Abstract
Airway hyperresponsiveness (AHR) is a hallmark feature in asthma characterized by exaggerated airway contractile response to stimuli due to increased airway sensitivity and chronic airway remodeling. We have previously shown that allergen-induced AHR in mice is associated with aberrant DNA methylation in the lung genome, suggesting that AHR could be epigenetically regulated, and these changes might predispose the animals to asthma. Previous studies demonstrated that overexpression of phosphodiesterase 4D (PDE4D) is associated with increased AHR. However, epigenetic regulation of this gene in asthmatic airway smooth muscle cells (ASMCs) has not been examined. In this study, we aimed to examine the relationship between epigenetic regulation of PDE4D and ASMC phenotypes. We identified CpG site–specific hypomethylation at PDE4D promoter in human asthmatic ASMCs. We next used methylated oligonucleotides to introduce CpG site–specific methylation at PDE4D promoter and examined its effect on ASMCs. We showed that PDE4D methylation decreased cell proliferation and migration of asthmatic ASMCs. We further elucidated that methylated PDE4D decreased PDE4D expression in asthmatic ASMCs, increased cAMP level, and inhibited the aberrant increase in Ca2+ level. Moreover, PDE4D methylation reduced the phosphorylation level of downstream effectors of Ca2+ signaling, including myosin light chain kinase and p38. Taken together, our findings demonstrate that gene-specific epigenetic changes may predispose ASMCs to asthma through alterations in cell phenotypes. Modulation of ASMC phenotypes by methylated PDE4D oligonucleotides can reverse the aberrant ASMC functions to normal phenotypes. This has provided new insight to the development of novel therapeutic options for this debilitative disease.
Keywords: airway hyperresponsiveness, airway smooth muscle cell, DNA methylation, methylated DNA oligonucleotides, phosphodiesterase 4D
Clinical Relevance
Prior epigenetic work has focused primarily on epigenetic regulation of pathways involved in the initiation of allergen sensitization in immune cells. However, few studies have investigated lung cell–specific epigenetic changes, which are directly linked to altered lung function. In the present study, we demonstrate that application of methylated DNA oligonucleotides can reverse the aberrant epigenetic changes in asthmatic airway smooth muscle cells and thus modify the airway smooth muscle responsiveness. Our findings may provide insight for the use of epigenetic modifiers in treating asthma through the reversible nature of epigenetic modulations.
Chronic asthma is characterized by persistent airway hyperresponsiveness (AHR), an exaggerated narrowing of the airway in response to a variety of physical and chemical stimuli due to increased airway sensitivity, inflammation, and remodeling (1, 2). Antiinflammatory therapy is currently the primary medication for asthma, and studies of AHR have been prevalently focused on inflammatory cells, mediators, and immune responses (3). However, accumulating evidence shows that AHR may also present in patients with asthma without inflammation, suggesting that airway inflammation cannot fully explain the mechanisms underlying AHR (4–6). Therefore, noninflammatory AHR mechanisms should not be overlooked, and require additional in-depth investigations.
Airway remodeling characterized by airway smooth muscle (ASM) hypertrophy and hyperplasia is a salient feature of asthma, and is linked to AHR (7, 8). In vitro studies have consistently shown that ASM cells (ASMCs) of subjects with asthma are hypercontractile and proliferative compared with nonasthmatic ASMCs (9, 10), indicating that the phenotypic changes persisted in cultured asthmatic ASMCs. Epigenetic modification via DNA methylation, histone modifications, or noncoding RNA is a possible mechanism for the persistent phenotypic changes in cells, tissues, and organs (11). Aberrant changes in histone modifications and microRNAs have been implicated in phenotypic switch in ASMCs (reviewed in Ref. 12). Specific histone modifications at inflammatory or growth factor genes may contribute to the increased production that modulates the ASMC phenotypes (13, 14). Asthmatic ASMCs showed increased histone H3K18 acetylation, and enhanced binding of p300 at the CXCL8 promoter as compared with nonasthmatic ASMCs. On the other hand, ASMCs showed alteration in expression of specific microRNAs in proinflammatory milieu, resulting in aberrant expression of genes modulating ASMC proliferation, hypertrophy, and contractility (15–18). Furthermore, DNA methylation is the best-studied epigenetic mechanisms in asthma studies. DNA methylation changes are associated with asthma (19, 20) and linked to specific triggers of asthma, such as pollutant exposures (21–24). We previously reported that acute exposure to house dust mite (HDM) induced AHR in a mouse model, and the AHR induction was associated with epigenetic modulations of genes related to ASMC proliferation and contraction (25, 26). Specifically, we found promoter demethylation of phosphodiesterase 4D (Pde4d) gene in tracheal ASMCs isolated from HDM-exposed mice. PDE4D is a cAMP-specific phosphodiesterase, which regulates intracellular cAMP level. Knockout of Pde4d abolishes the airway reactivity toward cholinergic stimulation and ovalbumin sensitization in mice (27). PDE4D, through regulation of intracellular cAMP level, modulates a variety of ASM functions, including cell death, cell proliferation, contraction, and migration (28–30). In fact, a genome-wide association analysis study has identified PDE4D as an asthma-susceptibility gene (31). Herein, we hypothesize that epigenetic regulation of PDE4D promoter methylation alters the expression of PDE4D, which leads to aberrant phenotypic changes of human asthmatic ASMCs. In this study, we aimed to: (1) examine the epigenetic alteration of PDE4D promoter occurring at human asthmatic ASMCs; (2) compare the phenotypes, including proliferation, migration, and agonist-induced Ca2+ response, of nonasthmatic and asthmatic ASMCs; and (3) investigate if modulation of PDE4D promoter methylation reverses the aberrant ASMC phenotypes in subjects with asthma. This study provides important information on the epigenetic regulation of PDE4D/cAMP signaling in asthmatic ASMCs, and may lead to a novel modality of treatment for this devastating disease.
