Abstract
The airway epithelium is a complex pseudostratified multicellular layer lining the tracheobronchial tree, functioning as the primary defense against inhaled environmental contaminants. The major cell types of the airway epithelium include basal, intermediate columnar, ciliated, and secretory. Basal cells (BCs) are the proliferating stem/progenitor population that differentiate into the other specialized cell types of the airway epithelium during normal turnover and repair. Given that cigarette smoke delivers thousands of xenobiotics and high levels of reactive molecules to the lung epithelial surface, we hypothesized that cigarette smoke broadly perturbs BC metabolism. To test this hypothesis, primary airway BCs were isolated from healthy nonsmokers (n = 11) and healthy smokers (n = 7) and assessed by global metabolic profiling by liquid chromatography–mass spectrometry. The analysis identified 52 significant metabolites in BCs differentially expressed between smokers and nonsmokers (P < 0.05). These changes included metabolites associated with redox pathways, energy production, and inflammatory processes. Notably, BCs from smokers exhibited altered levels of the key enzyme cofactors/substrates nicotinamide adenine dinucleotide, flavin adenine dinucleotide, acetyl coenzyme A, and membrane phospholipid levels. Consistent with the high burden of oxidants in cigarette smoke, glutathione levels were diminished, whereas 3-nitrotyrosine levels were increased, suggesting that protection of airway epithelial cells against oxidative and nitrosative stress is significantly compromised in smoker BCs. It is likely that this altered metabotype is a reflection of, and likely contributes to, the disordered biology of airway BCs consequent to the stress cigarette smoking puts on the airway epithelium.
Keywords: metabolic profiling, airway basal cells, progenitor cells, cigarette smoke, oxidative stress
Clinical Relevance
Based on the knowledge that cigarette smoke delivers high levels of xenobiotics and reactive components to the lung epithelial surface, we hypothesized that cigarette smoke perturbs the metabolism of human airway basal stem/progenitor cells, the cell type responsible for replenishing the mucociliary epithelium with turnover and repair. Using mass spectrometry–based global metabolite profiling, we identified a distinct metabotype in airway basal cells of smokers, with perturbations in molecules that serve as cofactors and intermediates for important cellular processes. We hypothesize that these metabolic alterations play a key role in disordering the biology of the airway epithelium of smokers, contributing to the development of lung disease.
The airway epithelium, a complex pseudostratified multicellular layer lining the tracheobronchial tree, plays a critical role in host defense against inhaled pathogens, particulates, and other environmental contaminants (1–3). The major cellular components of the epithelium are ciliated and secretory cells, the differentiated cells that mediate mucociliary clearance, and the basal stem/progenitor cells (BCs) and their daughter columnar intermediate cells that replenish the ciliated and secretory cells during normal turnover and after injury (1–8). The airway epithelium is the major disease-related target of cigarette smoking, the primary cause of chronic obstructive pulmonary disease (COPD) and lung cancer. The importance of BCs in the pathogenesis of COPD is highlighted by the observations that the first smoking-associated abnormalities in the lung are in the airway epithelium, characterized by a hyperplasia of the BCs and disordered BC differentiation that includes hyperplasia of mucus-producing secretory cells, decreased numbers of ciliated cells, shorter cilia, squamous cell metaplasia, and decreased epithelial barrier function (9–17). Relevant to these abnormalities, smoking induces changes in the BC transcriptome (18), and BCs of smokers are “fatigued” and less capable of differentiating to a competent mucociliary epithelium (19).
Based on the knowledge that BC biology is fundamental to the maintenance of a normal functioning airway epithelium and the known derangements to BCs associated with cigarette smoking, with its 4,000 xenobiotics and >1014 oxidants/puff (20), we hypothesized that smoking alters the metabolism of the BCs compared with that of nonsmokers. To assess this hypothesis and to gain insight into the molecular and biochemical derangements of airway BC biology induced by cigarette smoking, we applied mass spectrometry–based global metabolomics to compare the metabotype of primary BCs isolated from the airways of healthy smokers and healthy nonsmokers to provide a broad, unbiased readout of cellular metabolites associated with the stress of smoking. The data demonstrate that cigarette smoking is associated with significant changes in BC metabolites, including important cofactors and intermediates for key cellular processes that may contribute to aberrant BC differentiation and altered patterning of the airway epithelium.
Materials and Methods
Study Population
Healthy nonsmokers and healthy smokers were recruited from the general population in New York City under a protocol approved by the Weill Cornell Medical College Institutional Review Board (Table 1). The criteria for “healthy” were based on history, physical examination, complete blood count, coagulation studies, liver function tests, urine studies, chest X-ray, EKG, and pulmonary function tests (full inclusion/exclusion criteria are provided in the online supplement). All subjects were negative for HIV1 and had normal α1-antitrypsin levels. Smokers were defined as self-reported current smokers, and this status was verified by the measurement of urinary nicotine ≥30 ng/ml and/or cotinine ≥50 ng/ml. Nonsmokers were defined as self-reported life-long nonsmokers, with undetectable urine nicotine (<2 ng/ml) and cotinine (<5 ng/ml) levels.
Table 1.
Demographics of the Study Groups
| Parameter | Healthy Nonsmokers | Healthy Smokers | P Value* |
|---|---|---|---|
| Number of subjects | 11 | 7 | |
| Sex, M/F | 6/5 | 5/2 | 0.82 |
| Age, yr | 31.4 ± 11.3† | 42.1 ± 10.4 | 0.06 |
| Race (black/white/other) | 2/4/5 | 5/1/1 | 0.28 |
| Smoking history, pack-year | 0 | 30.2 ± 24.0 | |
| Urine nicotine, ng/ml | Negative | 2,482 ± 2,480 | |
| Urine cotinine, ng/ml | Negative | 2,106 ± 755 | |
| Pulmonary function parameters‡ | |||
| FVC | 100.7 ± 10.7 | 109.7 ± 16.2 | 0.23 |
| FEV1 | 98.5 ± 10.4 | 105.4 ± 18.1 | 0.38 |
| FEV1/FVC | 83.2 ± 7.1 | 77.9 ± 4.2 | 0.06 |
| TLC | 106.3 ± 16.2 | 100.9 ± 11.3 | 0.42 |
| DlCO | 92.5 ± 6.1 | 89.9 ± 9.0 | 0.52 |
Definition of abbreviations: DlCO, diffusing capacity of the lung for carbon monoxide; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; TLC, total lung capacity.
Two-tailed t test between samples assuming unequal variance or Chi-square test.
Data are presented as mean ± SD.
Pulmonary function testing parameters are given as % predicted value with the exception of FEV1/FVC, which is reported as % observed. FVC, FEV1, and FEV1/FVC are prebronchodilator values.
Culture of Primary Human Airway BCs
Primary airway BCs from nonsmokers (n = 11) and smokers (n = 7) were isolated using selective culture methods from large airway epithelial samples obtained by bronchoscopy as described previously (7). The BC phenotype of all cells was confirmed by positive staining for the BC markers KRT5 and CD151 (>99% positive cells) and negative staining for alternative differentiated airway epithelial cell types (secretory, ciliated cell, and neuroendocrine) as previously described (7). All cultures were seeded at 3,000 cells/cm2 into plastic flasks and maintained in BEGM medium (Lonza, Walkersville, MD) supplemented with 1% penicillin/streptomycin (GIBCO-Life Technologies, Grand Island, NY), 0.5% amphotericin B (GIBCO-Life Technologies), and 0.1% gentamicin (Sigma, St. Louis, MO) in a 5% CO2, 37°C humidified incubator with media replaced every 2 to 3 days. Each independent BC culture was passaged one round to obtain sufficient cell material for metabolic profiling. Once the cells reached 70% confluence, they were harvested by trypsinization in 0.05% EDTA (GIBCO-Life Technologies) for 5 minutes at 37°C, with the reaction stopped by addition of Hepes-buffered saline (Lonza) supplemented with 15% FBS (GIBCO-Life Technologies). The resulting dissociated cells were washed with PBS, pelleted, and stored at −80°C for subsequent metabolic profiling.
