Abstract
Owing to the extensive development of drug resistance in pathogens against the available antibiotic arsenal, antimicrobial resistance is now an emerging major threat to public healthcare. Anti-virulence drugs are a new type of therapeutic agent aiming at virulence factors rather than killing the pathogen, thus providing less selective pressure for evolution of resistance. One promising example of this therapeutic concept targets bacterial quorum sensing (QS), because QS controls many virulence factors responsible for bacterial infections. Marine sponges and their associated bacteria are considered a still untapped source for unique chemical leads with a wide range of biological activities. In the present study, we screened extracts of 14 sponge species collected from the Red and Mediterranean Sea for their quorum-quenching (QQ) potential. Half of the species showed QQ activity in at least 2 out of 3 replicates. Six out of the 14 species were selected for bacteria isolation, to test for QQ activity also in isolates, which, once cultured, represent an unlimited source of compounds. We show that ≈20% of the isolates showed QQ activity based on a Chromobacterium violaceum CV026 screen, and that the presence or absence of QQ activity in a sponge extract did not correlate with the abundance of isolates with the same activity from the same sponge species. This can be explained by the unknown source of QQ compounds in sponge-holobionts (host or symbionts), and further by the possible non-symbiotic nature of bacteria isolated from sponges. The potential symbiotic nature of the isolates showing QQ activity was tested according to the distribution and abundance of taxonomically close bacterial Operational Taxonomic Units (OTUs) in a dataset including 97 sponge species and 178 environmental samples (i.e., seawater, freshwater, and marine sediments). Most isolates were found not to be enriched in sponges and may simply have been trapped in the filtration channels of the sponge at the time of collection. Our results highlight potential for QQ-bioactive lead molecules for anti-virulence therapy both from sponges and the bacteria isolated thereof, independently on the symbiotic nature of the latter.
Keywords: anti-virulence, quorum sensing, porifera, biofilm inhibition, pyocyanin, Pseudomonas aeruginosa
Introduction
Overuse of antibiotics is one of the factors involved in emergence of drug resistant pathogens. The discovery of novel drugs is required to solve this emergent problem. The understanding of how inter-cellular microbial communication is involved in bacterial pathogenesis has revealed potential for alternative strategies to treat bacteria-mediated diseases (Rasko and Sperandio, 2010; LaSarre and Federle, 2013). Quorum sensing (QS) is a type of bacterial communication system that relies on secreted signaling molecules which regulate coordinated responses across a bacterial population (Lazdunski et al., 2004). In many cases, the responses elicited by QS signals contribute directly to pathogenesis through the synchronized production of virulence determinants, such as toxins and proteases (Rutherford and Bassler, 2012). It has been theorized that, if the signal communication that coordinates these pathogenic behaviors was blocked, bacteria would lose their ability to mount an organized assault on the host and thus their ability to form organized community structures with antibiotic resistance would be compromised (Cegelski et al., 2008). QS inhibitor compounds inactivate QS by different quorum quenching (QQ) mechanisms including enzymatic inactivation of the signal molecule (Dong et al., 2000, 2001), inhibition of signal biosynthesis (Hoang and Schweizer, 1999) and inhibition of signal detection (Yang et al., 2009; Ding et al., 2011). Although QS and QQ processes were both first discovered in marine organisms, information on QQ processes in the marine environment is lagging behind compared to the studies describing the wide presence of QQ activity in soil, plants and other terrestrial samples (e.g., Dong et al., 2000; Romero et al., 2011).
The majority of modern drugs either belongs to natural products or originate from them (Li and Vederas, 2009), with marine sponges being one of the most prolific sources of chemical diverse bioactive natural products (Blunt et al., 2015). These compounds have been studied in respect to chemical ecology and provide the sponge with defense against predation, fouling, and pathogen attack (e.g., Chu et al., 2013; Rohde et al., 2015). From a biotechnology perspective, these compounds can have a broad spectrum of action comprising anti-inflammatory, anticancer, and immune-modulating activities (e.g., Costantino et al., 2008, 2009; Mayer et al., 2010; Teta et al., 2013; Blunt et al., 2015; Kaufman et al., 2015). In this study we were interested in novel antimicrobials that are based on the QQ mechanism of action. We focused on sponges based on previous studies that demonstrated the potential of sponges as a promising source for QQ compounds (Skindersoe et al., 2008; Dobretsov et al., 2011; Pejin et al., 2014; Mai et al., 2015). The discovery and sustainable production of bioactive chemical compounds from sponges requires large amounts of tissue to be processed, which is often limited by ecological concerns (the finite availability of sponges in the sea). A growing body of evidence indicates that many of these chemical compounds are produced by the sponge-associated microbiome, which includes bacteria, archaea, fungi, and algae (Thomas et al., 2010). The isolation of bacteria responsible for production of metabolites of interest would circumvent the supply problem.
In the present study we screened 14 sponge species collected from the Red Sea and the Mediterranean Sea for QQ potential using reporter strain QSIS1 and Chromobacterium violaceum CV026. Eighty-six isolates deriving from six selected sponge species were further evaluated for QQ activity using C. violaceum CV026 and Agrobacterium tumefaciens NT1 (pZLR4) biosensors. Based on the data obtained from the QQ screening, extract from 17 isolates were screened further for inhibition of virulence factors. To assess whether QQ-active isolates are of likely symbiotic nature, and thus may be considered potential source of metabolites found in the sponge from which they were isolated, we determined the distribution and abundance of these 17 isolates in 97 sponge species vs. 178 environmental samples (including seawater, freshwater and marine sediments). Finally, out of the 17 QQ active isolates, four showing highest anti-virulence potential were used for metabolic profiling in view of future identification of the active molecules.
Materials and methods
Sponge sampling
A few cubic centimeters of sponge tissue were collected by scuba diving at 5–30 m depth from the Mediterranean (Achziv nature marine reserve) and Red Sea (Eilat, Israel). Sponges were identified morphologically following the Systema Porifera classification system (Hooper and Soest, 2002). For each sponge, samples were preserved in three different ways: (i) samples were placed individually in ziploc® plastic bags and then carried to the laboratory for bacterial isolation; (ii) samples were preserved in 90% ethanol as vouchers for DNA-based taxonomic identification (deposited in the Marine Microbiology Laboratory, Department of Marine Biology, University of Haifa, Israel); and (iii) samples were immediately frozen in liquid nitrogen upon surfacing from the dives, the frozen tissues were freeze dried using a lyophilizer, and the dried tissues were utilized for chemical extractions. Samples were collected in compliance with the 40246/2014 permit from the Israel Nature and National Parks Protection Authority.
Preparation of sponge extracts and screening for QQ activity
Lyophilized sponge tissue was macerated and extracted thrice with choloroform, chlorofom:methanol (1:1) and methanol with sonication and then filtered. All filtrates were combined and the solvents were evaporated under vacuum using a rotary vacuum evaporator (Heidolph, Germany). Extract stocks were prepared by dissolving with methanol to give a final concentration of 16 mg/mL. QQ screening of sponge extracts were performed following previously established protocols using the QSIS1 system (Rasmussen et al., 2005a) and the C. violaceum CV026 assay (McClean et al., 1997).
Isolation of heterotrophic bacteria
Samples collected in ziploc® plastic bags for bacterial isolation were washed with autoclaved calcium-magnesium free artificial sea water (ASW) to remove the loosely bound bacterial cells as well as debris. Approximately, 1 cm3 of the sponge tissue was cut with a sterile scalpel and immediately transferred to 9 mL of sterile ASW and grounded using mortar and pestle. Several dilutions (10−1 to 10−5) were prepared and 100 μL of each spread on different solid media. Culturing media used for the isolation of marine bacteria included: Marine agar (MA–Difco), International Streptomyces Project (ISP) 1 and 2, Starch Casein agar (SCA), Kuster's agar (KA) (Kumar and Kannabiran, 2010), M1 medium (M1) (Abdelmohsen et al., 2010), Casamino acid agar (CAA) (Webster et al., 2001), Luria-Bertani agar (LA-HiMedia), and Nutrient agar (NA-HiMedia) were prepared with 0.2 μm filtered natural seawater and 3 or 6% sea salt. All solid media contained Difco Bacto agar (20 g/L). Plates were incubated at 28°C and observed for growth after 2–4 weeks of incubation. Every 4 days, colonies were chosen according to their morphological characteristics and then transferred to slant culture to preserve at 4°C as well as to 20% (v/v) glycerol stock at −80°C.
Phylogenetic analysis of isolated strains
Molecular taxonomic characterization was performed using 16S rRNA sequencing. Isolation of DNA (Rainey et al., 1996), PCR primers and PCR conditions were adopted from previous studies (Jiang et al., 2006). Briefly, part of the 16S rRNA gene was amplified using MyTaq™ Red Mix (Bioline) and the primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-CGGTTACCTTGTTACGACTT-3′) (Sigma-Aldrich, Germany). PCR reactions were performed in a final volume of 50 μL in a Thermal Cycler (Bioer Technology), according to the following profile: 4 min at 94°C and 35 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C followed by 5 min at 72°C. The PCR product was purified and sequenced at Hylabs (Israel) using the forward primer 27F. Some strains were sequenced in addition with the reverse primer 1492R. In these cases, the two sequences were merged to give a near full length 16S rRNA gene sequence. The sequences were compared with those available in Genbank using BLASTn (Altschul et al., 1997). For the phylogenetic analysis, sequences of the closest described type strains were obtained from EzTaxon (Kim et al., 2012). Sequences of isolates and type strains were aligned using the SINA aligner verison 1.2.11 (Pruesse et al., 2012) and the alignment was manually improved. The final alignment contained 144 sequences and 693 positions.