Material and Methods
Detailed methods are described in the online supplement.
Cell Culture
Human ASMCs from subjects with and without asthma were either purchased (LONZA Inc., Walkersville, MD) or isolated from deceased donors using methods described previously (32).
Real-Time Quantitative PCR
Total RNA was extracted and reverse transcribed. mRNA levels were quantified by quantitative PCR (qPCR). The 2−ΔΔCt method was used to calculate the relative expression level of transcripts normalized to RPL19.
Bisulfite Genomic Sequencing
Bisulfite conversion of genomic DNA extracted from ASMCs was performed before PCR. PCR amplicon was subcloned into pCR2.1 vector. Three to four individual clones from each donor were sequenced.
Methylation-Specific PCR
CpG site–specific methylation of PDE4D was assayed by qPCR. Control DNAs (fully methylated and fully unmethylated) were mixed in various concentrations and served as quantification standards when determining the percentage of DNA methylation of samples from qPCR.
Transfection with Methylated Oligonucleotides
ASMCs were transfected with nontargeting control or DNA oligonucleotides specific for methylated PDE4D for 3 days. The phosphorothioate oligonucleotides were designed to replace the cytosines in CpG dinucleotides with methylated cytosine (33–35).
Measurement of the Intracellular cAMP Level
Transfected cells were incubated with new medium with 5% FBS for 24 hours before measurement. cAMP levels in cell supernatants were measured with a cAMP high-throughput screening Immunoassay (EMD Millipore, Billerica, MA).
alamarBlue Cell Viability Assay
Cell viability at 3 days after transfection was measured by alamarBlue assay (Invitrogen, Carlsbad, CA).
Cell Migration Assay
Cell migration was measured by QCM Chemotaxis assay (EMD Millipore, Billerica, MA).
Cytosolic Calcium Measurement
Transfected ASMCs were seeded onto glass coverslips overnight in growth medium and were then serum deprived for 48 hours before calcium measurements. Intracellular calcium concentration ([Ca2+]i) was monitored as described previously (36).
Western Blot Analysis
Transfected ASMCs were lysed, resolved by SDS-PAGE and electrotransferred onto a polyvinyl difluoride (PVDF) membrane. Standard Western blot procedures were followed.
Results
PDE4D mRNA was abundantly expressed in asthmatic ASMCs with a level fourfold higher than that in nonasthmatic ASMCs (Figure 1A). In silico analysis revealed that CpG islands (GC% >60%) at 5′ PDE4D encompassed the transcription and translation start sites (Figure 1B). The methylation status of a total of 99 CpG sites at the 5′ promoter region of PDE4D was examined by bisulfite sequencing, and the methylation status of individual CpG site (CpG sites 26–99) at the 5′ PDE4D (−98 to +608, including the 5′ untranslated exon and first exon) is shown in Figure 1C. There was no significant difference in methylation status of CpG sites 1–25 between nonasthmatic and asthmatic ASMCs (data not shown). By contrast, PDE4D promoter was demethylated significantly in asthmatic ASMCs comparing the average percent methylation of CpG sites 26–99 with nonasthmatic ASMC (P = 0.02). Furthermore, a cluster of CpG sites (boxed region, CpG sites 87–90; Figure 1C) showing differential methylation (P < 0.01) was identified in asthmatic ASMCs. These results indicate that PDE4D expression was increased in asthmatic ASMCs, and that the increased mRNA transcription is associated with demethylation in a specific CpG cluster of PDE4D promoter.
Figure 1.
Difference in gene expression and promoter methylation level of phosphodiesterase 4D (PDE4D) between nonasthmatic and asthmatic airway smooth muscle cells (ASMCs). (A) mRNA expression level of PDE4D in asthmatic ASMCs (five donors) relative to nonasthmatic ASMCs (five donors) assayed by quantitative polymerase chain reaction (qPCR). Values are mean ± SD. **P < 0.01. (B) Schematic diagram of CpG dinucleotide (CG) content (%) in the 5′ promoter region of PDE4D. In silico analysis identified the CpG islands (shaded in gray in the genomic DNA sequence) based on the CG content greater than 60% with an observed:expected ratio of 0.6 (MethPrimer). ATG, translational start site; TSS, transcription start site; UTR, untranslated region. PCR amplicon generated by bisulfite sequencing PCR (BSPCR) and methylation-specific qPCR (MSPCR) indicated by the regions bounded by arrows. (C) Methylation status of individual CpG site at the PDE4D promoter in nonasthmatic (three donors) and asthmatic ASMCs (three donors) was assayed by bisulfite sequencing. Unmethylated (open circles) or methylated (solid circles) CpGs are indicated. Each row of circles represents an individual clone sequenced. Three to four individual clones from each donor were picked for sequencing. Boxed area illustrates the specific CpG sites showing differential methylation in asthmatic ASMCs (P < 0.01).