Metabolite Extraction from Basal Cell Pellets
Cell pellets (∼1 × 106 cells/pellet) from all subjects were extracted simultaneously in 80/20 methanol/deionized H2O (−70°C) to rapidly quench cell metabolism and extract metabolites for ratiometric analysis. Individual pellets were homogenized using a stainless steel bead beater (Tissue-Lyser II; Qiagen, Valencia, CA). The resulting metabolite extracts were centrifuged for 20 minutes (16,000× g) to pellet precipitated protein. Supernatants from three successive methanol/deionized H2O (−70°C) extractions of each BC pellet were pooled and vacuum centrifuged until dry (Savant SPD121P; Thermo Scientific, Asheville, NC). To account for variations in BC pellet size, samples were normalized based on total protein concentration in each cell pellet after methanolic extraction. For protein quantification, 100 μl of 0.2 N NaOH was added to each extracted cell pellet and heated for 20 minutes at 95°C, followed by a DC protein assay (Bio-Rad, Hercules, CA). Based on calculated total protein in each extracted pellet, all dried metabolite extracts were later resuspended to give an equivalent concentration (1 μg/μl). On the day of analysis, dried metabolite extracts were resuspended in 70/30 acetonitrile/deionized H2O with 0.2% ammonium hydroxide and transferred to autosampler vials for analysis by liquid chromatography/mass spectrometry (LC/MS).
LC/MS Data Acquisition and Metabolite Profiling
Global metabolite profiling was performed on a system that comprises a high-pressure LC separation system using two binary Agilent 1200 pumps, one isocratic pump (for continuous spray of calibrant ion), coupled to a dual electrospray ionization source and an accurate mass time-of-flight mass spectrometer (Model 6538; Agilent Technologies, Santa Clara, CA) (21). Metabolites were chromatographically separated using aqueous neutral phase (ANP) on silica hybrid resin (2.1 × 150 mm, 3.5 μm particle size) (Microsolv Technology Corp., Eatontown, NJ) and analyzed in two different detection modes (positive and negative ion monitoring).
Acquired raw data files were processed with Agilent MassHunter Qualitative Analysis software (version B06). Comparative between-group analyses were performed using Agilent MassProfiler professional software (MPP, version B12.01) and Agilent MassHunter Profinder Software (version B06.00), as described previously (21). MPP analysis was used to generate metabolite features based on elution profiles of identical mass and retention times within a defined mass accuracy (<5 ppm). Aligned metabolite features within and between groups were directly applied for statistical comparisons including unsupervised pattern recognition algorithms, principal component analysis, and hierarchical clustering analysis. Principal component analysis data were exported into Partek software for visualization (Partek Genomic Suite version 6.6). MassHunter Profinder was used to examine raw data chromatograms for accuracy and to confirm trends seen in statistical analysis by MPP. MS data were analyzed by unpaired Student’s t test for between-group statistical differences in molecular expression with the Benjamini-Hochberg correction applied to correct for multiple group comparisons (P < 0.05).
Differentially Expressed Metabolite Identification and Pathway Analysis
Metabolites found to be differentially expressed between groups with P values <0.05 and with confident empirical formulae were searched against the Metlin and HMDB metabolite databases for structural identification. Structural assignments were made only in cases where validation could be affirmed by matching of chromatographic retention time and/or fragmentation patterns with pure molecule standards from an expanding in-house database. Based on the set of structurally identified differentially expressed metabolites, Metaboanalyst software (MetaboAnalyst 3.0, www.metaboanalyst.ca) (22) was used to perform metabolite enrichment and impact analysis with the goal of identifying metabolic pathways that are most perturbed in BC from cigarette smokers versus nonsmokers. Enrichment was assessed using the MSEA (Metabolite Set Enrichment Analysis) library containing 88 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways and the over-representation analysis module (ORA). ORA was used to determine if identified and structurally validated metabolites are represented more than expected by chance. The P value from ORA indicates the probability of seeing a number of identified metabolites in a given compound list. Pathways were considered enriched at P < 0.05.
Results
Metabolomic Profiling of Nonsmokers versus Healthy Smokers
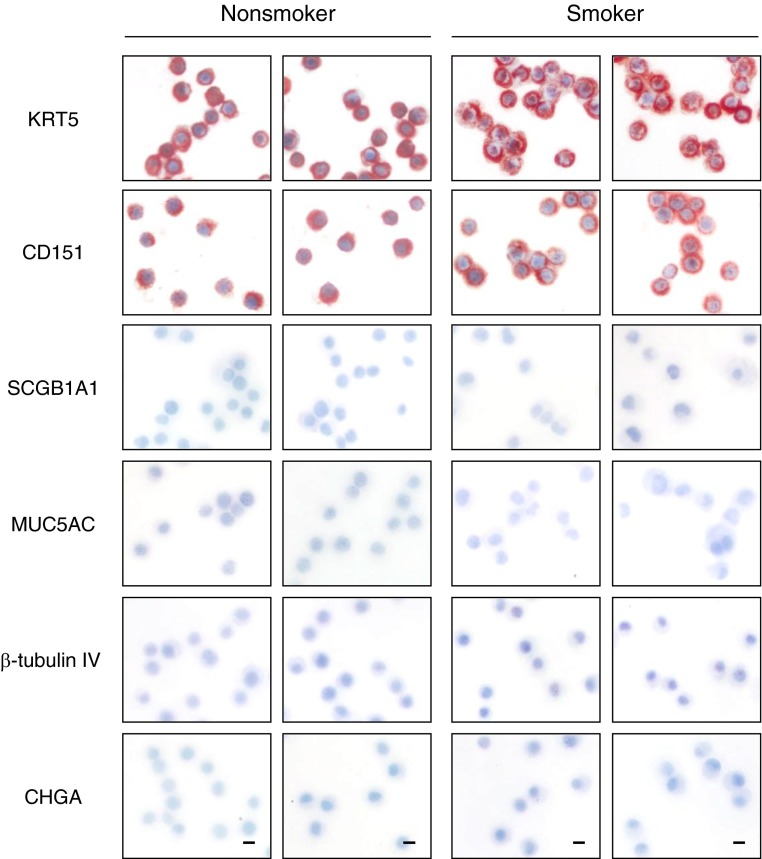
To assess the effect of cigarette smoking on the metabolism of human airway epithelial basal stem/progenitor cells (BCs), primary BCs were isolated from healthy smokers and nonsmokers using selective culture methods from large airway epithelial samples obtained by bronchoscopy (Table 1). BC phenotype of each culture was confirmed by positive staining for BC-specific markers (KRT5, CD151) and negative staining for markers characterizing alternative differentiated airway epithelial cell types (i.e., secretory, ciliated, and neuroendocrine cells) (Figure 1).
Figure 1.
Characterization of primary human nonsmoker and smoker airway basal cells. Immunohistochemical characterization of cytopreps of primary human airway basal cells isolated using selective culture methods from large airway epithelial samples obtained by bronchoscopy from healthy nonsmokers and healthy smokers. Shown are cytokeratin 5 (KRT5; basal cell), tetraspanin-24 (CD151; basal cell), secretoglobin family 1A member 1 (SCGB1A1; secretory cell), mucin 5AC (MUC5AC; secretory cell), β-tubulin IV (ciliated cell), and chromogranin A (CHGA; neuroendocrine cell). Scale bar, 20 μm. Data shown are representative images from a single primary donor sample obtained for each phenotype.