A maximum likelihood phylogenetic tree based on the alignment was constructed using Mega 6.06 (Tamura et al., 2013). The Kimura-two-parameter model with Gamma distributed rate variation among sites and a proportion of invariant sites (+G +I) was the best substitution model for the data according to the Bayesian information criterion (BIC) of the model test implemented in MEGA6.06. The initial tree for the heuristic search with the Nearest Neighbor Interchange (NNI) algorithm was a Neighbor-Joining tree base on pairwise distances using the maximum composite likelihood approach. Validation of reproducibility of the branching patterns was made by bootstrap based on 100 re-samplings.
Bacterial extract preparation and screening
Isolates were grown in 50 mL of AM3 production medium (Bacterial peptone 15 g/L, soybean powder 5 g/L, soluble starch 15 g/L, glycerol 15 g/L, CaCO3 2 g/L, and Sea salt 3%, adjusted to pH 7.2) in a 250 mL Erlenmeyer flask and incubated for 7 days in an incubator shaker (200 rpm) at 28°C. After incubation, equal volume of butanone was added to each culture flask and then sonicated for 30 min. The organic phase was separated and concentrated in a rotary vacuum to obtain the crude extract. Dried crude extracts were used to prepare stock solutions using methanol to obtain the final concentration of 16 mg/mL. QQ activity was tested with two biosensors C. violaceum CV026 following a pre-established protocol (McClean et al., 1997) and with an adaptation of the thin layer chromatography (TLC) overlay technique using A. tumefaciens NT1 (pZLR4) (Farrand et al., 2002; Steindler and Venturi, 2007). In brief, A. tumefaciens NT1 (pZLR4) was cultured in AB broth medium [containing 6% (w/v) K2HPO4, 2% (w/v) KH2PO4, 2% (w/v) NH4Cl, 0.6% (w/v) MgSO4·7H20, 0.3% (w/v) KCl, 0.02% (w/v) CaCl2, and 0.005% (w/v) FeSO4·7H2O], supplemented with 30 μg/mL gentamicin and 0.7% (w/v) glucose. Aliquots (160 μg) of crude extracts were spotted on reverse-phase C18-TLC plate (RP-C18 TLC), air-dried, overlaid with AB medium supplemented with 0.7% agar, X-Gal (40 μg/mL), C10 HSL (100 nM), and the biosensor A. tumefaciens NT1 (pZLR4), and incubated overnight at 30°C. Extracts presenting potential QQ activity in both assays were further tested by an additional violacein quantification assay in which inhibition of violacein synthesis was monitored in parallel to growth inhibition in 96 well plates, with 0.8 mg of extract, following an established protocol (Martinelli et al., 2004). Briefly, an overnight culture of C. violaceum CV026, supplemented with OC6-HSL (0.125 μg/mL) was diluted with LB medium to an OD600 of 1.2 and 180 μL were added to each well followed by the addition of 0.8 mg (20 μL) of crude extracts. The 96-well plate was incubated at 28°C in an incubator shaker. After 16 h, 96-well plates were allowed to dry at 60°C until all medium had evaporated (around 6 h to overnight). DMSO (200 μL) was added onto each well and placed in a shaker until all the violacein was solubilized. The absorbance of each well was read at 590 nm using TriStar Multimode Microplate reader (Berthold Technologies GmbH& Co. KG, Germany). Turbidity at OD660 nm was used for growth control. This was done to test whether the detected QQ activity was not related to growth inhibition of C. violaceum CV026. Penicillic acid (0.025 mg/mL; Rasmussen et al., 2005b) and ampicillin (3 μg/mL) were used as the positive controls for violacein and growth inhibition assays respectively. Inhibition of violacein production or growth was calculated as percentage inhibition compared to the inhibition by the negative control (methanol).
Antibacterial activity
The antibacterial activity of extracts was tested by agar well diffusion assay against three strains: P. aeruginosa PAO1 (PAO1), Bacillus subtilis CU1050 (BS), and Escherichia coli GM1655 (EC) as previously described (Saurav and Kannabiran, 2012). Briefly, 50 μL from the crude extract stock (16 mg/mL) were added in each well and the plates were incubated at 37°C for 24 h, after which activity was evidenced by the presence of a zone of inhibition surrounding the well. Each test was repeated three times and the antibacterial activity was expressed as the mean diameter of the inhibition zones (mm) produced by the tested extracts. Streptomycin was used as positive control for PAO1, and BS and ampicillin for EC. Methanol (solvent, in which the extracts were re-suspended) was used as a negative control. Further, minimum inhibitory concentration (MIC) was determined by broth 2-fold micro dilution method (CLSI M100-S20; Clinical and Laboratory Standards Institute, 2007) for 17 QQ positive isolate extracts. Briefly, the crude extracts were serially diluted (2000-4 μg/mL) using DMSO in Muller- Hinton broth. The inoculum [≈5 × 105 CFU/mL (final concentration)] was prepared from an overnight culture and was added to each well containing the extract. After incubating 96-well flat bottomed plates aerobically at 37.8°C for 24 h, the OD was measured using a spectrophotometer (600 nm) to determine MIC values. Negative (Culture + DMSO) and positive controls (Ampicillin) were also included.
Inhibition of virulence factor production-pyocyanin and protease
The assays were performed on the 17 isolates that showed QQ activity based on C. violaceum CV026 test. Inhibitory activity of pyocyanin production by Pseudomonas aeruginosa PAO1 was performed as described earlier (Pejin et al., 2014). Briefly, an overnight culture of P. aeruginosa PAO1 was diluted with LB medium to an OD600 of 0.2 and 4.5 mL were transferred to a 20 mL test tube. The diluted cultures were supplemented with 250 μL of test-extracts. Methanol and penicillic acid were used as negative and positive controls, respectively. After overnight incubation at 37°C and 200 rpm, 3 mL of chloroform were added to each test tube and mixed vigorously. The organic layer was collected by centrifugation (2000 × g) and transferred to a fresh tube. One milliliter of 0.2 M hydrochloric acid was added to the organic layer and the absorbance was measured at 520 nm. The experiments were performed in triplicate. Pyocyanin concentration (μg/mL) was calculated as:
| (1) |
where OD is optical density value obtained at 520 nm, 17.072 is the extinction coefficient to obtain the value in μg/mL, and 1.5 is the dilution factor (3 mL from initial 4.5 mL of chloroform were used; El-Fouly et al., 2015).
For assessing protease activity assay, 2 mL of LB broth were supplemented with extracts at non-inhibitory concentration (NIC) and with 1% (20 μL) of PAO1 O/N culture (OD600 of 0.4). DMSO and penicillic acid were used as negative and positive controls, respectively. After 18 h of incubation at 37°C, the protease activity was determined by a skim milk plate assay (Chu et al., 2013). Briefly, 100 μL of cell free supernatant of treated (extracts and penicillic acid) and untreated (negative control) PAO1 was loaded in each well containing Muller Hinton Agar with 2% skim milk and was incubated at 37°C for 24 h. Protease activity was evidenced by the presence of a zone of casein hydrolysis surrounding the well. Each test was performed in duplicates and its activity was expressed as the mean diameter of the hydrolysis zones (mm) produced by the tested extracts.
Biofilm inhibition assay
Inhibitory effects of extracts on biofilm formation by P. aeruginosa PAO1 (PAO1), Bacillus subtilis (BS), and Escherichia coli (EC) were tested by static microtiter 96 well plate assays as previously described (Sankar Ganesh and Rai Vittal, 2015). Briefly, 100 μL of LB medium were inoculated with an equal volume (100 μL) of an overnight culture (OD600 of 0.1) of each strain (PAO1, BS, and EC). 0.8 mg of test-extracts was added to each well. To facilitate biofilm formation, cells were incubated statically for 18 h at 37°C. Loosely bound bacteria and medium were discarded, and the plates were air-dried for 15 min and stained with 100 μL of 1% (v/v) crystal violet for 45 min. After discarding the stain, the stained biofilms were washed with distilled water and the bound stain was extracted by the addition of 200 μL of ethanol (95%, v/v). The resulting solution (200 μL) was transferred to a clean microtiter well plate to record the absorbance at 590 nm. Experiments were performed in triplicate. Streptomycin (32 μg/mL) was used as positive control for PAO1 and BS, whereas penicillic acid (8 μg/mL) was used for EC. Methanol (solvent in which the extracts were re-suspended) was used as a negative control.
Distribution and abundance of QQ active isolates in sponge vs. environmental samples
To determine distribution and abundance of bacteria with QQ activity in sponge vs. environmental samples (seawater, marine sediments, and fresh water), we used the Sponge Microbiome Project dataset (SMP), a recently published large dataset of 16S rRNA amplicon sequencing data deriving from sponge species and environmental samples, and part of the Earth Microbiome Project (EMP, www.earthmicrobiome.org). Samples from the SMP project were taken and processed according to standard operating procedures to ensure maximum comparability. Each sponge species was sampled at least three times and samples were collected using sterile equipment. Sample processing, sequencing and core amplicon data analysis were performed by the EMP project and all metadata was made public through the data portal (www.microbio.me/emp) (Gilbert et al., 2014). The V4 region of the 16S rRNA gene was amplified using the primers 515F and 806R and sequenced using the HiSeq2500 platform (Illumina) (Caporaso et al., 2011). Sequencing data are publicly available through the Qiita website (http://qiita.ucsd.edu/) under Project ID 1740.