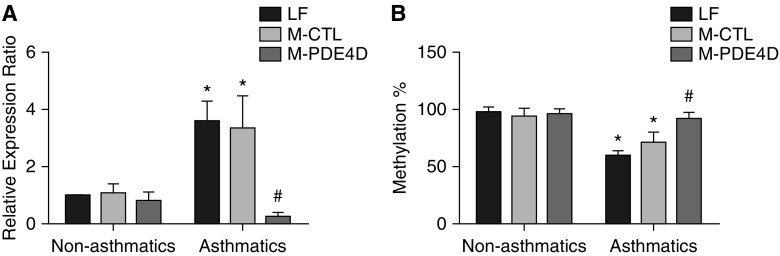
Methylated DNA oligonucleotides were designed to introduce DNA methylation at the specific CpG sites on PDE4D promoter (boxed region). Synthetic methylated oligonucleotides can modify targeted cytosine residue to 5-methyl-cytosine and bind to one strand of the gene to generate a hemimethylated DNA. This hemimethylated DNA allows binding of DNA methyltransferase 1, which catalyzes methylation at the complementary strand, resulting in fully methylated target CpG sites in both DNA strands (33). Asthmatic ASMCs transfected with methylated PDE4D oligonucleotides (M-PDE4D) showed an eightfold reduction in mRNA level of PDE4D compared with those treated with nontargeting methylated DNA oligonucleotides (M-CTL) and lipofectamine (LF) control, whereas PDE4D expression in nonasthmatic ASMCs was unaffected (Figure 2A). In concord with the increased PDE4D expression, the percentage of promoter methylation of PDE4D was significantly lower in the asthmatic ASMCs (Figure 2B). Treatment of asthmatic ASMCs with M-PDE4D completely reversed the reduction of CpG site–specific methylation, as assayed by methylation-specific PCR (Figure 2B). M-PDE4D increased the percentage methylation of PDE4D by approximately 30% (to fully methylated) and a corresponding eightfold reduction in PDE4D mRNA level in asthmatic ASMCs. This suggests that PDE4D was silenced successfully in asthmatic ASMCs by M-PDE4D. The absence of additional silencing effect of the methylated oligonucleotides on PDE4D expression in nonasthmatic ASMCs suggests that their respective CpG sites were already fully methylated.
Figure 2.
Effect of methylated oligonucleotides on gene expression and promoter methylation of PDE4D in nonasthmatic and asthmatic ASMCs. (A) mRNA expression level of PDE4D and (B) CpG site–specific methylation (%) of PDE4D (+366 to +592) assayed by MSPCR in nonasthmatic (five donors) and asthmatic (five donors) ASMCs transfected with PDE4D-targeting (M-PDE4D) or nontargeting methylated oligonucleotide control (M-CTL). β-actin was used as the reference gene to normalize the amount of bisulfite-treated DNA template. Methylated DNA standards (0, 25, 50, 75, and 100% methylated DNA) were used when determining percentage of DNA methylation of samples from qPCR. *P < 0.05 in comparison to lipofectamine (LF) control of nonasthmatic ASMCs; #P < 0.05 in comparison to M-CTL of asthmatic ASMCs.
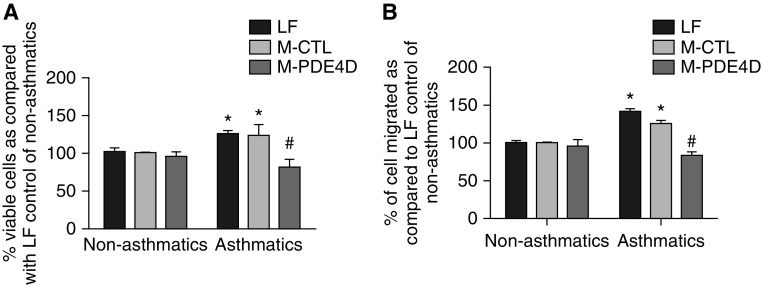
The effect of epigenetic modulation of PDE4D on cell phenotypes was gauged by determining growth and migration of ASMCs transfected with M-PDE4D, M-CTL, or LF. The number of viable asthmatic ASMCs in the M-CTL or LF control was noticeably higher than that of the corresponding nonasthmatic ASMCs, suggesting enhanced proliferation in asthmatic ASMCs. Transfection of M-PDE4D normalized the increase in the viable asthmatic ASMCs, but had no effect on the nonasthmatic ASMCs (Figure 3A). Chemotaxis assay showed that asthmatic ASMCs had a higher migration activity, which was inhibited by M-PDE4D (Figure 3B), but M-PDE4D had no effect on the migration of nonasthmatic ASMCs. These data demonstrate that epigenetic modulation of PDE4D regulates the cell proliferation and migration of asthmatic ASMCs.
Figure 3.
Methylated PDE4D inhibited cell proliferation and migration of asthmatic ASMCs. (A) Cell viability assayed by alamarBlue and (B) cell migration examined by chemotaxis assay in nonasthmatic (three donors) and asthmatic (three donors) ASMCs treated with LF with or without PDE4D-targeting (M-PDE4D) or nontargeting methylated oligonucleotide control (M-CTL). *P < 0.05 in comparison to LF control of nonasthmatic ASMCs; #P < 0.05 in comparison to M-CTL of asthmatic ASMCs.