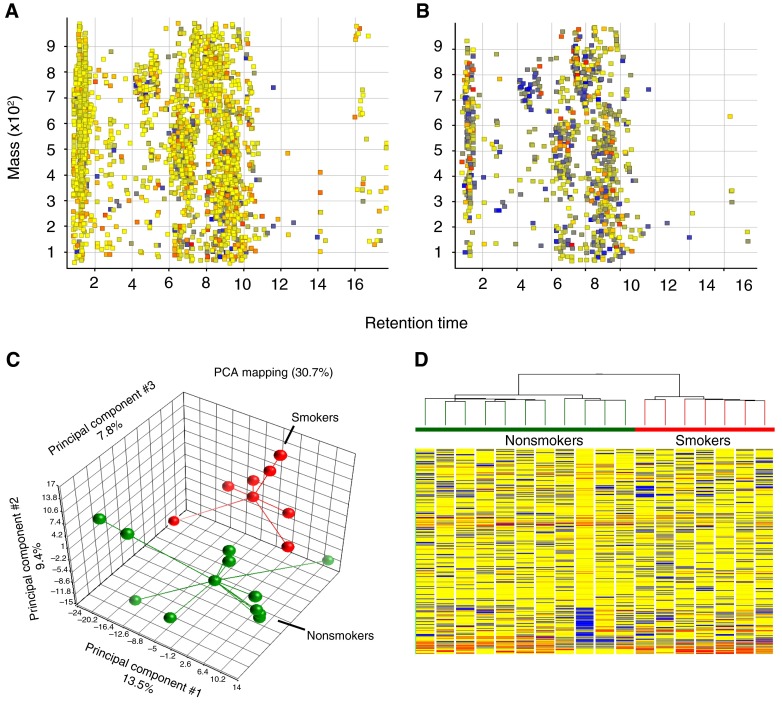
Using a multimode LC/MS-based analytical platform, we analyzed small molecule (50–1,000 Da) differences in extracts from BCs obtained from nonsmokers and smokers. Each acquisition mode contributed substantially to feature coverage, with ANPNEG LC/MS providing quantification for 3,388 features and ANPPOS LC/MS providing quantification for 2,920 features. To eliminate potential artifacts among the 6,308 total features observed in the 18 BC samples, we considered only those features that were found and quantified in at least 50% of samples from one or both groups. Using this filter, of the 2,920 features detected in ANPPOS LC/MS, 952 features were quantified in at least 50% of BCs from nonsmokers and smokers (Figures 2A and 2B). Without any preconception about sample grouping, principal component analysis demonstrated differences in levels among these 952 metabolites (Figure 2C). Hierarchical clustering analysis showed patterns of within-group similarities (smokers and nonsmokers) and between-group differences (smokers versus nonsmokers) (Figure 2D).
Figure 2.
Untargeted liquid chromatography–mass spectrometry metabolite profiling of basal cells from nonsmokers and smokers. (A) Retention time versus mass depicting 2,920 aligned features detected with positive ion monitoring and quantified across all basal cell (BC) samples. (B) Retention time versus mass depicting 952 features observed in at least 50% of BC samples from at least one group. (C) Principal component analysis (PCA) showing that BCs from the nonsmoking group (green) cluster separately from the smoking group (red). Each point corresponds to a single sample. Each sample is connected by a vector to the groups’ centroid, representing the geometric center of each axis for all samples in that group. (D) Unsupervised hierarchical clustering analysis, with samples color-coded by phenotype, displays expression patterns and clustering of 952 quantified BC metabolites. Each column represents a single subject BC sample; horizontal bars represent feature intensity depicted as a heat map, ranging from cold (blue) to hot (red).
Metabolic Changes in BCs Associated with Cigarette Smoking
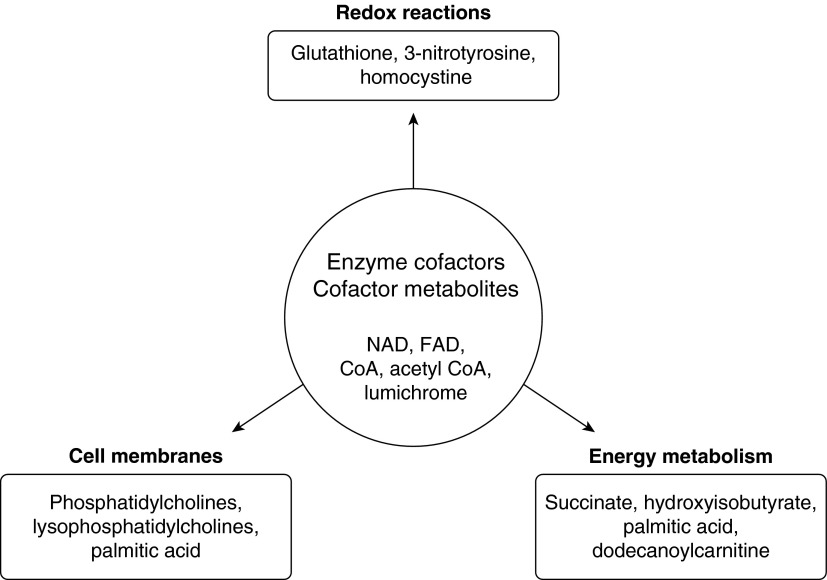
Of the features discovered in >50% of the 18 BC samples, 400 molecules were structurally identified based on a consideration of their accurate masses and retention time matches to a database of 610 metabolite reference standards. Among these 400 distinct molecules, there were 52 metabolites with significantly altered levels in BCs from smokers versus nonsmokers (P < 0.05) (Table 2). These 52 differentially expressed BC metabolites were grouped into five broad categories: (1) enzyme cofactors, cofactor metabolites, and citric acid cycle intermediates; (2) amino acid and protein metabolism; (3) lipid, fatty acid, and steroid metabolism; (4) amino sugar and nucleotide metabolism; and (5) other metabolites. To assess the potential biological significance of the 52 differentially expressed metabolites, MetaboAnalyst tools were used to map these metabolites into pathways. Enrichment analysis of the smoking-associated metabolites, based on a consideration of KEGG pathways, highlighted differences in metabolites relevant to the metabolism of lipids, amino acids, and the citric acid cycle as well as essential cofactors for enzymes that assume multiple roles in diverse metabolic pathways. A schematic diagram based on pathway analysis of the differentially expressed metabolites between smokers and nonsmokers shows dominant pathways that are affected by cigarette smoking (Figure 3).
Table 2.