Illumina reads were processed in mothur v.1.31.2 (Schloss et al., 2009). Firstly, quality-filtered, demultiplexed fastq sequences were trimmed according to quality (using thetrim.seqscommand: parameters qwindowaverage = 30, qwindowsize = 5, maxambig = 0, maxhomop = 8, minlength = 100). To minimize computational effort, files were reduced to non-identical sequences (unique.seqsand count.seqs). Non-redundant sequences were aligned (align.seqs: flip = t) to a trimmed reference bacterial database, SILVA 102 (Quast et al., 2013; pcr.seqs:start = 11894, end = 25319, keepdots = F), which was provided by mothur (Quast et al., 2013). Only sequences that aligned to the expected position were kept (screen.seqs:start = 1968, end = 4411; filter.seqs: vertical = T, trump =). Aligned reads were reduced to non-redundant sequences (unique.seqs). Chimeric sequences were detected using Uchime (chimera.uchime: dereplicate = t) (Edgar et al., 2011), and filtered out (remove.seqs). Pairwise distances between aligned sequences were calculated (dist.seqs: cutoff = 0.05) and were used for clustering. Prior to clustering, aligned sequences were phylogenetic classified based on the trimmed SILVA database (classify.seqs) (Wang et al., 2007). Sequences were clustered (cluster.split: fasta =, count =, taxonomy =, splitmethod = classify, taxlevel = 4, cutoff = 0.03, hard = t, method = furthest) and converted to .shared file format (make.shared: list =, count =, label = 0.03). Finally, OTU representative sequences were retrieved based on the distance among the cluster sequences (get.oturep: list =, label = 0.03, fasta =, count =, method = abundance) and were further classified based on SILVA, Greengenes, and RDP taxonomies (classify.seqs: fasta =, template =, taxonomy =, cutoff = 60; DeSantis et al., 2006; Cole et al., 2009; Quast et al., 2013). Furthermore, Fastq sequences from additional samples (n = 340) that were generated at a later time point were processed with the same pipeline. These sequences were integrated into the shared file using QIIME 1.8 (Caporaso et al., 2010), based on their similarity to the OTU representative sequences (parallel_pick_otus_uclust_ref.py: –similarity 0.985 –optimal_uclust). Sequences that were not similar to the OTU representative sequences were separately clustered with mothur and integrated into the previous files (.shared and taxonomy files). The integrated OTU table (.shared file) was filtered to remove low-abundance sequences (sequences less than 0.001% across the whole dataset) and chloroplasts (according to SILVA or Greengenes). Additionally, counts from seawater-like OTUs (>0.01 % across all seawater samples) were removed from sponge samples. File manipulation and processing was carried out with python scripts (http://www.python.org). A subset of this dataset was used for downstream analyses, and included only samples with at least 2 replicates (Table S1).
16S rRNA gene sequences derived from 17 QQ-active bacteria were blasted (Camacho et al., 2009) against a subset of the Sponge Microbiome dataset consisting all the samples of minimum 2 replicates and total number of reads higher than 3000, and included 937 adult sponge samples (97 sponge species), 85 sponge larvae samples, 142 seawater samples, 33 marine sediment samples, and 3 fresh water samples. The OTUs with bit score higher than 172 and sequence identity ≥98% to the sequences of our isolates were used to determine the relative abundance to closely related bacteria among different sponge species and environmental samples. Relative abundance (%) was calculated as:
| (2) |
where x is the sum of reads in the SMP matching to all the relevant OTUs (OTUs with bit score >172 and sequence identity ≥98% to our isolates) and y is the total number of reads in each sample.
The graph showing the relative abundance of OTUs in sponge vs. environmental samples was generated using an aligned dot plot graph in GraphPad Prism 5.02. Data is expressed as mean ± standard error.
Bioassay-guided fractionation and metabolic profiles of selected strains
Extracts from 4 strains were selected for metabolic profiling using LC-HRMS/MS based on the QQ and virulence activity. To ensure that the active compound elutes from the column, a bioassay guided fractionation was performed. Crude extracts of the four selected isolates were dissolved in methanol at a concentration of 25 mg/mL. A Waters spherisorb S10 ODS2 column (4.6 × 200 mm) was used for the separation and attached to the HPLC. Two solvents were used as mobile phase: Phase A, water:acetonitrile (90:10) with 0.1% formic acid, and Phase B, acetonitrile:water (90:10). The elution procedure consisted of a linear gradient from 5 to 100% of phase B over 23 min, and an isocratic profile over 5 min with 100% phase B, followed by a isocratic profile for 2.5 min with 5% of Phase A. The volume of sample injected was 40 μl of extract. A biassay guided fractionation was followed to check the successful elution of the active compound. Fractions were collected at 7 min time intervals (fraction 1: 0–7 min, fraction 2: 7.01–14 min, fraction 3: 14.01–21 min and fraction 4: 21.01–30 min). For each extract three injections were done in order to collect sufficient amount for QQ assay (well diffusion assay using C. violaceum CV026, described previously).
LC-HRMS/MS experiments were carried out on a maXis ESI–QTOF Instant Expertise™, coupled to a Thermo Scientific Dionex UltiMate 3000 Rapid Separation LC (RSLC) system equipped with a UV/Vis detector acquiring at 215 and 290 nm. The four selected samples were separated on the same column as used for the bioassay-guided fractionation, using the same solvents and gradients. Samples were acquired using the following parameters: MS1 and MS2 were acquired at 1 Hz. MS1 scan was limited between 50 and 1500 m/z and the top 3 ions were selected for fragmentation in CID (collision induced dissociation). Acquired precursor was excluded for 30 s after two MS2 events. Data was recorded and analyzed using the software Bruker compass Data Analysis 4.2.
Raw LC-MS data was initially converted to mzXML format by Mass Convert tool Proteo Wizard (ASFPHA. 2015. Available: http://proteowizard.sourceforge.net/). The converted data (mzXML format) was then imported onto XCMS Online (Tautenhahn et al., 2012; Macintyre et al., 2014). Parameters were selected to match the instrument setup (UPLC/Q-TOF pos) with slight modifications. Various parameters were set and modified for feature detection (Tautenhahn et al., 2008), retention time correction (Prince and Marcotte, 2006) alignment (Smith et al., 2006) and annotation (Kuhl et al., 2012) to obtain the best results, for example under feature detection mode, m/z tolerance deviation of 40 ppm was set and mass tolerance level for the identification step was set to 50 ppm. All results obtained are directly linked to the METLIN metabolite database such that metabolite details and MS/MS data (if available) are displayed when a database hit is selected. Further, compounds were identified with the aid of existing high resolution MS from in-house database AntiMarin (Tawfike et al., 2013) for marine secondary metabolites. Compounds with a match were further evaluated for their UV and exact mass from previous reports. Components with no match in the databases were considered as putative novel metabolites.
Data deposition
The sequences of the isolated bacteria used in this study were deposited in GenBank (NCBI) with the serial accession numbers KU196823-KU196855, KU206727-KU206761, and KU206763-KU206780. Sequences of the SMP/EMP OTUs with 98–100% identity to the 16S rRNA gene sequences of the 17 selected isolates can be found at the Qiita website (https://qiita.ucsd.edu/) under project ID 1740.
Results
QQ activity in sponge extracts
QQ activity was found in half of the sponge species tested. Out of 14 marine sponge species collected, each in triplicate, the extract of seven species showed QQ activity based on the QSIS1 assay (species were considered positive if activity was detected in at least two replicates). In the CV026 assay four of the 14 sponge species showed QQ activity, all four were found QQ-active also according the QSIS1 assay. These four sponge species included two Red Sea sponges, Suberites clavatus and Negombata magnifica, and two Mediterranean Sea species, Ircinia variabilis, and Sarcotragus sp. (Table 1). Amphimedon chloros extract showed QQ activity in the QSIS1 assay, but due to presence of high antimicrobial activity against CV026, its QQ activity could not be confirmed via CV026 assay. The extracts of the Red Sea sponges Diacarnus erythraenus and Theonella swinhoei were active only based on the QSIS1 system.
Table 1.
Quorum quenching (QQ, based on reporter strain QSIS1 and C. violaceum CV026) and antimicrobial activity (AM, against C. violaceum CV026) of sponge extracts from 14 different sponge species.
| Sponges | CV026 | QSIS1 | Origin of sponge | |
|---|---|---|---|---|
| AM | ||||
| Amphimedon chloros | ND | + | + | RS |
| Axinella verrucosa | − | − | − | MS |
| Chondrosia reniformis | − | − | − | MS |
| Crella cyathophora | − | − | − | RS |
| Diacarnus erythraenus | − | − | + | RS |
| Ircinia variabilis | + | + | + | MS |
| Negombata magnifica | + | + | + | RS |
| Petrosia ficiformis | − | − | − | MS |
| Phorbas sp. | − | − | − | MS |
| Pione vastifica | − | − | − | RS |
| Sarcotragus sp. | + | + | + | MS |
| Siphonochalina siphonella | − | − | − | RS |
| Suberites clavatus | + | + | + | RS |
| Theonella swinhoei | − | − | + | RS |
A species was considered positive if at least 2 out of 3 replicates were positive for the tested activity. Abbreviation used: RS, Red Sea; MS, Mediterranean Sea; ND, Not Determined.