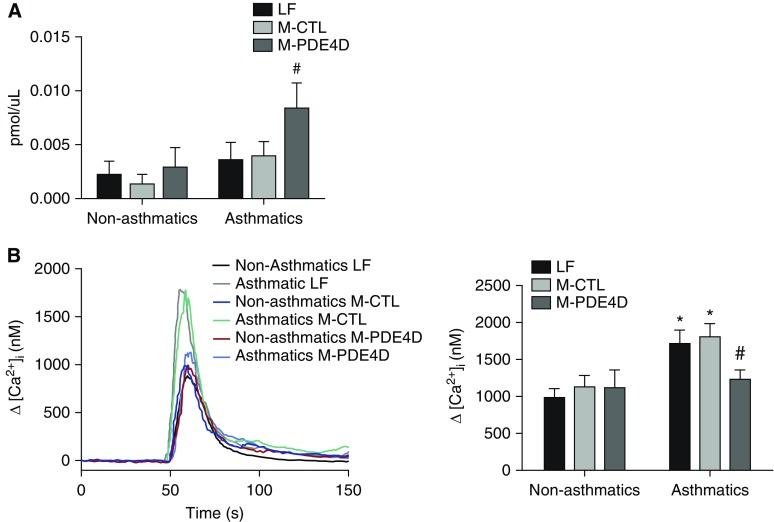
Increase in intracellular cAMP concentration ([cAMP]i) alters ASMC proliferation, migration, and contraction via modulation of the [Ca2+]i (37). To investigate the mechanism underlying the epigenetic effect of PDE4D on the ASMC phenotypes, the change in [cAMP]i and [Ca2+]i in M-PDE4D–transfected ASMCs was measured. There was no significant difference in basal [cAMP]i between nonasthmatic and asthmatic ASMCs, but [cAMP]i was elevated significantly in asthmatic ASMCs after M-PDE4D transfection (Figure 4A). The Ca2+ response elicited by 3 μM histamine was significantly higher in asthmatic ASMCs, compared with the nonasthmatic ASMCs (Figure 4B). M-PDE4D transfection decreased the histamine-induced Ca2+ response by 40% in asthmatic ASMCs to the same level of the nonasthmatic ASMCs. This inhibitory effect of M-PDE4D was not observed in nonasthmatic ASMCs. Our findings indicate that the enhanced Ca2+ response to histamine in asthmatic ASMCs could be related to the epigenetic alteration of PDE4D.
Figure 4.
Methylated PDE4D altered intracellular cAMP and Ca2+ level in asthmatic ASMCs. (A) cAMP level (pmol/μl) was measured by cAMP high-throughput screening immunoassay in nonasthmatic (five donors) and asthmatic (five donors) ASMCs transfected with PDE4D-targeting (M-PDE4D) or nontargeting methylated oligonucleotide (M-CTL), or transfection control (LF). (B) Averaged Ca2+ transient traces showing histamine-induced Ca2+ response in LF, M-CTL, or M-PDE4D–transfected nonasthmatic (five donors) and asthmatic (five donors) ASMCs. Peak Ca2+ responses were quantified as means ± SEM (right panel; a total of 8–18 measurements per treatment group). *P < 0.05 in comparison to LF control of nonasthmatic ASMCs; #P < 0.05 in comparison to M-CTL of asthmatic ASMCs. cAMP, cyclic adenosine monophosphate.
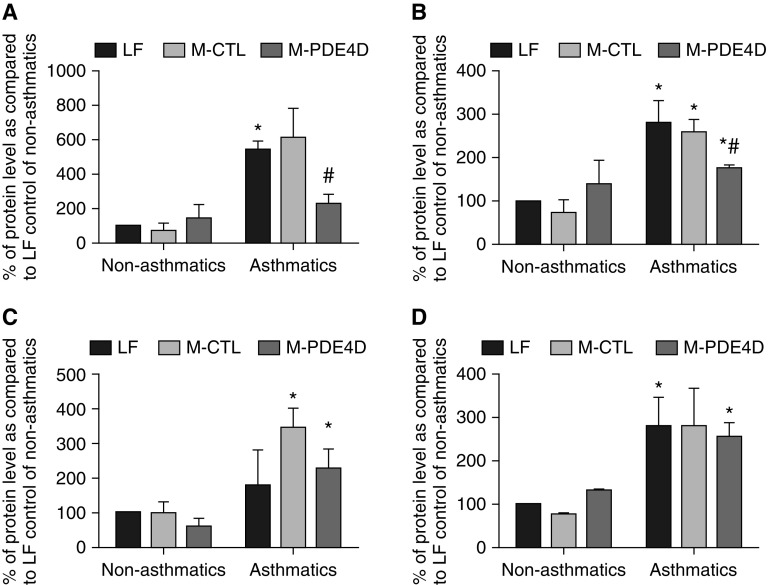
To further investigate if alterations in Ca2+ mobilization via gene silencing of PDE4D leads to aberrant asthmatic ASMC phenotypes, we examined the effect of M-PDE4D on the phosphorylation of myosin light chain kinase (MLCK), p38, and extracellular signal–regulated kinase (ERK) 1/2, which are the downstream effectors of Ca2+-induced signaling (Figure 5). Asthmatic ASMCs showed increased levels of phosphorylated MLCK, p38, and ERK2 proteins as compared with that of nonasthmatic ASMCs. Furthermore, M-PDE4D caused a 120 and 50% decrease in phosphorylation of MLCK and p38, respectively, in asthmatic ASMCs, but had no effect on nonasthmatic ASMCs (Figures 5A and 5B). The expression levels of phosphorylated ERK1/2 in both nonasthmatic and asthmatic cells treated with M-PDE4D were unchanged. Taken together, we provide evidence that epigenetic alteration of PDE4D can modulate the Ca2+ homeostasis and activation of MLCK and p38 signaling, which may contribute to the aberrant ASMC phenotypes observed in subjects with asthma.