Basal Cell Metabolites Significantly Different in Smokers versus Nonsmokers
| Metabolite/Pathway | Molecular Formula | Neutral Mass | Retention time (min) | Detection mode | Fold Change, Smoker versus Nonsmoker* | P Value† |
|---|---|---|---|---|---|---|
| Enzyme cofactors, cofactor metabolites, and citric acid cycle intermediates | ||||||
| Coenzyme A | C21H36N7O16P3S | 767.115 | 9.4 | ANPNEG | 2.3 | 0.030 |
| Lumichrome | C12H10N4O2 | 242.080 | 1.4 | ANPPOS | −1.6 | 0.020 |
| Pantothenic acid | C9H17NO5 | 219.111 | 4.2 | ANPPOS | −1.6 | 0.040 |
| Succinate | C4H6O4 | 118.029 | 7.0 | ANPNEG | −1.5 | 0.004 |
| Flavin adenine dinucleotide | C27H33N9O15P2 | 785.156 | 6.9 | ANPPOS | 1.4 | 5.0 × 10−4 |
| Succinic anhydride | C4H4O3 | 100.015 | 7.1 | ANPNEG | −1.3 | 0.006 |
| Nicotinamide | C6H6N2O | 122.048 | 2.1 | ANPPOS | −1.3 | 0.010 |
| Acetyl CoA | C23H38N7O17P3S | 809.128 | 7.3 | ANPPOS | −1.3 | 0.010 |
| Nicotinamide adenine dinucleotide | C21H27N7O14P2 | 663.109 | 9.1 | ANPNEG | −1.1 | 0.002 |
| Amino acid and protein metabolism | ||||||
| 3-Nitrotyrosine | C9H10N2O5 | 226.056 | 6.8 | ANPPOS | 2.3 | 0.010 |
| Ophthalmic acid | C11H19N3O6 | 289.129 | 9.1 | ANPNEG | −1.5 | 0.010 |
| Citrulline | C6H13N3O3 | 175.082 | 9.6 | ANPNEG | −1.5 | 0.020 |
| Homovanillic acid | C9H10O4 | 182.058 | 1.5 | ANPNEG | −1.3 | 5.0 × 10−4 |
| 1-Pyrroline-2-carboxylic acid | C5H7NO2 | 113.047 | 8.7 | ANPNEG | −1.3 | 0.007 |
| Phenylacetic acid | C8H8O2 | 136.052 | 1.5 | ANPNEG | −1.3 | 0.003 |
| Cysteamine | C2H7NS | 77.029 | 2.1 | ANPPOS | −1.3 | 0.020 |
| Pipecolinic acid | C6H11NO2 | 129.079 | 8.3 | ANPPOS | −1.3 | 0.020 |
| 3-Hydroxyanthranilic acid | C7H7NO3 | 153.042 | 1.2 | ANPNEG | −1.3 | 0.030 |
| Kynurenin | C10H12N2O3 | 208.109 | 6.2 | ANPPOS | −1.3 | 0.040 |
| 3-Methylhistidine | C7H11N3O2 | 169.085 | 11.1 | ANPPOS | −1.3 | 0.040 |
| Glutathione | C10H17N3O6S | 307.083 | 8.5 | ANPPOS | −1.2 | 0.002 |
| Formylmethionine | C6H11NO3S | 177.049 | 2.0 | ANPNEG | −1.2 | 0.004 |
| Hydroxyisobutyric acid | C4H8O3 | 104.047 | 1.8 | ANPNEG | −1.2 | 0.030 |
| Homocystine | C8H16N2O4S2 | 268.054 | 10.2 | ANPNEG | −1.2 | 0.040 |
| Methylguanidine | C2H7N3 | 73.064 | 9.8 | ANPNEG | 1.1 | 0.003 |
| Tryptamine | C10H12N2 | 160.100 | 9.3 | ANPNEG | −1.1 | 0.020 |
| Lipid, fatty acid, and steroid metabolism | ||||||
| Butenylcarnitine | C11H19NO4 | 229.131 | 10.1 | ANPNEG | 1.8 | 0.020 |
| Retinol | C20H30O | 286.229 | 1.2 | ANPNEG | −1.8 | 0.030 |
| Deoxycorticosterone acetate | C23H32O4 | 372.230 | 1.1 | ANPNEG | −1.6 | 0.050 |
| Lysophosphatidylcholine (16:1) | C24H48NO7P | 493.320 | 9.5 | ANPPOS | −1.5 | 0.003 |
| Dodecanoylcarnitine | C19H37NO4 | 343.275 | 9.2 | ANPNEG | 1.5 | 0.050 |
| Lysophosphatidylcholine (18:2) | C23H44NO7P | 519.339 | 9.4 | ANPPOS | −1.4 | 0.005 |
| Lysophosphatidylcholine (16:0) | C24H50NO7P | 495.330 | 9.4 | ANPPOS | −1.3 | 0.005 |
| Lysophosphatidylcholine (18:3) | C26H48NO7P | 517.325 | 9.4 | ANPPOS | −1.3 | 0.003 |
| Palmitic acid | C16H32O2 | 256.239 | 1.2 | ANPNEG | −1.3 | 0.020 |
| Lauric acid | C12H24O2 | 200.177 | 1.2 | ANPNEG | −1.3 | 0.030 |
| Stearic acid | C18H36O2 | 284.271 | 1.1 | ANPNEG | −1.3 | 0.030 |
| Cytidine diphosphate choline | C14H26N4O11P2 | 488.107 | 12.4 | ANPPOS | −1.3 | 0.040 |
| Lysophosphatidylethanolamine (18:2) | C23H44NO7P | 477.287 | 6.2 | ANPNEG | −1.2 | 0.010 |
| Phosphatidylcholine (35:2) | C43H82NO8P | 771.599 | 7.8 | ANPPOS | −1.2 | 0.030 |
| Phosphatidylcholine (36:4) | C44H80NO8P | 781.578 | 7.4 | ANPPOS | −1.2 | 0.040 |
| Taurodeoxycholic acid | C26H45NO6S | 499.296 | 1.1 | ANPNEG | −1.1 | 0.006 |
| Amino sugar and nucleotide metabolism | ||||||
| Succinyladenosine | C14H17N5O8 | 383.112 | 7.8 | ANPPOS | −1.8 | 0.020 |
| Thymidine | C10H14N2O5 | 242.088 | 1.4 | ANPPOS | −1.6 | 0.020 |
| ADP-ribose | C15H23N5O14P2 | 559.069 | 7.2 | ANPPOS | −1.4 | 0.005 |
| Glucosamine | C6H13NO5 | 179.079 | 9.6 | ANPNEG | 1.2 | 0.020 |
| Other metabolites | ||||||
| Fucose | C6H12O5 | 164.089 | 1.5 | ANPPOS | −2.6 | 0.040 |
| Sedoheptulose 5-phosphate | C7H15O10P | 290.054 | 9.3 | ANPNEG | −1.9 | 0.030 |
| Oxalic acid | C2H2O4 | 89.994 | 7.5 | ANPNEG | −1.4 | 0.020 |
| Erucic acid | C22H42O2 | 338.318 | 1.2 | ANPPOS | −1.5 | 0.030 |
| 5-Valerolactone | C5H8O2 | 100.052 | 2.1 | ANPPOS | −1.3 | 0.010 |
| Arachidic acid | C20H40O2 | 312.303 | 1.1 | ANPNEG | −1.3 | 0.010 |
Definition of abbreviations: Acetyl CoA, acetyl coenzyme A; ADP-ribose, adenosine diphosphate ribose; ANPNEG, aqueous neutral phase chromatography with negative ion monitoring; ANPPOS, aqueous neutral phase chromatography with positive ion monitoring.
Mean smokers versus mean nonsmokers. Metabolites in each category are ordered based on relative fold-change in smokers versus nonsmokers from greatest to least.
Two-tailed t test between samples assuming unequal variance.
Figure 3.
Smoking-affected metabolic pathways. Schematic diagram based on metabolic pathway analysis of differentially expressed metabolites between smokers and nonsmokers using Metaboanalyst software. Dominant pathways affected include redox reactions, cell membrane metabolism, energy metabolism, and enzyme cofactors and cofactor metabolites. CoA, coenzyme A; FAD, flavin adenine dinucleotide; NAD, nicotinamide adenine dinucleotide.