Isolation and identification of cultivable bacteria
Six of the 14 sponge species screened for QQ activity were selected for bacteria isolation and determination of QQ potential in their extracts. These were: A. chloros, Crella cyathophora, D. erythraenus, Pione vastifica, and S. clavatus from the Red Sea and Sarcotragus sp. from the Mediterranean Sea. Sponge species selected included both sponges with and without QQ activity, to test whether more isolates with QQ activity could be recovered from sponges that originally showed QQ activity in their extract. All the different media used for isolation (see Materials and Methods) resulted in the isolation of bacterial strains, with Marine agar providing the highest diversity. A total of 86 representative isolates deriving from these six sponge species were selected based on morphological characteristics and RFLP analysis (data not shown) for 16S rRNA gene sequencing, and for phylogenetic analysis. Based on BLASTn and phylogenetic analysis of the partial 16S rRNA gene (>600bp), 61% of the isolated bacteria belonged to the phylum Proteobacteria, 21% to Firmicutes, 14% to Actinobateria, and 4% to Bacteroidetes (Figures 1, 2, Table 2). All the sponge species used for the isolation yielded at least one putative novel taxon with a total of 17 strains showing 16S rRNA identities ≤ 98% to previously described type strains and therefore likely representing new taxa (Table 3).
Figure 1.
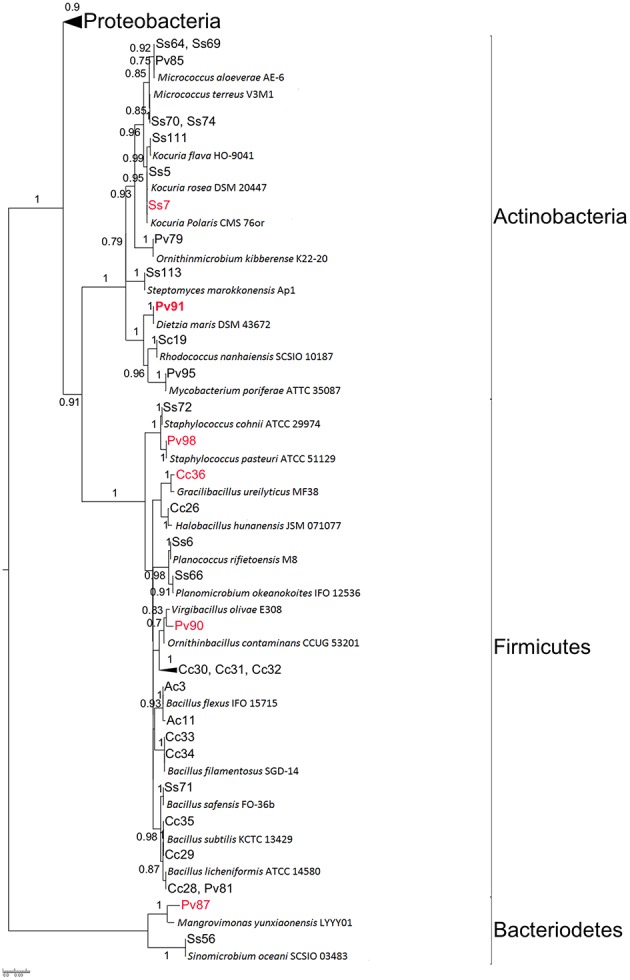
Maximum likelihood tree of isolates and their closest EzTaxon hits. The detailed view on the Proteobacteria is shown in Figure 2. Isolates with QQ activity are highlighted in red. Isolates in bold were used for further chemical analysis by LC-MS/MS. Bootstrap values > 70% (0.7) are shown. Strain ID provides information on the sponge source of each isolate: Ac, Amphimedon chloros; Cc, Crella cyathophora; Pv, Pione vastifica; Ss, Sarcotragus sp.; Sc, Suberites clavatus.
Figure 2.
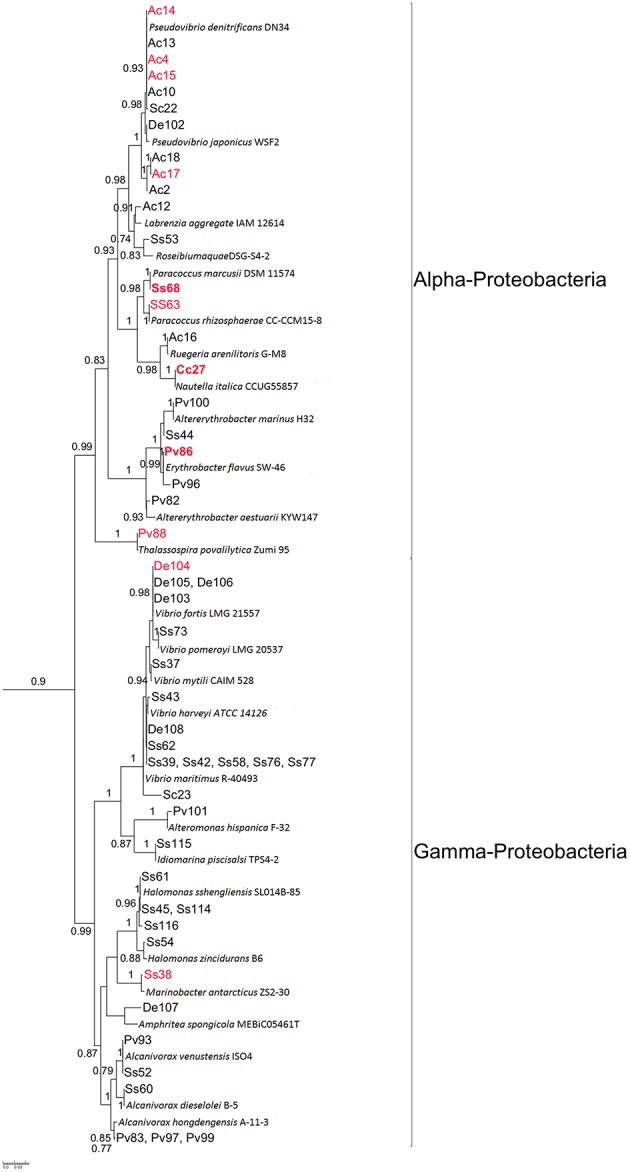
Detailed view on Proteobacteria sub-tree from the maximum likelihood tree shown in Figure 1. Isolates with QQ activity are highlighted in red. Isolates in bold were used for further chemical analysis by LC-MS/MS. Bootstrap values >70% (0.7) are shown. Strain ID provides information on the sponge source of each isolate: Ac, Amphimedon chloros; Cc, Crella cyathophora; Pv, Pione vastifica; Ss, Sarcotragus sp.; Sc, Suberites clavatus.
Table 2.
Overview of sponge isolates screened for quorum quenching activity (QQ) and antibacterial activity.
| Isolates | Similarity | Closest related | Accession number | Class | Genera | QQ assay | Antimicrobial activity | ||
|---|---|---|---|---|---|---|---|---|---|
| CV026 | PAO1 | BS | EC | ||||||
| Ac10 | 100 | Pseudovibrio denitrificans UST4-50 | KM196102 | Alphaproteobacteria | Pseudovibrio | − | − | − | − |
| Ac11 | 99 | Bacillus flexus SBANSCa10 | KR186190 | Bacilli | Bacillus | − | − | − | − |
| Ac12 | 98 | Labrenzia sp. FGH455 | JQ342690 | Alphaproteobacteria | Labrenzia | − | − | + | − |
| Ac13 | 100 | Alphaproteobacterium 5PC-32 | EF657795 | Alphaproteobacteria | − | − | + | − | |
| Ac14 | 99 | Alphaproteobacterium GUDS1146 | KF282463 | Alphaproteobacteria | + | − | − | − | |
| Ac15 | 99 | Alphaproteobacterium 5PC-32 | EF657795 | Alphaproteobacteria | + | − | + | − | |
| Ac16 | 100 | Bacterium 2D802 | JF411487 | Alphaproteobacteria | − | − | − | − | |
| Ac17 | 96 | Pseudovibrio japonicus U1021 | AB734990 | Alphaproteobacteria | Pseudovibrio | + | − | − | − |
| Ac18 | 97 | Bacterium 1H203 | JF411464 | Alphaproteobacteria | − | − | − | − | |
| Ac2 | 97 | Pseudovibrio japonicus M4B33 | LN812991 | Alphaproteobacteria | Pseudovibrio | − | − | − | − |
| Ac3 | 100 | Bacillus flexus P9 | KR935237 | Bacilli | Bacillus | − | − | − | − |
| Ac4 | 100 | Alteromonadales bacterium GUDS1275 | KF282544 | Gammaproteobacteria | Alteromonas | + | − | − | − |
| Cc26 | 99 | Halobacillus virgiliensis S22 | AM161502 | Bacilli | Halobacillus | − | − | − | − |
| Cc27 | 99 | Nautella sp. ZJ2706 | KP301105 | Alphaproteobacteria | Nautella | + | − | + | − |
| Cc28 | 100 | Bacillus aerius rawirorabr 15 | LC092833 | Bacilli | Bacillus | − | − | − | − |
| Cc29 | 100 | Bacillus subtilis ASC4 | GU227615 | Bacilli | Bacillus | − | − | − | − |
| Cc30 | 98 | Oceanobacillus sp. Ma-21 | KF958190 | Bacilli | Oceanobacillus | − | − | − | − |
| Cc31 | 98 | Oceanobacillus sp. Ma-21 | KF958190 | Bacilli | Oceanobacillus | − | − | − | − |
| Cc32 | 98 | Oceanobacillus sp. Ma-21 | KF958190 | Bacilli | Oceanobacillus | − | − | − | − |
| Cc33 | 100 | Bacillus endophyticus HSL6 | KP866218 | Bacilli | Bacillus | − | − | − | − |
| Cc34 | 100 | Bacillus endophyticus HSL6 | KP866218 | Bacilli | Bacillus | − | − | − | − |
| Cc35 | 100 | Bacillus subtilis Pls8 | KR709146 | Bacilli | Bacillus | − | − | − | − |
| Cc36 | 99 | Gracilibacillus sp. IBP-VN3 | HM587330 | Bacilli | Gracilibacillus | + | − | + | − |
| De102 | 100 | Alpha proteobacterium 9PC-7 | EF657826 | Alphaproteobacteria | − | − | − | − | |
| De103 | 100 | Vibrio fortis | KM041184 | Gammaproteobacteria | Vibrio | − | − | − | − |
| De104 | 100 | Vibrio fortis CAIM 1769 | HM584115 | Gammaproteobacteria | Vibrio | + | − | + | − |
| De105 | 100 | Vibrio pelagius | KM041188 | Gammaproteobacteria | Vibrio | − | − | − | − |
| De106 | 100 | Vibrio fortis M1C_12m_09 | KM041184 | Gammaproteobacteria | Vibrio | − | − | − | − |
| De107 | 97 | Hahellaceae bacterium Ez249 | HE818298 | Gammaproteobacteria | Hahellaceae | − | − | − | − |
| De108 | 99 | Vibrio sp. S2-26 | KM216237 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Pv100 | 99 | Altererythrobacter marinus H32 | NR_116432 | Alphaproteobacteria | Altererythrobacter | − | − | − | − |
| Pv101 | 100 | Alteromonas sp. PRIM-21 | KJ210054 | Gammaproteobacteria | Alteromonas | − | − | + | − |
| Pv79 | 99 | Bacterium RB26DO | KP684372 | Actinobacteria | − | − | − | − | |