Figure 5.
Methylated PDE4D altered phosphorylation of myosin light chain kinase (MLCK) and p38, but not extracellular signal–regulated kinase (ERK) 1/2 in asthmatic ASMCs. Expression of the phosphorylated (A) MLCK, (B) p38, (C) ERK1, and (D) ERK2 proteins in nonasthmatic (three donors) and asthmatic (three donors) ASMCs transfected with PDE4D-targeting (M-PDE4D) or nontargeting methylated oligonucleotide (M-CTL), or transfection control (LF) was quantitated as percent of band intensity as compared with LF control of nonasthmatic ASMCs. *P < 0.05 in comparison to LF control of nonasthmatic ASMCs; #P < 0.05 in comparison to M-CTL of asthmatic ASMCs.
Discussion
The present study demonstrates that PDE4D promoter demethylation contributes to the increased PDE4D gene expression in asthmatic ASMCs. This is in agreement with our previous study showing that HDM-induced AHR was associated with the epigenetic alterations of Pde4d (25). Furthermore, we showed that the epigenetic alteration of PDE4D was associated with the abnormal increase in cell proliferation, migration, and histamine-induced Ca2+ response in asthmatic ASMCs. By introducing PDE4D promoter methylation by methylated oligonucleotides, the aberrant phenotypes in asthmatic ASMCs could be reversed. Our results, hence, suggest that the CpG site–specific demethylation of PDE4D may account, at least in part, for the altered phenotypes of asthmatic ASMCs that are responsible for the airway remodeling and AHR in patients with asthma. Strikingly, a moderate change in CpG site–specific methylation at the PDE4D promoter by methylated oligonucleotides caused a dramatic decrease in PDE4D expression. By in silico search of the TRANSFAC database (38), we identified transcriptional factors (TFs), Sp1, Sp3, E2f-1, and p300, may potentially bind to the specific CpG sites (boxed area, Figure 1C) at the PDE4D promoter. De novo methylation has been shown to be associated with the recognition site of Sp1 and E2f-1 (39, 40). Sp3 is shown to interact with histone deacetylases (41), whereas p300 histone acetyltransferase modulates gene transcription via chromatin remodeling (42). Given the fact that DNA methylation occurring at recognition sites of TFs may suppress the gene transcription by either inhibiting the binding of TFs to the promoter and/or hindering the chromatin stability by further recruiting DNA methyltransferases and histone modification enzymes to the promoter, it is possible that moderate or slight changes in methylation pattern at specific CpGs may contribute to a larger change in gene expression level. Although future studies are needed to examine the regulation of the PDE4D transcriptional activity, our data provide an insight in to the development of efficient and specific inhibitors for PDE4D.
PDE4D belongs to the PDE4 family, and is the major PDE subtype in human ASMCs. It is a cAMP-specific phosphodiesterase for the degradation of cAMP for lowering [cAMP]i. cAMP plays important roles in various physiological functions of ASMCs. β2-Agonist inhibits ASMC proliferation and contraction through cAMP production mediated by G protein–coupled receptors and adenylyl cyclase (43, 44). Trian and colleagues (28) reported that β2 adrenergic receptor-mediated cAMP generation is dysregulated in asthmatic ASMCs by the increased cAMP degradation by PDE4, may be partly due to the increased PDE4D expression. They further propose the intrinsic abnormality seen in asthmatic ASMCs in the absence of proinflammatory milieu may be due to the genetic and/or epigenetic predisposition. Herein, we provide the evidence that PDE4D is overexpressed via promoter demethylation in asthmatic ASMCs. Increased PDE4D expression enhances hydrolysis of cAMP that inhibits activity of protein kinase A. Reduction in protein kinase A activity inhibits the sarcoplasmic reticulum (SR) Ca2+ release from inositol trisphoshate receptor and extracellular Ca2+ entry, and lessens the amelioration of SR Ca2+ uptake via sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) (45). Eventually, the intracellular Ca2+ level is increased, which may up-regulate MLCK and p38, leading to enhanced ASMC contraction, proliferation, and migration (46–49). Hence, PDE4D up-regulation can suppress the cAMP-dependent signaling pathways, leading to aberrant ASMC phenotypes. Our data support the hypothesis that the aberrant cAMP production is “programmed” via epigenetic regulation of PDE4D, and it may predispose the ASMCs to be hyperresponsive. On the other hand, gene silencing of PDE4D via DNA methylation contributes to the decreased Ca2+-dependent activities through cAMP signaling to inhibit cell growth and migration, suppress ASM contraction, and shift ASM tone to relaxation. We summarized the possible mechanisms underlying epigenetic regulation of ASMCs via PDE4D methylation in Figure 6.
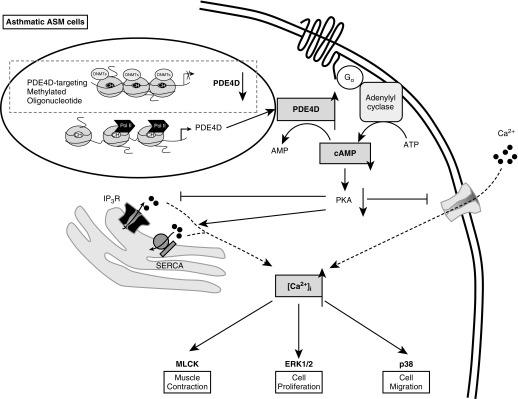
Figure 6.