Differences in the levels of enzyme cofactors and their metabolites essential for support of numerous enzyme-catalyzed redox reactions and the generation of key biosynthetic intermediates were detected in smokers versus nonsmokers included nicotinamide adenine dinucleotide (NAD), nicotinamide, pantothenic acid, flavin adenine dinucleotide (FAD), coenzyme A (CoA), and acetyl CoA (Table 2). A significant decrease in NAD was observed in BCs from smokers, concurrent with decreases in levels of the NAD precursor kynurenin (derived from tryptophan metabolism) and nicotinamide. NAD-dependent ADP-ribosylation is an important metabolic checkpoint that regulates the repair of DNA damage (23). Cigarette smoking was associated with a decrease in BC stores of both NAD and ADP-ribose (carbohydrate metabolism). Levels of FAD, also a redox cofactor and prosthetic group for essential enzymatic machinery, were significantly increased in BCs from smokers. CoA, acetyl CoA, and its precursor pantothenate, all central cytosolic intermediates involved in energy metabolism and acetylation reactions involving both proteins and small molecules, were also altered in BCs from smokers, as shown by an increase in CoA, along with decreased pantothenate and acetyl CoA levels.
Among metabolites in the citric acid cycle, succinate was significantly decreased in BCs from smokers. Evidence of disruption of the full cycle is suggested by significantly altered levels of coenzymes NAD, FAD, and acetyl CoA. A diminished level of the NO synthase coproduct citrulline, along with increases in 3-nitrotyrosine (3-NT) is consistent with a dysregulation of NO synthesis and an elevated level of nitrosative stress in BCs from smokers.
Membrane phospholipid levels were significantly perturbed in BCs from smokers. Changes mostly affected phosphatidylcholines and lysophosphatidylcholines, as manifest by decreased levels in BCs from smokers relative to nonsmokers. Notably, phosphatidylcholine is a cellular osmolyte and surfactant that lines the airway epithelium, and lysophospholipid moieties are products of phospholipid hydrolysis (24–26). Evidence of a potential disruption in fatty acid and ketone body metabolism is suggested in BCs from smokers based on significant observed decreases in the levels of acetyl CoA and hydroxyisobutyrate. Levels of fatty acids and their metabolic and oxidation products, including palmitic acid and dodecanoylcarnitine, were also significantly altered in BCs from smokers. Regarding energy metabolism, phosphatidylcholine homeostasis and 1-carbon metabolism, including methionine synthesis, are linked (27).
Influence of Cigarette Smoke on the Redox Status of Airway BCs
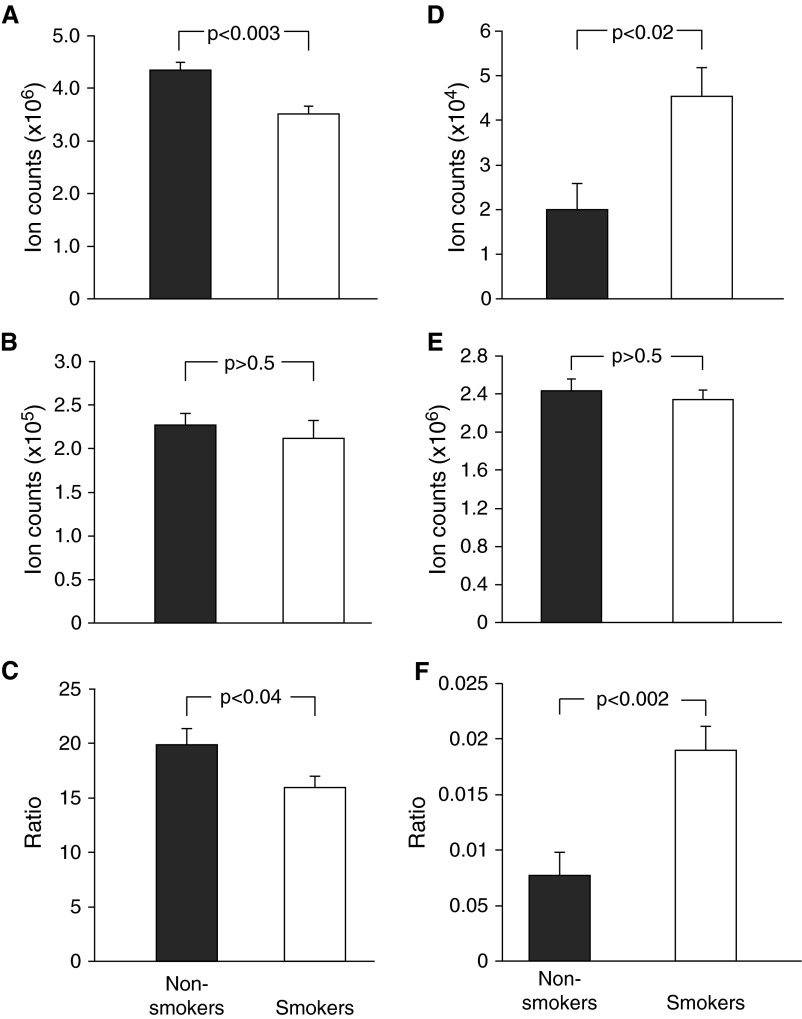
Based on the knowledge that cigarette smoke exposure provides a heavy oxidant burden to the airway epithelium (20), we examined the ratios of reduced to oxidized glutathione (GSH/GSSG) and 3-NT/tyrosine (Tyr) ratio within BCs as markers of oxidative and nitrosative stress, respectively (28, 29). Levels of total GSH were significantly decreased in BCs from smokers relative to nonsmokers (Figure 4A), and, whereas relative levels of GSSG remained unchanged (Figure 4B), the ratio of GSH/GSSG was significantly lower in BCs from smokers versus nonsmokers (Figure 4C). Additionally, the level of ophthalmic acid, a GSH analog, was also diminished in BCs from smokers (Table 2). In addition to oxidative stress, evidence of nitrosative stress was also apparent in BCs from smokers. The levels of nitrosative stress hallmark 3-NT were significantly elevated in the smoker BCs (Figure 4D). Unmodified tyrosine levels in BCs were not influenced by smoking (Figure 4E), but the 3-NT/Tyr ratio was significantly higher in BCs from smokers versus nonsmokers (Figure 4F).
Figure 4.
Basal cell redox status expressed as glutathione/oxidized glutathione (GSH/GSSG) and 3-nitrotyrosine/tyrosine (3-NT/Tyr) ratios. (A) Relative amounts of GSH accumulated in BCs from smokers compared with nonsmokers. (B) Relative amounts of GSSG accumulated in BCs from smokers compared with nonsmokers. (C) GSH/GSSG ratio comparing smokers with nonsmokers was calculated on a subject-by-subject basis by measuring relative ion counts for each molecule in each subject. (D) Relative amounts of 3-NT accumulated in BCs from smokers compared with nonsmokers. (E) Relative amounts of unmodified tyrosine accumulated in BCs from smokers compared with nonsmokers. (F) The 3-NT/Tyr ratio comparing smokers to nonsmokers was calculated on a subject-by-subject basis by measuring relative ion counts for each molecule in each subject. For all panels, data are presented as mean ± SEM, with statistics calculated by an unpaired Student’s t test.