| Pv81 | 99 | Bacillus sp. NCIM2637 | KT291161 | Bacilli | Bacillus | − | − | − | − |
| Pv82 | 98 | Erythrobacter sp. HME6855 | HQ449708 | Alphaproteobacteria | Erythrobacter | − | − | − | − |
| Pv83 | 98 | Alcanivorax sp. E46 | KJ364611 | Gammaproteobacteria | Alcanivorax | − | − | − | − |
| Pv85 | 100 | Micrococcus luteus CL10 | KR780393 | Actinobacteria | Micrococcus | − | − | − | − |
| Pv86 | 100 | Erythrobacter sp. CUA-870 | KJ732928 | Alphaproteobacteria | Erythrobacter | + | − | − | − |
| Pv87 | 94 | Formosa sp. CZ-BI2 | JX306765 | Flavobacteriia | Mangrovimonas | + | − | + | − |
| Pv88 | 99 | Marine bacterium SE96 | AY038924 | Gammaproteobacteria | Halomonadaceae | + | − | + | − |
| Pv90 | 97 | Bacillus nitritophilus IB-256 | AJ309562 | Bacilli | Bacillus | + | − | − | − |
| Pv91 | 100 | Dietzia maris IHBB 9296 | KR085794 | Actinobacteria | Dietzia | + | − | + | − |
| Pv93 | 99 | Alcanivorax sp. JXH-324 | KR012312 | Gammaproteobacteria | Alcanivorax | − | − | + | − |
| Pv95 | 100 | Mycobacterium sp. 13-17-1 | KM886164 | Actinobacteria | Mycobacterium | − | − | + | − |
| Pv96 | 98 | Erythrobacter sp. 32NM FL03 | KM357374 | Alphaproteobacteria | Erythrobacter | − | − | − | − |
| Pv97 | 98 | Alcanivorax sp. 2A75 | AB435642 | Gammaproteobacteria | Alcanivorax | − | − | − | − |
| Pv98 | 100 | Staphylococcus pasteuri SMJ33 | KT036409 | Bacilli | Staphylococcus | + | − | − | − |
| Pv99 | 99 | Alcanivorax sp. 2A17 | AB435641 | Gammaproteobacteria | Alcanivorax | − | − | − | − |
| Sc19 | 99 | Rhodococcus nanhaiensis SCSIO 10197 | JN582176 | Actinobacteria | Rhodococcus | − | − | + | − |
| Sc22 | 95 | Alphaproteobacterium D21 | DQ399723 | Alphaproteobacteria | − | − | − | − | |
| Sc23 | 96 | Vibrio ponticus H078 | KJ577073 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss111 | 99 | Kocuria sp. MNmon19 SNSum1 | KP639593 | Actinobacteria | Kocuria | − | − | − | − |
| Ss113 | 99 | Streptomyces sp. 1_C7_48 | EF540458 | Actinobacteria | Streptomyces | − | − | − | − |
| Ss114 | 99 | Halomonas sp. JSM 102013 | KR109186 | Gammaproteobacteria | Halomonas | − | − | − | − |
| Ss115 | 99 | Idiomarina sp. JL974 | DQ985042 | Gammaproteobacteria | Idiomarina | − | − | − | − |
| Ss116 | 99 | Halomonas alimentaria HQB612 | KT758546 | Gammaproteobacteria | Halomonas | − | − | − | − |
| Ss37 | 100 | Vibrio tubiashii 04/002-1T2 | KF270470 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss38 | 99 | Marinobacter sp. LV10R510-8 | KF384121 | Gammaproteobacteria | Marinobacter | + | − | − | − |
| Ss39 | 99 | Vibrio owensii HQB622 | KT758552 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss42 | 100 | Vibrio sp. BL0118 | KT731385 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss43 | 99 | Vibrio harveyi isolate M1B39 | LN812999 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss44 | 99 | Erythrobacter vulgaris Nordv6 | KC462903 | Alphaproteobacteria | Erythrobacter | − | − | − | − |
| Ss45 | 99 | Halomonas sp. JSM 102013 | KR109186 | Gammaproteobacteria | Halomonas | − | − | − | − |
| Ss5 | 100 | Kocuria rosea 0112ALTP5 | LN867187 | Actinobacteria | Kocuria | − | − | − | − |
| Ss52 | 100 | Alcanivorax sp. JXH-324 | KR012312 | Gammaproteobacteria | Alcanivorax | − | − | − | − |
| Ss53 | 97 | Roseibium denhamense M2B5 | LN812987 | Alphaproteobacteria | Roseibium | − | − | − | − |
| Ss54 | 99 | Chromohalobacter sp. Sa11 | AB305300 | Gammaproteobacteria | Chromohalobacter | − | − | − | − |
| Ss56 | 100 | Sinomicrobium oceani AAl04, | KJ754140 | Flavobacteriia | Sinomicrobium | − | − | − | |
| Ss58 | 100 | Vibrio harveyi ATCC 33843 | CP009467 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss6 | 99 | Planococcus rifietoensis CCMM B654 | FR695449 | Bacilli | Planococcus | − | − | + | − |
| Ss60 | 99 | Alcanivorax sp. NCCP-902 | AB970645 | Gammaproteobacteria | Alcanivorax | − | − | − | − |
| Ss61 | 100 | Halomonas shengliensis RB 40 | KJ939459 | Gammaproteobacteria | Halomonas | − | − | − | − |
| Ss62 | 99 | Bacterium SCSIO13137 | KJ802238 | Gammaproteobacteria | − | − | − | − | |
| Ss63 | 100 | Paracoccus sp. PaH2.06a | GQ391948 | Alphaproteobacteria | Paracoccus | + | − | − | − |
| Ss64 | 100 | Micrococcus sp. SK57 | LC068964 | Actinobacteria | Micrococcus | − | − | − | − |
| Ss66 | 99 | Planomicrobium okeanokoites NA-7 | KC967059 | Bacilli | Planomicrobium | − | − | − | |
| Ss68 | 100 | Paracoccus sp. JSM 2185010 | KR109058 | Alphaproteobacteria | Paracoccus | + | − | − | − |
| Ss69 | 100 | Micrococcus sp. SK57 | LC068964 | Actinobacteria | Micrococcus | − | − | − | − |
| Ss7 | 99 | Kocuria polaris XJB-YJ7-2 | KM186611 | Actinobacteria | Kocuria | + | − | − | − |
| Ss70 | 99 | Bacterium RH70DO | KP684403 | Actinobacteria | − | − | − | − | |
| Ss71 | 99 | Bacillus sp. 9RB4 | KT150223 | Bacilli | Bacillus | − | − | − | − |
| Ss72 | 100 | Staphylococcus cohnii RCB1038 | KT261250 | Bacilli | Staphylococcus | − | − | − | − |
| Ss73 | 100 | Vibrio sp. NB0080 | KP770094 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss74 | 99 | Bacterium RH70DO | KP684403 | Actinobacteria | − | − | − | − | |
| Ss76 | 99 | Vibrio sp. PB43 | KT731397 | Gammaproteobacteria | Vibrio | − | − | − | − |
| Ss77 | 99 | Vibrio harveyi NS131631 | KR003734 | Gammaproteobacteria | Vibrio | − | − | + | − |
Strain ID provides information on the sponge source of each isolate (Ac, Amphimedon chloros; Cc, Crella cyathophora; De, Diacarnus erythraenus; Pv, Pione vastifica; Ss, Sarcotragus sp. and Sc, Suberites clavatus). Closest 16S rRNA sequence based on % identity, and accession numbers were retrieved from NCBI database using BLASTn, and did not include uncultured/environmental sample sequences. Abbreviations used for isolates indicate the source sponge species: (Ac), Amphimedon chloros; (Cc), Crella cyathophora; (De), Diacarnus erythraenus; (Pv), Pione vastifica; (Ss), Sarcotragus sp. and (Sc), Suberites clavatus.
Table 3.