Proposed mechanisms of how epigenetic regulation of PDE4D via promoter methylation alters ASMC phenotypes. In asthmatic ASMCs, PDE4D promoter is aberrantly unmethylated, and hence promotes PDE4D gene expression. Overexpression of PDE4D enhances the hydrolysis of cAMP and lowers the intracellular level of cAMP that could lead to decreased activity of protein kinase A (PKA). Reduction in PKA activity inhibits the sarcoplasmic reticulum (SR) Ca2+ release from inositol trisphoshate receptor (IP3R) and extracellular Ca2+ entry, and lessens the amelioration of SR Ca2+ uptake via sacro/endoplasmic reticulum Ca2+ ATPase (SERCA). Eventually, the intracellular Ca2+ concentration ([Ca2+]i) is increased, which may up-regulate MLCK, p38, and/or ERK1/2, leading to enhanced cell contraction, proliferation, and migration. Aberrant ASMC phenotypes may predispose the ASMCs to be hyperresponsive. Introduction of CpG site–specific methylation at PDE4D promoter by methylated oligonucleotide results in suppression of PDE4D expression to the level of the nonasthmatic ASMCs and ultimately reverses the aberrant cell phenotypes seen in asthmatic ASMCs. CH, methyl group at 5' cytosine; DNMT, DNA methyltransferase; Gα, G-protein alpha; Pol II, RNA polymerase II.
The important role of PDE4D in asthma has been implicated in a genome-wide association analysis study that identified PDE4D as an asthma-susceptibility gene. Multiple PDE4D single-nucleotide polymorphisms (SNPs) were strongly associated with patients with asthma of different ethnicities. However, to date, there has been no direct study on the effect of PDE4D SNPs on PDE4D function. Our present study indicates that, in addition to the SNPs, PDE4D is epigenetically modulated in asthmatic ASMCs. However, how the persistent epigenetic alteration of PDE4D is established in asthmatic ASMCs remains unknown. We previously demonstrated the changes in mRNA level of the DNA methylation modulators, including Dnmt3A, methyl-CpG–binding domain proteins (Mbd2 and Mbd3), and ten-eleven translocation proteins (Tet1) in mouse chronically exposed to house allergen (26). It will be informative to further examine how PDE4D is epigenetically regulated by these DNA methylation modulators, and how these modifications persist in the presence or absence of stimuli as in our cultured asthmatic ASMCs. AHR is often considered an epiphenomenon of airway inflammation. However, recent studies revealed that ASMCs showed asthmatic phenotypes, even in the absence of the inflammation (4, 5, 6). Our findings show that asthmatic ASMCs are epigenetically modulated, and such epigenetic alterations could be the consequence of prior allergen exposures, inflammation, or some as-yet unidentified mechanisms that predispose the ASMC to being highly proliferative and contractile.
Pharmacological PDE inhibitors have been recently developed for the treatment of asthma because of their bronchodilator and antiinflammatory effects (50). Although the causal relationship between epigenetic regulation of PDE4D and airway inflammation has not yet been studied, we here demonstrate the successful modification of the CpG site–specific methylation of PDE4D in asthmatic ASMCs using methylated DNA oligonucleotides, which may provide an alternative method of epigenetic therapy for asthma. It is exciting to speculate as to the possibility of reversing the predisposed asthmatic phenotype in ASMCs to normal status with gene-specific methylated DNA oligonucleotides.
Acknowledgments
Acknowledgments
We would like to thank Drs. Steven An (Johns Hopkins University) and Reynold Panettieri (University of Pennsylvania Medical Center) for providing us the human ASM cells and guidance on culturing the human ASM cells.
Footnotes
This work was supported by National Institute of Environmental Health Sciences grant ES016887 (W.-y.T.) and National Heart, Lung, and Blood Institute grant HL10342 (W.M.); A.H.Y.L. was supported by a postdoctoral fellowship from the American Lung Association, and Y.S. was supported by National Natural Science Foundation of China grant 81000006.