Transcriptomic Changes of Metabolic Related Genes in BCs Associated with Cigarette Smoking
To assess whether the smoking dependent metabolic changes observed in BCs result from smoking-induced expression changes in genes that encode enzymes associated with production of these metabolites, we analyzed our previously published RNA seq dataset comparing smoker versus nonsmoker primary BCs (18). Even though the gene expression data are generated from an independent dataset and a different passage of cells, they will provide a good estimation of the correlation between smoking-induced gene expression and metabolic changes. Using KEGG pathway analysis, we identified a total of 475 unique enzymes (see the online supplement for complete list) involved in the biochemical pathways that generate the 52 smoking dysregulated metabolites (Table 2). From these 475 genes, we observed a significant (P < 0.05) difference in expression of 80 genes between smokers versus nonsmokers, which was associated with the production of 18 metabolites, including acetyl CoA, coenzyme A, FAD, glutathione, lysophosphatidylethanolamine(18:2), nicotinamide, NAD, phosphatidylcholine(35:2), and thymidine (see Table E1 in the online supplement). Interestingly, for the majority of the 80 genes (67.5%), the direction of expression change between smokers and nonsmokers was concordant with the metabolite data. For example, we observed a significant decrease in the levels of acetyl CoA in smokers versus nonsmokers and a significant decrease in expression of six of eight genes involved in the acetyl CoA pathway (NAA20, KAT2B, ACSS2, KAT7, ACAA2, and ACACB). In addition, for glutathione we observed a significant decrease in the levels of smokers versus nonsmokers and a significant decrease in expression of six of eight genes involved in the glutathione pathway (MGST1, GSTO2, GPX8, GSTK1, ESD, and MGST2). A similar trend was observed for genes linked to the production of other metabolites, including coenzyme A, nicotinamide, NAD and succinate. However, for some genes the direction of expression change between smokers and nonsmokers was discordant with the metabolite data. Examples include genes linked to production of citrulline, lysophosphatidylcholine (16:1), and thymidine. These data suggest that for these metabolites enzyme activity is regulated independent of the transcript level and may involve alternative mechanisms including allosteric control and post-translational modifications.
Discussion
Cigarette smoking, the major risk factor for COPD and lung cancer (30, 31), delivers to the airway epithelium massive amounts of reactive molecules (20). This oxidative burden can overwhelm antioxidant defenses in the lung, leading to lung injury by various mechanisms, likely contributing to a disordered BC function (9–17). To gain insights into the molecular and biochemical mechanisms by which cigarette smoke perturbs BC biology, we used mass spectrometry–based global metabolite profiling to compare the metabolomes of BCs from healthy smokers versus nonsmokers. The LC/MS platform allowed for broad metabolite coverage and analytical sensitivity, uncovering significant smoking-associated perturbations in the levels of molecules that contribute to key metabolic circuits and signaling pathways.
Untargeted metabolite profiling identified 52 metabolites that exhibit significantly altered levels in BCs from healthy smokers relative to nonsmokers. These metabolites include species that are important enzyme cofactors, metabolites of cofactors, contributors to lipid and amino acid metabolism, the citric acid cycle, and cell redox state. Accordingly, the data demonstrated that cigarette smoking alters the BC concentrations of pivotal molecules in intermediary metabolism.
Smoking-affected molecules in BCs include pantothenate (vitamin B5, the essential precursor to coenzyme A, notable for its role in the synthesis and oxidation of fatty acids and oxidation of pyruvate in the citric acid cycle), lumichrome (an oxidation product of vitamin B2, riboflavin), FAD (a redox-active coenzyme derived from riboflavin), and NAD (a redox-active coenzyme derived from niacin). Together, these coenzymes mediate fundamental metabolic reactions and cell signaling events to allow for cell adaptation to a changing environment (23, 32). Mitochondrial ATP synthesis for cell homeostasis is driven by the oxidation of reduced pyridine nucleotides NADH and FADH2, which in turn derive from the beta-oxidation of lipids, glycolysis, and the citric acid cycle (33). A smoking-associated aberration in BC beta-oxidation is consistent with the observation that nonacylated CoA as well as acyl-carnitines (butenyl and dodecanoyl) are significantly increased, whereas acetyl CoA (the proximal “fuel” for beta-oxidation) is significantly diminished. Other nonredox functions for NAD involve NAD-dependent protein deacetylation reactions that contribute to epigenetic regulation and processes such as DNA repair, telomere maintenance, and aging (23, 33). The observation of diminished levels of both NAD and ADP-ribose in BCs from smokers further suggests a possible link between cigarette smoking and altered DNA repair mechanisms that involve NAD-dependent ADP-ribosylation (23). Although the brunt of acetyl CoA generated by glycolysis in most cells is used by mitochondria for ATP generation, cytosolic acetyl CoA is also a driver for protein acetylation, including histone-mediated epigenetic modifications, and contributes to the regulation of autophagy (23, 34). Thus, the decreased levels of acetyl CoA observed in BCs from cigarette smokers can potentially affect mitochondrial respiration, gene transcription, and organellar catabolic pathways.
GSH is a cysteine-containing tripeptide that constitutes the major pool of low-molecular-weight thiols and thereby functions as the pivotal source of intracellular reducing equivalents. Importantly, GSH is essential for the reversal of many oxidative reactions in cells via electron donation, resulting in a concomitant oxidation to glutathione disulfide (GSSG) (35, 36). Glutathione reductase, a constitutive enzyme that is also inducible by oxidative stress, serves to restore GSH from GSSG at the expense of NADPH. Thus, the ratio of GSH to GSSG within cells is a telling biomarker of cell redox status, and an accumulation of GSSG owing to excess oxidant exposure can be inferred from a decrease in the ratio of GSH to GSSG (35, 36). Metabolite profiling revealed markedly diminished levels of GSH in BCs from smokers. Whereas GSSG levels appeared unaffected, the ratio of GSH to GSSG was significantly decreased in BCs from smokers in accord with a decrease in redox potential and a predicted reciprocal increase in oxidative modifications involving proteins and lipids. This finding is in accord with previous mass spectrometric and metabolomic profiling studies reporting decreased GSH levels in bronchial epithelial cells exposed to cigarette smoke components (37, 38). One explanation for the lowered GSH content in BCs from smokers is an increase in protein glutathionylation, not directly quantified in the present study but which can result in altered cell signaling pathway activity owing to protein function perturbations (35, 36).
A significantly elevated level of 3-NT was observed in BCs from smokers, along with an increased 3-NT/Tyr ratio. 3-NT accumulation in airway BCs from smokers is a footprint for the increased formation of reactive nitrogen species in the airways (28). Superoxide reacts at a near diffusion-limited rate with nitric oxide, also a free radical, to form cytotoxic peroxynitrite (ONOO−), a powerful oxidant that directly oxidizes numerous proteins, lipids, and DNA (39–41). The amino acid tyrosine is susceptible to nitration by ONOO−; thus, tyrosine nitration is an indicator of nitrosative stress in numerous pathologies, including conditions involving the respiratory tract (28, 42). Of note, whereas 3-NT formation is increased in BCs from smokers, free tyrosine levels are relatively unchanged. This smoking-associated increase in the 3-NT/Tyr ratio not only serves as a footprint of nitrosative stress, but 3-NT modification of proteins has often been shown to have deleterious effects on cell functions, including the induction of airway hyperresponsiveness and inhibition of pulmonary surfactant activity (43).
One consequence of an increase in NO-derived 3-NT is a diminished availability of NO bioactivity that can perturb physiological airway functions. One important role for NO in the airways may be to maintain ciliary beating rhythmicity, critical for airway mucus clearance, a process that is characteristically defective in smokers with COPD (44). In ciliated airway epithelial cells, NO is selectively produced by neuronal NOS in the axoneme of cilia (45), and this NO production is essential for the preservation of rhythmicity and directionality of beating (46, 47). In addition to 3-NT accumulation as an indicator of diminished NO bioactivity in BCs, we further observed a significant and marked decrease in the NOS co-product citrulline. Thus, taken together, the metabolic findings suggest a diminished level of NO bioactivity in BCs from smokers, with the potential to contribute to initiation or progression of smoking-induced airway disturbances.
