TSPO genetic subgroup (HAB or MAB) be applied to the entire AD cohort? J Nucl Med 2015; 56: 707–713. [DOI] [PubMed] [Google Scholar]
- 77.Stocco DM. The role of PBR/TSPO in steroid biosynthesis challenged. Endocrinology 2014; 155: 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morohaku K, Pelton SH, Daugherty DJ, et al. Translocator protein/peripheral benzodiazepine receptor is not required for steroid hormone biosynthesis. Endocrinology 2014; 155: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tu LN, Morohaku K, Manna PR, et al. Peripheral benzodiazepine receptor/translocator protein global knockout mice are viable with no effects on steroid hormone biosynthesis. J Biol Chem 2014; 289: 27444–27454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Papadopoulos V, Amri H, Boujrad N, et al. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. Steroids 1997; 62: 21–28. [DOI] [PubMed] [Google Scholar]
- 81.Swardfager W, Lanctôt K, Rothenburg L, et al. A meta-analysis of cytokines in Alzheimer’s disease. Biol Psychiatry 2010; 68: 930–941. [DOI] [PubMed] [Google Scholar]
- 82.Mantovani A, Biswas SK, Galdiero MR, et al. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol 2013; 229: 176–185. [DOI] [PubMed] [Google Scholar]
- 83.Prinz M, Priller J, Sisodia SS, et al. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci 2011; 14: 1227–1235. [DOI] [PubMed] [Google Scholar]
- 84.Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol 2011; 11: 775–787. [DOI] [PubMed] [Google Scholar]
- 85.Goldmann T, Prinz M. Role of microglia in CNS autoimmunity. J Immunol Res 2013; 2013: e208093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider JA, Arvanitakis Z, Bang W, et al. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology 2007; 69: 2197–2204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.