List of isolates with ≤98%sequence identity to 16S rRNA genes of the closest type strains in EzTaxon.
| Isolates | Closest type strain (EzTaxon) | Phylum | Similarity (%) | Accession no. of closest type strain | Base pair sequenced (bp) |
|---|---|---|---|---|---|
| Ac2 | Pseudovibrio japonicus WSF2 | Proteobacteria | 97.7 | AB246748 | 724 |
| Ac12 | Labrenzia aggregata IAM 12614 | Proteobacteria | 96.9 | AAUW01000001 | 615 |
| Ac17 | Pseudovibrio japonicus WSF2 | Proteobacteria | 96.5 | AB246748 | 747 |
| Ac18 | Pseudovibrio denitrificans DN34 | Proteobacteria | 97.5 | AY486423 | 1333 |
| Sc23 | Vibrio maritimus R-40493 | Proteobacteria | 96.8 | GU929925 | 541 |
| Cc26 | Halobacillus hunanensis JSM 071077 | Firmicutes | 97.5 | FJ425898 | 642 |
| Cc30 | Ornithinibacillus contaminans CCUG 53201 | Firmicutes | 97.3 | FN597064 | 1404 |
| Cc31 | Ornithinibacillus contaminans CCUG 53201 | Firmicutes | 95.5 | FN597064 | 680 |
| Cc32 | Ornithinibacillus contaminans CCUG 53201 | Firmicutes | 95.9 | FN597064 | 747 |
| Cc36 | Gracilibacillus ureilyticus MF38 | Firmicutes | 97.6 | EU709020 | 1438 |
| Ss53 | Roseibium aquae DSG-S4-2 | Proteobacteria | 96.0 | KC762314 | 765 |
| Ss54 | Halomonas zincidurans B6 | Proteobacteria | 98.0 | JQ781698 | 1361 |
| Pv82 | Altererythrobacter aestuarii KYW147 | Proteobacteria | 97.2 | FJ997597 | 710 |
| Pv87 | Mangrovimonas yunxiaonensis LYYY01 | Bacteroidetes | 95.4 | JQ937283 | 1354 |
| Pv90 | Virgibacillus salarius SA-Vb1 | Firmicutes | 96.9 | AB197851 | 1418 |
| Pv97 | Alcanivorax hongdengsis A-11-3 | Proteobacteria | 95.3 | EU438901 | 690 |
| De107 | Amphritia spongicola MEBiC05461T | Proteobacteria | 92.0 | GU289646 | 708 |
Abbreviations used for isolates indicate the source sponge species: (Ac), Amphimedon chloros; (Cc), Crella cyathophora; (De), Diacarnus erythraenus; (Pv), Pione vastifica; (Ss), Sarcotragus sp. and (Sc), Suberites clavatus.
Quorum quenching in bacterial isolates
The butanone extracts of the fermentation broths of 86 isolates were tested for QQ activity with two bioassays (i) a well diffusion assay using biosensor C. violaceum CV026 and (ii) a TLC overlay assay using A. tumefaciens NT1 (pZLR4) as biosensor. A total of 17 crude extract (~20% of the isolates) showed QQ activity according to both biosensors used. Most isolates with QQ activity (n = 9) belonged to the Alphaproteobacteria, followed by Firmicutes (n = 3), Gammaproteobacteria (n = 2), Actinobacteria (n = 2), and Bacteroidetes (n = 1) (Table 2, Figures 1, 2). The most active strains (Ac4, Ac17, Cc27, Ss68, Pv86) were classified on the basis of their zone of violacein production inhibition and belonged to the genera Alteromonas, Nautella, Paracoccus, and Pseudovibrio. Activity was also confirmed by inhibition of violacein production based on microtiter plate assay monitored in parallel to growth inhibitory activity. It was observed that none of the extracts from the selected strains showed any significant inhibitory activity against C. violaceum CV026 except for extracts Pv88 and Pv98 with mild inhibition of 11.9 and 17.3% respectively, when compared with the negative control (Figure S1).
Antibacterial activity
Seven of the extracts showed antibacterial activity against the Gram-positive strains B. subtilis, with extract Cc27 showing the largest zone of inhibition against B. subtilis (21 mm), none exhibited significant antagonistic activity against the two Gram-negative strains (P. aeruginosa PAO1 and E. coli; Table 1). In order to confirm the growth inhibitory activity, MIC was determined for the 17 QQ positive strains and the results were tabulated in Table S2. In accordance with well diffusion assay, none of the extract exhibited significant inhibitory activity against P. aeruginosa PAO1 except for the extract De103 with a MIC value of 125 μg/mL.
Inhibition of virulence factor production
All the experiments were run at NIC, to make sure that the growth of microbes is unaffected by the presence of the test extract. Extracts that were positive for QQ activity were screened for inhibition of two virulence factors that are controlled by QS in P. aeruginosa PAO1 (pyocyanin and protease production). Six extracts (Ac4, Ac17, Cc27, De103, Ss63, and Pv95) reduced pyocyanin production by more than 70% (Figure 3). Six extracts reduced protease production (Ac4, Ss7, Cc27, Ss68, Pv91, and De103; Table S3). Activities of De103 may be related to its growth inhibitory activity rather than QQ (see above). Protease inhibition was measured as zone of casein hydrolysis on milk agar plates. The highest zone of inhibition was observed with the extract of Ss68 (15 mm), when compared to the negative (20 mm) and positive control (16 mm).
Figure 3.
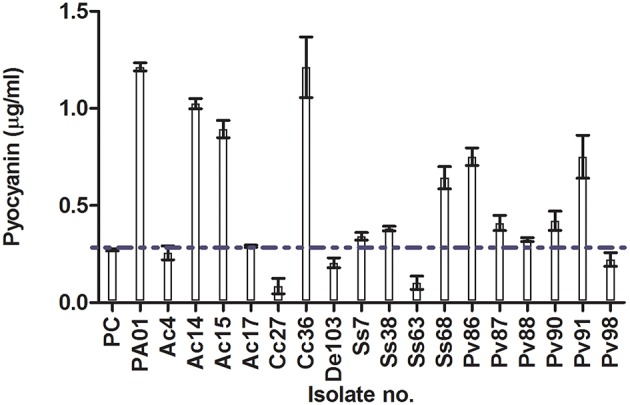
Inhibition of pyocyanin production (μg/mL) by extracts of 17 isolated strains selected based on QQ activity. Taxonomic information on strains can be found in Table 2. PC, Positive control (penicillic acid); PAO1, P. aeruginosa PAO1 grown with methanol (solvent used to dissolve extracts). Horizontal blue dotted line shows 70% of pyocyanin inhibition. Data is expressed as mean ± standard error.
Biofilm inhibition
Eight extracts (Ac4, Cc27, Pv86, Pv88, Pv90, Pv91, Ss7, and Ss68) reduced biofilm formation by more than 50% in at least one of three tested bacterial strains (P. aeruginosa PAO1, B. subtilis and E. coli; Figure 4, Figure S2). Four strain extracts were selected for metabolic profiling based on their quorum quenching potential: three Alphaproteobacteria, Ss68 and Pv86 for showing biofilm inhibition against P. aeruginosa PAO1, Cc27 for showing pyocyanin inhibition and protease inhibition in P. aeruginosa PAO1 and also biofilm inhibition against PAO1, BS, and EC. Cc27 also showed antimicrobial potential against the Gram-positive bacteria BS, thus its biofilm inhibition of BS may have been related to antimicrobial activity. Further, the actinobacterium Pv91 was selected for its vioalcein inhibition (96 well plates QQ assay), protease inhibition in P. aeruginosa PAO1, and its antimicrobial activity against BS.
Figure 4.
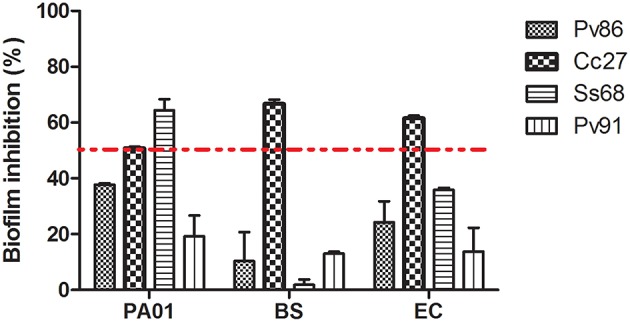
Inhibition of biofilm formation by four selected isolates (Pv86, Cc27, Ss68, and Pv91) calculated as inhibition of biofilm formation by the isolate extract compared to that of the negative control (methanol). Results for biofilm inhibition in (1) P.aeruginosa PAO1 (PA01), (2) Bacillus subtilis (BS), and (3) E. coli (EC). Red dotted line represents 50% biofilm inhibition. Results for extracts from additional strains are shown in Supplemental Material (Figure S2). Note that some activities may have resulted from growth inhibition. Specifically extracts Ss68, Pv91, and Ss7 showed growth inhibition against BS and extract Ac4 inhibited growth of EC (see Table S2 for details on MIC). Data is expressed as mean ± standard error.
Distribution and abundance of QQ active isolates in sponge vs. seawater and sediment samples
To test for a potential symbiotic nature of isolates with QQ activity, we analyzed the distribution and abundance of the QQ active isolates in the SMP dataset in sponge samples vs. seawater, freshwater and marine sediment samples. Unfortunately, this SMP dataset does not yet include the sponge species used for bacteria isolation in this study, whilst it will in the future. To account for small divergence expected in closely related symbionts found in different sponge species, we enabled some mismatches when performing the BLAST search, and included in downstream analysis any OTUs from the dataset which passed 98% identity and a bit score of 172 to our isolates. Using this criterion, out of 17 isolates tested, 15 had hits in the SMP dataset used. The OTUs are shown in Table S4.