It has been previously observed that altered glycerophospholipid metabolism is affected by cigarette smoking (48, 49). In support of this, our metabolomic analysis demonstrated that the levels of most detected glycerophospholipids were significantly diminished in BCs from smokers. Phosphatidylcholine is a major component of cell membranes that confers fluidity to these structures and plays a role in membrane-mediated cell signaling events (50). However, in the lung, phosphatidylcholines also play an important role as cellular osmolytes and pulmonary surfactants (24, 25, 51) that maintain cellular volume and lung tissue elasticity. Although phosphatidylcholine represents 80% of the pulmonary surfactant phospholipids and the bulk of synthesis and secretion takes place in type II alveolar cells (25, 49), it is reported that other cells that line the airway epithelium also produce phospholipid surfactants that help regulate fluid volume and tissue structure (52). The decreased levels of phosphatidylcholine in BCs from smokers could indicate a compromise in their ability to maintain cell volume and the transport of these metabolites. Although sphingolipids have been implicated in lung diseases and recent work by Bowler and colleagues (53) has demonstrated an aberration in plasma sphingolipid metabolism from subjects with COPD, this was not observed in BCs from smokers. The mechanistic basis and functional significance of the observed disturbance in BC phospholipid levels from smokers remains to be determined.
In summary, this study demonstrates that cigarette smoking is associated with significant changes in the metabolic profile of human airway BCs, a cell type that is critical to the maintenance of a normal functioning airway epithelium, with differences in the levels of multiple metabolites, including important cofactors and intermediates for key cellular processes that may contribute to aberrant BC function. The altered metabotype of healthy smoker BCs provides insight into the molecular and biochemical derangements induced by cigarette smoking and serves as a platform for further studies to identify the mechanisms whereby alteration of metabolic pathways affect BC function.
Acknowledgments
Acknowledgments
The authors thank B.-G. Harvey, R. J. Kaner, A. E. Tilley, and M. R. Staudt for help in coordinating and obtaining the large airway epithelium samples for basal cell culture; V. Arbelaez, F. Zhang for technical assistance; and N. Mohamed for help in preparing this manuscript.
Footnotes
This work was supported by National Institutes of Health (NIH) grants HL107882, HL113443, HL087062, UL1 TR000457, and UL1 RR024143 and by the Family Smoking Prevention and Tobacco Control Act. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Food and Drug Administration.
Author Contributions: Conception and design: R.S.D., M.S.W., S.S.G., and R.G.C. Analysis and interpretation: R.S.D., M.S.W., Y.S.-B., Q.C., S.S.G., and R.G.C. Drafting the manuscript for important intellectual content: R.S.D., M.S.W., S.S.G., and R.G.C.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0055OC on July 10, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8:432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 2.Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam A, Wadsworth S, Dorscheid D, Man SF, Sin DD. The airway epithelium: more than just a structural barrier. Ther Adv Respir Dis. 2011;5:255–273. doi: 10.1177/1753465810396539. [DOI] [PubMed] [Google Scholar]
- 4.Evans MJ, Van Winkle LS, Fanucchi MV, Plopper CG. Cellular and molecular characteristics of basal cells in airway epithelium. Exp Lung Res. 2001;27:401–415. doi: 10.1080/019021401300317125. [DOI] [PubMed] [Google Scholar]
- 5.Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells. 2007;25:139–148. doi: 10.1634/stemcells.2006-0288. [DOI] [PubMed] [Google Scholar]
- 6.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackett NR, Shaykhiev R, Walters MS, Wang R, Zwick RK, Ferris B, Witover B, Salit J, Crystal RG. The human airway epithelial basal cell transcriptome. PLoS One. 2011;6:e18378. doi: 10.1371/journal.pone.0018378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auerbach O, Forman JB, Gere JB, Kassouny DY, Muehsam GE, Petrick TG, Smolin HJ, Stout AP. Changes in the bronchial epithelium in relation to smoking and cancer of the lung: a report of progress. N Engl J Med. 1957;256:97–104. doi: 10.1056/NEJM195701172560301. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy SM, Elwood RK, Wiggs BJ, Paré PD, Hogg JC. Increased airway mucosal permeability of smokers: relationship to airway reactivity. Am Rev Respir Dis. 1984;129:143–148. doi: 10.1164/arrd.1984.129.1.143. [DOI] [PubMed] [Google Scholar]
- 11.Lumsden AB, McLean A, Lamb D. Goblet and Clara cells of human distal airways: evidence for smoking induced changes in their numbers. Thorax. 1984;39:844–849. doi: 10.1136/thx.39.11.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wistuba II, Gazdar AF. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1:331–348. doi: 10.1146/annurev.pathol.1.110304.100103. [DOI] [PubMed] [Google Scholar]
- 13.Shaykhiev R, Otaki F, Bonsu P, Dang DT, Teater M, Strulovici-Barel Y, Salit J, Harvey BG, Crystal RG. Cigarette smoking reprograms apical junctional complex molecular architecture in the human airway epithelium in vivo. Cell Mol Life Sci. 2011;68:877–892. doi: 10.1007/s00018-010-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heijink IH, Brandenburg SM, Postma DS, van Oosterhout AJ. Cigarette smoke impairs airway epithelial barrier function and cell-cell contact recovery. Eur Respir J. 2012;39:419–428. doi: 10.1183/09031936.00193810. [DOI] [PubMed] [Google Scholar]
- 15.Hessel J, Heldrich J, Fuller J, Staudt MR, Radisch S, Hollmann C, Harvey BG, Kaner RJ, Salit J, Yee-Levin J, et al. Intraflagellar transport gene expression associated with short cilia in smoking and COPD. PLoS One. 2014;9:e85453. doi: 10.1371/journal.pone.0085453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crystal RG. Airway basal cells: the “smoking gun” of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190:1355–1362. doi: 10.1164/rccm.201408-1492PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaykhiev R, Crystal RG. Early events in the pathogenesis of chronic obstructive pulmonary disease: smoking-induced reprogramming of airway epithelial basal progenitor cells. Ann Am Thorac Soc. 2014;11:S252–S258. doi: 10.1513/AnnalsATS.201402-049AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan DM, Vincent TL, Salit J, Walters MS, Agosto-Perez F, Shaykhiev R, Strulovici-Barel Y, Downey RJ, Buro-Auriemma LJ, Staudt MR, et al. Smoking dysregulates the human airway basal cell transcriptome at COPD risk locus 19q13.2. PLoS One. 2014;9:e88051. doi: 10.1371/journal.pone.0088051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staudt MR, Buro-Auriemma LJ, Walters MS, Salit J, Vincent T, Shaykhiev R, Mezey JG, Tilley AE, Kaner RJ, Ho MW, et al. Airway basal stem/progenitor cells have diminished capacity to regenerate airway epithelium in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190:955–958. doi: 10.1164/rccm.201406-1167LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pryor WA, Stone K. Oxidants in cigarette smoke: radicals, hydrogen peroxide, peroxynitrate, and peroxynitrite. Ann N Y Acad Sci. 1993;686:12–27, discussion 27–28. doi: 10.1111/j.1749-6632.1993.tb39148.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen Q, Park HC, Goligorsky MS, Chander P, Fischer SM, Gross SS. Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS One. 2012;7:e37149. doi: 10.1371/journal.pone.0037149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 23.Green DR, Galluzzi L, Kroemer G. Cell biology: metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang F. Mechanisms and significance of cell volume regulation. J Am Coll Nutr. 2007;26(5, suppl):613S–623S. doi: 10.1080/07315724.2007.10719667. [DOI] [PubMed] [Google Scholar]