Author Contributions: Conception and design—A.H.Y.L., J.S.K.S., W.-y.T.; analysis and interpretation—A.H.Y.L., Y.S., J.S.K.S., and W.-y.T.; reviewing the manuscript for important intellectual content—A.H.Y.L., W.M., J.S.K.S., and W.-y.T.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0079OC on July 16, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Dekkers BG, Maarsingh H, Meurs H, Gosens R. Airway structural components drive airway smooth muscle remodeling in asthma. Proc Am Thorac Soc. 2009;6:683–692. doi: 10.1513/pats.200907-056DP. [DOI] [PubMed] [Google Scholar]
- 2.Nakagome K, Nagata M. Pathogenesis of airway inflammation in bronchial asthma. Auris Nasus Larynx. 2011;38:555–563. doi: 10.1016/j.anl.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Barnes PJ. Anti-inflammatory therapy for asthma. Annu Rev Med. 1993;44:229–242. doi: 10.1146/annurev.me.44.020193.001305. [DOI] [PubMed] [Google Scholar]
- 4.Baroffio M, Barisione G, Crimi E, Brusasco V. Noninflammatory mechanisms of airway hyper-responsiveness in bronchial asthma: an overview. Ther Adv Respir Dis. 2009;3:163–174. doi: 10.1177/1753465809343595. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes DJ, Mitchell RW, Lakser O, Dowell M, Stewart AG, Solway J. Do inflammatory mediators influence the contribution of airway smooth muscle contraction to airway hyperresponsiveness in asthma? J Appl Physiol (1985) 2003;95:844–853. doi: 10.1152/japplphysiol.00192.2003. [DOI] [PubMed] [Google Scholar]
- 6.Grainge CL, Lau LC, Ward JA, Dulay V, Lahiff G, Wilson S, Holgate S, Davies DE, Howarth PH. Effect of bronchoconstriction on airway remodeling in asthma. N Engl J Med. 2011;364:2006–2015. doi: 10.1056/NEJMoa1014350. [DOI] [PubMed] [Google Scholar]
- 7.Al Heialy S, McGovern TK, Martin JG. Insights into asthmatic airway remodelling through murine models. Respirology. 2011;16:589–597. doi: 10.1111/j.1440-1843.2011.01974.x. [DOI] [PubMed] [Google Scholar]
- 8.Bentley JK, Hershenson MB. Airway smooth muscle growth in asthma: proliferation, hypertrophy, and migration. Proc Am Thorac Soc. 2008;5:89–96. doi: 10.1513/pats.200705-063VS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto H, Moir LM, Oliver BG, Burgess JK, Roth M, Black JL, McParland BE. Comparison of gel contraction mediated by airway smooth muscle cells from patients with and without asthma. Thorax. 2007;62:848–854. doi: 10.1136/thx.2006.070474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelaia G, Renda T, Gallelli L, Vatrella A, Busceti MT, Agati S, Caputi M, Cazzola M, Maselli R, Marsico SA. Molecular mechanisms underlying airway smooth muscle contraction and proliferation: implications for asthma. Respir Med. 2008;102:1173–1181. doi: 10.1016/j.rmed.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Tang WY, Ho SM. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8:173–182. doi: 10.1007/s11154-007-9042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clifford RL, Singer CA, John AE. Epigenetics and miRNA emerge as key regulators of smooth muscle cell phenotype and function. Pulm Pharmacol Ther. 2013;26:75–85. doi: 10.1016/j.pupt.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clifford RL, Patel JK, John AE, Tatler AL, Mazengarb L, Brightling CE, Knox AJ. CXCL8 histone H3 acetylation is dysfunctional in airway smooth muscle in asthma: regulation by BET. Am J Physiol Lung Cell Mol Physiol. 2015;308:L962–L972. doi: 10.1152/ajplung.00021.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol. 2012;189:819–831. doi: 10.4049/jimmunol.1103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42:506–513. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3β. J Biol Chem. 2010;285:29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jude JA, Dileepan M, Subramanian S, Solway J, Panettieri RA, Jr, Walseth TF, Kannan MS. miR-140-3p regulation of TNF-α–induced CD38 expression in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2012;303:L460–L468. doi: 10.1152/ajplung.00041.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry MM, Baker JE, Gibeon DS, Adcock IM, Chung KF. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am J Respir Cell Mol Biol. 2014;50:7–17. doi: 10.1165/rcmb.2013-0067OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC. DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol. 2008;28:139–146. doi: 10.1007/s10875-007-9148-1. [DOI] [PubMed] [Google Scholar]
- 20.Brand S, Kesper DA, Teich R, Kilic-Niebergall E, Pinkenburg O, Bothur E, Lohoff M, Garn H, Pfefferle PI, Renz H. DNA methylation of TH1/TH2 cytokine genes affects sensitization and progress of experimental asthma. J Allergy Clin Immunol. 2012;129:1602–10.e6. doi: 10.1016/j.jaci.2011.12.963. [DOI] [PubMed] [Google Scholar]
- 21.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, Miller RL, Perera F, Ho SM. Maternal exposure to polycyclic aromatic hydrocarbons and 5′-CpG methylation of interferon-γ in cord white blood cells. Environ Health Perspect. 2012;120:1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sofer T, Baccarelli A, Cantone L, Coull B, Maity A, Lin X, Schwartz J. Exposure to airborne particulate matter is associated with methylation pattern in the asthma pathway. Epigenomics. 2013;5:147–154. doi: 10.2217/epi.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadeau K, McDonald-Hyman C, Noth EM, Pratt B, Hammond SK, Balmes J, Tager I. Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol. 2010;126:845–852.e10. doi: 10.1016/j.jaci.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Rager JE, Bauer RN, Müller LL, Smeester L, Carson JL, Brighton LE, Fry RC, Jaspers I. DNA methylation in nasal epithelial cells from smokers: identification of ULBP3-related effects. Am J Physiol Lung Cell Mol Physiol. 2013;305:L432–L438. doi: 10.1152/ajplung.00116.