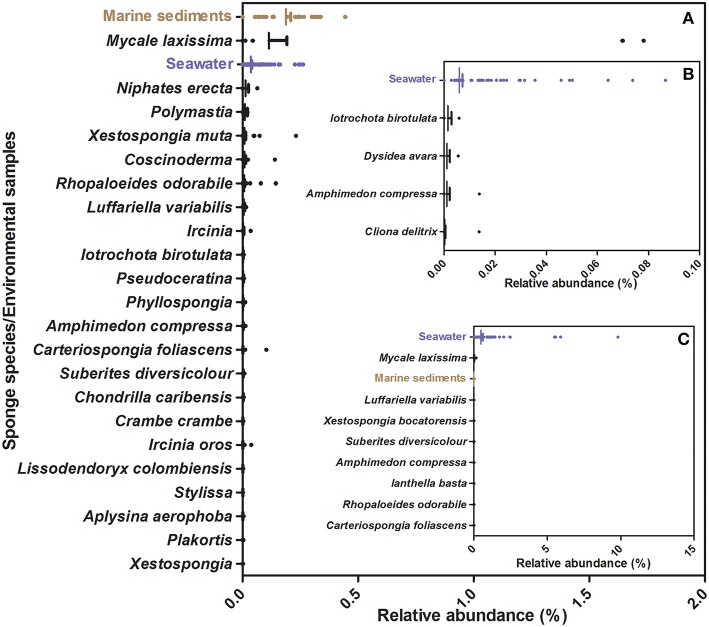
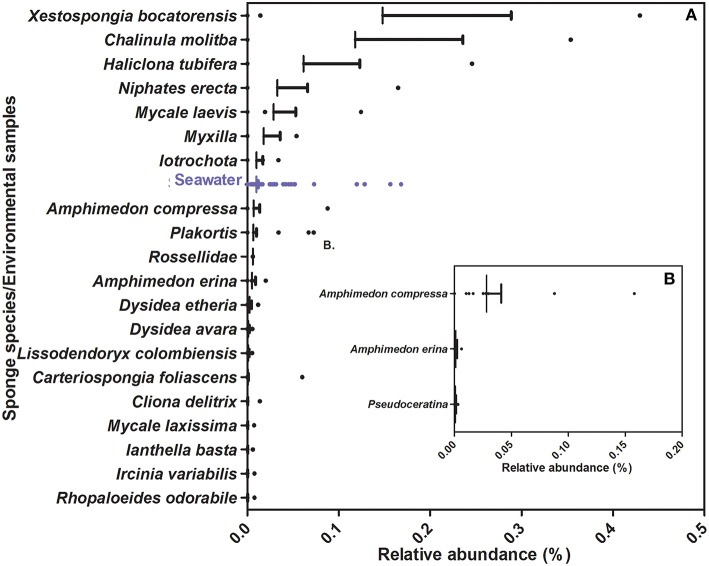
Of these, most seems to be more common in environmental (seawater/marine sediments) than in sponge samples (e.g., Figure 5, Figures S3, S4B, S5), suggesting they may have been food-bacteria trapped in the sponge filtration system at the time of sampling. Several isolates seem to be facultative sponge-associated bacteria, found at low abundances both in sponge species and in environmental samples [e.g., Ss68 (Figure 6A), Ac4, Ac14, Ac15, (Figure S4A), Pv98 (Figure S6)]. One isolate (Ac17), from the sponge Amphimedon chloros, and belonging to the genus Pseudovibrio, showed 100% sequence identity to an OTU from the SMP dataset (OTU51717), which was present at low abundance in only three sponge species (Figure 6B), and absent from the 143 seawater and 34 sediment samples, suggesting it may be an obligatory sponge symbiont. Furthermore, 2 of 3 species in which OTU51717 was found belong to the genus Amphimedon (A. compressa and A. erina, Figure 6B) as for the isolation source of Ac17, A. chloros, suggesting a narrow host sponge range for this likely sponge-symbiont.
Figure 5.
Relative abundance of OTUs from the Sponge Microbiome Project with ≥98% identity to 16S rRNA sequences from strains isolated in this study. (A) Information relative to OTUs closely affiliated to isolate Cc27, (B) Information relative to OTUs closely affiliated with isolate Pv91. (C) Information relative to OTUs closely affiliated with isolates Pv86. Vertical bar represents the mean, the hinge represents SEM (Standard Error of Mean), and dots represent outlier values beyond mean.
Figure 6.
Relative abundance of OTUs from the Sponge Microbiome Project with ≥98% identity to 16S rRNA sequences from strains isolated in this study. (A) Information relative to OTUs closely affiliated to isolate Ss68, (B) Information relative to OTUs closely affiliated with isolate Ac17. Vertical bar represents the mean, the hinge represents SEM (Standard Error of Mean) and dots represent outlier values beyond mean.
Bioactive guided fractionation and metabolic profile for the four selected strains
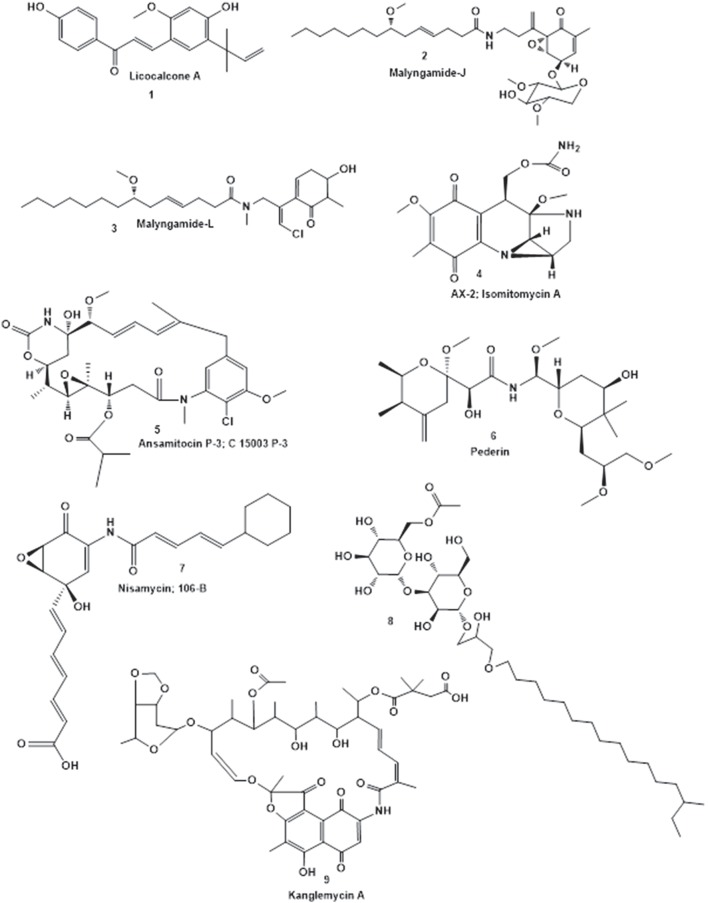
Bioassay-guided fractionation resulted in the determination of active fractions on retention time scale. Fraction 2 for Cc27, fraction 3 and 4 for Ss68, fraction 4 for Pv86 and fraction 3 for Pv91 were found to be positive for QQ activity. LC-HRMS/MS profiles of the crude extracts of the four selected strains (Cc27, Ss68, Pv86, and Pv91) were further analyzed on XCMS online. Retention time drifts between the samples were compensated. An overlay of all total ion chromatograms (TICs) acquired is shown before (Figure S7A) and after (Figure S7B) retention time correction. Cloud plots were generated with 855 features (a molecular entity with a unique m/z and retention time, p ≤ 0.001) (Figure S7C), along-with location of MS/MS scans (Figure S7D). Additionally, an interactive version of the cloud plot was generated using METLIN database, in which statistics and putative identities are displayed in a pop-up window when mouse is scrolled over the circles in the plot. The shade of color is used to represent p-value, with brighter circles having lower p-values. The circles representing features with hits in the METLIN database are shown with a black filled circle (Figure S8). Selected features with their corresponding mzmed, rtmed, adduct, and METLIN MS/MS spectra are tabulated in Tables S5A–D. The most relevant chemical structures for selected metabolites were identified on the basis of their putative METLIN assignment, fragmentation pattern in mass spectra coupled with dereplication using AntiMarin database (compounds 1-9, Figure 7).
Figure 7.
Chemical structures of representative secondary metabolites putatively identified by LC-HRMS/MS in crude extracts of Cc27 (1-3), Ss68, Pv86 (4-6), and Pv91 (7-9) using AntiMarin database.
Discussion
Sponges are a recognized source for secondary metabolites, and reports of bioactive compounds are available for part of the sponge genera or species here analyzed: Sarcotragus spp. (Liu et al., 2001, 2002, 2003; Wang et al., 2008; Abed et al., 2011; Bisio et al., 2014), Amphimedon spp. (Watanabe et al., 1998; Ovenden et al., 1999; Matsunaga et al., 2004; Takekawa et al., 2006; Takahashi et al., 2009), Crella cyathophora (D'Auria et al., 1998; Ma et al., 2009), Diacarnus sp. (El Sayed et al., 2001; Youssef et al., 2001; Youssef, 2004; Ibrahim et al., 2008) and Suberites sp. (Mitova et al., 2004; Liu et al., 2009; Takahashi et al., 2011; Flemer et al., 2012; Hwang et al., 2013). However, none of these species have been previously investigated in terms of QQ activity. It is also known that space and time can affect sponge-symbiont associations and the quality and quantity of the bioactive compounds (Thomas et al., 2010) further justifying analysis of specimens of the same species when collected from new sites and different seasons. This is the first study testing for bioactive compounds in the boring clionaid sponge Pione vastifica and its associated bacteria. Further, this is the first study to isolate bacteria from Suberites clavatus and Crella cyathophora.