- 25.Akella A, Deshpande SB. Pulmonary surfactants and their role in pathophysiology of lung disorders. Indian J Exp Biol. 2013;51:5–22. [PubMed] [Google Scholar]
- 26.Zhuge Y, Yuan Y, van Breemen R, Degrand M, Holian O, Yoder M, Lum H. Stimulated bronchial epithelial cells release bioactive lysophosphatidylcholine 16:0, 18:0, and 18:1. Allergy Asthma Immunol Res. 2014;6:66–74. doi: 10.4168/aair.2014.6.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeisel SH. Metabolic crosstalk between choline/1-carbon metabolism and energy homeostasis. Clin Chem Lab Med. 2013;51:467–475. doi: 10.1515/cclm-2012-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baraldi E, Giordano G, Pasquale MF, Carraro S, Mardegan A, Bonetto G, Bastardo C, Zacchello F, Zanconato S. 3-Nitrotyrosine, a marker of nitrosative stress, is increased in breath condensate of allergic asthmatic children. Allergy. 2006;61:90–96. doi: 10.1111/j.1398-9995.2006.00996.x. [DOI] [PubMed] [Google Scholar]
- 29.Zitka O, Skalickova S, Gumulec J, Masarik M, Adam V, Hubalek J, Trnkova L, Kruseova J, Eckschlager T, Kizek R. Redox status expressed as GSH:GSSG ratio as a marker for oxidative stress in paediatric tumour patients. Oncol Lett. 2012;4:1247–1253. doi: 10.3892/ol.2012.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adcock IM, Caramori G, Barnes PJ. Chronic obstructive pulmonary disease and lung cancer: new molecular insights. Respiration. 2011;81:265–284. doi: 10.1159/000324601. [DOI] [PubMed] [Google Scholar]
- 31.Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer. 2013;13:233–245. doi: 10.1038/nrc3477. [DOI] [PubMed] [Google Scholar]
- 32.Yun J, Johnson JL, Hanigan CL, Locasale JW. Interactions between epigenetics and metabolism in cancers. Front Oncol. 2012;2:163. doi: 10.3389/fonc.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heikal AA. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomarkers Med. 2010;4:241–263. doi: 10.2217/bmm.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder S, Pendl T, Zimmermann A, Eisenberg T, Carmona-Gutierrez D, Ruckenstuhl C, Mariño G, Pietrocola F, Harger A, Magnes C, et al. Acetyl-coenzyme A: a metabolic master regulator of autophagy and longevity. Autophagy. 2014;10:1335–1337. doi: 10.4161/auto.28919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend DM, Tew KD, Tapiero H. The importance of glutathione in human disease. Biomed Pharmacother. 2003;57:145–155. doi: 10.1016/s0753-3322(03)00043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lushchak VI. Glutathione. J Amino Acids. 2012;2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Toorn M, Smit-de Vries MP, Slebos DJ, de Bruin HG, Abello N, van Oosterhout AJ, Bischoff R, Kauffman HF. Cigarette smoke irreversibly modifies glutathione in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1156–L1162. doi: 10.1152/ajplung.00081.2007. [DOI] [PubMed] [Google Scholar]
- 38.Vulimiri SV, Misra M, Hamm JT, Mitchell M, Berger A. Effects of mainstream cigarette smoke on the global metabolome of human lung epithelial cells. Chem Res Toxicol. 2009;22:492–503. doi: 10.1021/tx8003246. [DOI] [PubMed] [Google Scholar]
- 39.Tuo J, Liu L, Poulsen HE, Weimann A, Svendsen O, Loft S. Importance of guanine nitration and hydroxylation in DNA in vitro and in vivo. Free Radic Biol Med. 2000;29:147–155. doi: 10.1016/s0891-5849(00)00324-5. [DOI] [PubMed] [Google Scholar]
- 40.Deeb RS, Hao G, Gross SS, Lainé M, Qiu JH, Resnick B, Barbar EJ, Hajjar DP, Upmacis RK. Heme catalyzes tyrosine 385 nitration and inactivation of prostaglandin H2 synthase-1 by peroxynitrite. J Lipid Res. 2006;47:898–911. doi: 10.1194/jlr.M500384-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrissey BM, Schilling K, Weil JV, Silkoff PE, Rodman DM. Nitric oxide and protein nitration in the cystic fibrosis airway. Arch Biochem Biophys. 2002;406:33–39. doi: 10.1016/s0003-9861(02)00427-7. [DOI] [PubMed] [Google Scholar]
- 43.Haddad IY, Ischiropoulos H, Holm BA, Beckman JS, Baker JR, Matalon S. Mechanisms of peroxynitrite-induced injury to pulmonary surfactants. Am J Physiol. 1993;265:L555–L564. doi: 10.1152/ajplung.1993.265.6.L555. [DOI] [PubMed] [Google Scholar]
- 44.Tilley AE, Walters MS, Shaykhiev R, Crystal RG. Cilia dysfunction in lung disease. Annu Rev Physiol. 2015;77:379–406. doi: 10.1146/annurev-physiol-021014-071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson CL, Lucas JS, Walker WT, Owen H, Premadeva I, Lackie PM. Neuronal NOS localises to human airway cilia. Nitric Oxide. 2015;44:3–7. doi: 10.1016/j.niox.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Jain B, Rubinstein I, Robbins RA, Leise KL, Sisson JH. Modulation of airway epithelial cell ciliary beat frequency by nitric oxide. Biochem Biophys Res Commun. 1993;191:83–88. doi: 10.1006/bbrc.1993.1187. [DOI] [PubMed] [Google Scholar]
- 47.Jiao J, Wang H, Lou W, Jin S, Fan E, Li Y, Han D, Zhang L. Regulation of ciliary beat frequency by the nitric oxide signaling pathway in mouse nasal and tracheal epithelial cells. Exp Cell Res. 2011;317:2548–2553. doi: 10.1016/j.yexcr.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Moré JM, Voelker DR, Silveira LJ, Edwards MG, Chan ED, Bowler RP. Smoking reduces surfactant protein D and phospholipids in patients with and without chronic obstructive pulmonary disease. BMC Pulm Med. 2010;10:53. doi: 10.1186/1471-2466-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal AR, Yin F, Cadenas E. Short-term cigarette smoke exposure leads to metabolic alterations in lung alveolar cells. Am J Respir Cell Mol Biol. 2014;51:284–293. doi: 10.1165/rcmb.2013-0523OC. [DOI] [PubMed] [Google Scholar]
- 50.van Meer G, Voelker DR, Feigenson GW, van MG. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu SH, Possmayer F. Lipid compositional analysis of pulmonary surfactant monolayers and monolayer-associated reservoirs. J Lipid Res. 2003;44:621–629. doi: 10.1194/jlr.M200380-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Khubchandani KR, Snyder JM. Surfactant protein A (SP-A): the alveolus and beyond. FASEB J. 2001;15:59–69. doi: 10.1096/fj.00-0318rev. [DOI] [PubMed] [Google Scholar]
- 53.Bowler RP, Jacobson S, Cruickshank C, Hughes GJ, Siska C, Ory DS, Petrache I, Schaffer JE, Reisdorph N, Kechris K. Plasma sphingolipids associated with chronic obstructive pulmonary disease phenotypes. Am J Respir Crit Care Med. 2015;191:275–284. doi: 10.1164/rccm.201410-1771OC. [DOI] [PMC free article] [PubMed] [Google Scholar]