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang Y, Das S, Rabold R, Sham JS, Mitzner W, Tang WY. Epigenetic alterations by DNA methylation in house dust mite–induced airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2013;49:279–287. doi: 10.1165/rcmb.2012-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng RY, Shang Y, Limjunyawong N, Dao T, Das S, Rabold R, Sham JS, Mitzner W, Tang WY. Alterations of the lung methylome in allergic airway hyper-responsiveness. Environ Mol Mutagen. 2014;55:244–255. doi: 10.1002/em.21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen G, Jin S, Umetsu DT, Conti M. Absence of muscarinic cholinergic airway responses in mice deficient in the cyclic nucleotide phosphodiesterase PDE4D. Proc Natl Acad Sci USA. 2000;97:6751–6756. doi: 10.1073/pnas.97.12.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trian T, Burgess JK, Niimi K, Moir LM, Ge Q, Berger P, Liggett SB, Black JL, Oliver BG. β2-Agonist induced cAMP is decreased in asthmatic airway smooth muscle due to increased PDE4D. PLoS One. 2011;6:e20000. doi: 10.1371/journal.pone.0020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Méhats C, Jin SL, Wahlstrom J, Law E, Umetsu DT, Conti M. PDE4D plays a critical role in the control of airway smooth muscle contraction. FASEB J. 2003;17:1831–1841. doi: 10.1096/fj.03-0274com. [DOI] [PubMed] [Google Scholar]
- 30.Kolosionek E, Savai R, Ghofrani HA, Weissmann N, Guenther A, Grimminger F, Seeger W, Banat GA, Schermuly RT, Pullamsetti SS. Expression and activity of phosphodiesterase isoforms during epithelial mesenchymal transition: the role of phosphodiesterase 4. Mol Biol Cell. 2009;20:4751–4765. doi: 10.1091/mbc.E09-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tirumurugaan KG, Jude JA, Kang BN, Panettieri RA, Walseth TF, Kannan MS. TNF-α induced CD38 expression in human airway smooth muscle cells: role of MAP kinases and transcription factors NF-κB and AP-1. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1385–L1395. doi: 10.1152/ajplung.00472.2006. [DOI] [PubMed] [Google Scholar]
- 33.Yao X, Hu JF, Daniels M, Shiran H, Zhou X, Yan H, Lu H, Zeng Z, Wang Q, Li T, et al. A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma. J Clin Invest. 2003;111:265–273. doi: 10.1172/JCI15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, McNeal JE, Ho SM. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Wu M, Xiao H, Lee MT, Levin L, Leung YK, Ho SM. Methylation of a single intronic CpG mediates expression silencing of the PMP24 gene in prostate cancer. Prostate. 2010;70:765–776. doi: 10.1002/pros.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang YL, Lin AH, Xia Y, Lee S, Paudel O, Sun H, Yang XR, Ran P, Sham JS. Nicotinic acid adenine dinucleotide phosphate (NAADP) activates global and heterogeneous local Ca2+ signals from NAADP- and ryanodine receptor-gated Ca2+ stores in pulmonary arterial myocytes. J Biol Chem. 2013;288:10381–10394. doi: 10.1074/jbc.M112.423053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozier A, Allard B, Bara I, Girodet PO, Trian T, Marthan R, Berger P. The pivotal role of airway smooth muscle in asthma pathophysiology. J Allergy. 2011;2011:742710. doi: 10.1155/2011/742710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 39.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 40.Saadeh H, Schulz R. Protection of CpG islands against de novo DNA methylation during oogenesis is associated with the recognition site of E2f1 and E2f2. Epigenetics Chromatin. 2014;7:26. doi: 10.1186/1756-8935-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunes MJ, Milagre I, Schnekenburger M, Gama MJ, Diederich M, Rodrigues E. Sp proteins play a critical role in histone deacetylase inhibitor–mediated derepression of CYP46A1 gene transcription. J Neurochem. 2010;113:418–431. doi: 10.1111/j.1471-4159.2010.06612.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3–lysine 4–specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 43.Hakonarson H, Grunstein MM. Regulation of second messengers associated with airway smooth muscle contraction and relaxation. Am J Respir Crit Care Med. 1998;158:S115–S122. doi: 10.1164/ajrccm.158.supplement_2.13tac700. [DOI] [PubMed] [Google Scholar]
- 44.Stewart AG, Tomlinson PR, Wilson JW. β2-Adrenoceptor agonist–mediated inhibition of human airway smooth muscle cell proliferation: importance of the duration of β2-adrenoceptor stimulation. Br J Pharmacol. 1997;121:361–368. doi: 10.1038/sj.bjp.0701128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nuttle LC, Farley JM. Frequency modulation of acetylcholine-induced oscillations in Ca++ and Ca(++)-activated Cl− current by cAMP in tracheal smooth muscle. J Pharmacol Exp Ther. 1996;277:753–760. [PubMed] [Google Scholar]
- 46.Janssen LJ, Tazzeo T, Zuo J. Enhanced myosin phosphatase and Ca2+-uptake mediate adrenergic relaxation of airway smooth muscle. Am J Respir Cell Mol Biol. 2004;30:548–554. doi: 10.1165/rcmb.2003-0212OC. [DOI] [PubMed] [Google Scholar]
- 47.Pfitzer G. Invited review: regulation of myosin phosphorylation in smooth muscle. J Appl Physiol (1985) 2001;91:497–503. doi: 10.1152/jappl.2001.91.1.497. [DOI] [PubMed] [Google Scholar]
- 48.Burgess JK, Lee JH, Ge Q, Ramsay EE, Poniris MH, Parmentier J, Roth M, Johnson PR, Hunt NH, Black JL, et al. Dual ERK and phosphatidylinositol 3-kinase pathways control airway smooth muscle proliferation: differences in asthma. J Cell Physiol. 2008;216:673–679. doi: 10.1002/jcp.21450. [DOI] [PubMed] [Google Scholar]
- 49.Gerthoffer WT. Migration of airway smooth muscle cells. Proc Am Thorac Soc. 2008;5:97–105. doi: 10.1513/pats.200704-051VS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beghè B, Rabe KF, Fabbri LM. Phosphodiesterase-4 inhibitor therapy for lung diseases. Am J Respir Crit Care Med. 2013;188:271–278. doi: 10.1164/rccm.201301-0021PP. [DOI] [PubMed] [Google Scholar]