The main focus of our work was to search for inhibitors of QS as non-antibiotics based therapeutic agents against QS mediated bacterial infections. Sponges were chosen as the model animal for the search of QQ compounds for several reasons. It has been shown that bacteria inside sponges likely co-operate using QS signal molecules, the most studied being N-acyl homoserine lactones (Taylor et al., 2004; Mohamed et al., 2008; Gardères et al., 2012; Zan et al., 2012; Britstein et al., 2016). It can thus be expected that sponge symbionts will also compete through the quenching of QS systems, making sponges a promising source of QSI compounds. This is corroborated by a previous screen performed in the Great Barrier Reef that found QSI compounds in marine organisms, with sponges and soft corals providing the greatest numbers of QQ active extracts (Skindersoe et al., 2008). Here, we performed a screen using the same screening system (QSIS1) for sponge species collected at different geographic sites. Our results correspond to those previously reported, 46.9% of sponge species were shown to be positive for QQ in the Great Barrier Reef, and 50% were found positive in our study from the Mediterranean and the Red Sea. Our parallel screen with the CV026 system showed only 28.6% QQ-positive sponge species, however the positive sponge species were common to both QQ assays (CV026 and QSIS1), providing evidence that the CV026 is a good and fast screen method, but may result in under-representation of positive QQ samples. The discrepancy between results from the two QQ detection systems was in part related to strong antimicrobial activity of one extract against CV026 that rendered the assay impossible (extract from Amphimedon chloros), and likely because of intrinsic differences between the two QQ-reporter systems, QSIS1 utilizing the LuxR from Vibrio fischeri and C. violaceum CV026 using CviR.
By comparing the presence of QQ activity in extract from different sponge species and the QQ activity in extracts of bacteria isolated from the same sponge species, we could test if results from sponge extracts could be a good predictor of results from sponge isolates. For this purpose six sponge species were selected, two that had been found positive for QQ in both assays, two found positive only with QSIS1 and two negative for QQ according to both assays. The number of isolates that we identified per sponge species was variable (3–35), yet it was evident that sponges showing no QQ activity by both assay-system could provide isolates with QQ activity (e.g., P. vastifica, 37.5% of isolates (n = 16) being positive for QQ) and that sponges positive for QQ activity with both systems did not necessarily provide a higher number of isolates with QQ activity [Sarcotragus sp., 12.5% of the isolates (n = 35) being positive for QQ]. It should be noted that for testing isolate-extracts for QQ activity we only used the CV026 assay, and thus there may be an underestimation of the number of QQ active isolates. The lack of match between QQ results obtained from sponge extracts vs. extract from their bacterial isolates can be explained by the lack of knowledge on the source of the QQ compounds in complex holobiont systems, such as sponges. Both the sponge host and the isolates may produce QQ compounds and the regulation of their production is also not understood. Further, many of the isolates may not represent true sponge-associated bacteria, and thus the compounds produced by them may not be characteristic of compounds found in the sponge, from which they were isolated. The discrepancy between the natural microbial community associated to sponges as described by culture-independent studies and the cultivable microbial community has been reported before (e.g., Webster et al., 2001), hence a comparison between our isolates with QQ activity and sponge-associated bacteria as known per culture-independent techniques is informative on the likely nature (symbiotic or free-living) of our isolates. Our results on the distribution and abundance of bacteria closely related to our isolates in a wide dataset of sponge and environmental samples corroborate the likely non-symbiotic nature of most isolates that showed QQ activity. One exception being Ac17, a Pseudovibrio sp. that appeared present only in three sponge species and absent from all tested environmental samples (Figure 6B). Ac17 also showed 96.3% 16S rRNA identity to Pseudovibrio sp. POLY-S9, a strain recently isolated from an intertidal sponge of the Atlantic (Alex and Antunes, 2015). The genome of Pseudovibrio sp. POLY-S9 showed typical symbiotic features (e.g., ankyrin repeat proteins and tetratricopeptide repeats domain-encoding proteins) and was suggested to produce active secondary metabolites based on the presence of biosynthetic gene clusters such as polyketide synthases and nonribosomal peptide synthetases (Alex and Antunes, 2015). The potential ability of the genus Pseudovibrio to produce bioactive compounds has been reported before, and presence of antimicrobial activity was shown for several members of this genus (e.g., Kennedy et al., 2008; Margassery et al., 2012; Graca et al., 2013; Crowley et al., 2014; Harrington et al., 2014), whilst the potential of Pseudovibrio species as producers of QS-inhibitor molecules still needs further investigation. Among our 86 selected isolates 10 belonged to the genus Pseudovibrio and 4 showed QQ activities indicating at least a potential that warrants further investigations. Whilst most of our QQ active isolates may not represent true sponge symbionts, anti-virulence activity in their extracts and the fact that isolates represent an unlimited source of compounds, makes their non-symbiotic nature less relevant for biotechnological purposes.
Quorum sensing regulates several bacterial phenotypes essential for the successful establishment of symbiotic, pathogenic, or commensal relationships with eukaryotic hosts, including motility, exo-polysaccharide production, biofilm formation, and toxin production. P. aeruginosa, an opportunistic human pathogen is responsible for both acute and chronic infections. Beyond its virulence factors (e.g., pyocyanin production) its ability to form biofilm renders it more resistant to clearance by immune defense systems and by antibiotic therapy. Our screen for pyocyanin inhibition and protease activity showed that several of the extracts from QQ-active strains reduced also these QS-regulated phenotypes. Whether the observed inhibition of pyocyanin production and protease activity result from the presence of a quorum-quenching compound in the extracts used, remains to be validated. In the case of the extracts of isolates Ac4 and Cc27, the inhibition of both pyocyanin production and protease activity in P. aeruginosa PAO1 suggests that the mechanism of action may indeed relate to QQ compounds.
Based on the activities here described, four strains were chosen for a tentative identification of the metabolic profile of their extracts. LC-HRMS/MS analysis and comparison with existing high resolution MS records from online (METLIN) and in-house databases (AntiMarin), was used to putatively identify the presence of known and to detect the presence of unknown compounds. The extract from isolate Cc27, from C. cyathophora, appeared to contain a polyphenol, Licocalcone A (1) and two lipopeptides, Malyngamide J (2) and L (3). Licocalcone A (Fu et al., 2004) and Malyngamides (Ainslie et al., 1985) are well-known natural products, and Malyngamides have been previously isolated from marine cyanobacterium Lyngbya majuscule (Wu et al., 1997). Further, 8 potentially new metabolites were detected in the extract of Cc27. This isolate has 99% identity at the 16S rRNA level to Nautella sp. R11, a pathogen of the macroalga Delisea pulchra. Interestingly, this pathogen harbors a luxR-solo homolog (a luxR devoid of cognate luxI, varR) that is involved in colonization and virulence of the macroalga (Gardiner et al., 2015), and the macroalga is also known to produce a QS inhibitor compound (Manefield et al., 2001). LC-HRMS/MS analysis of two selected strains isolated from Pione vastifica, Pv86, and Pv91 showed that almost all the selected peaks in the extract of Pv86 could be putatively identified using online and in house databases (4-6), whereas only three of 13 selected metabolites were putatively identified from the extract of Pv91 (7-9). LC-HRMS/MS analysis of the extract of Paracoccus sp. Ss68 resulted in the putative identification of three known metabolites based on the METLIN database (METLIN ID 20991, 44851, and 18061), none of eight additional selected peaks showed matches in the AntiMarin database, suggesting this strain may produce a large number of yet unknown compounds, one of which may be the relevant one for the QQ activity observed in its extract. This study supports that QQ is a common attribute among marine sponges, and that culturable sponge associated bacteria are additional source for these compounds. Ongoing work is focused on the isolation, identification and structural elucidation of the potential QQ lead molecules produced by the selected strains.
Author contributions
KS, LS conceived and designed the experiments; KS, RB performed the experiments; KS, MH, IB, GO, VC, DM, and LS analyzed the data; KS wrote the paper.
Funding
This study was supported by the Bi-lateral Italy-Israel R&D grant, MOST grant#10705-3 to L. Steindler entitled: “A novel approach to fight antibiotic-resistant pathogens: acquisition of quorum sensing inhibitors from marine sponges” (by the Ministry of Science and Technology, Israel). The work was partially supported by the European Union's Seventh Framework Programme (FP7) 2007–2013 under Grant Agreement No. 311848 (Bluegenics). KS had a post-doctoral fellowship from the Israeli Council for Higher Education (VATAT) and University of Haifa, MH received a post-doctoral fellowship by the Helmsley Trust.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the Inter-University Institute (IUI) in Eilat for their help during the course of this study. Samples were collected in compliance with the 40246/2014 permit from the Israel Nature and National Parks Protection Authority.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00416
References
- Abdelmohsen U. R., Pimentel-Elardo S. M., Hanora A., Radwan M., Abou-El-Ela S. H., Ahmed S., et al. (2010). Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs 8, 399–412. 10.3390/md8030399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed C., Legrave N., Dufies M., Robert G., Guerineau V., Vacelet J., et al. (2011). A new hydroxylated nonaprenylhydroquinone from the Mediterranean marine sponge Sarcotragus spinosulus. Mar Drugs 9, 1210–1219. 10.3390/md9071210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainslie R. D., Barchi J. J., Kuniyoshi M., Moore R. E., Mynderse J. S. (1985). Structure of malyngamide C. J. Organ. Chem. 50, 2859–2862. [Google Scholar]
- Alex A., Antunes A. (2015). Whole genome sequencing of the symbiont Pseudovibrio sp. from the intertidal marine sponge Polymastia penicillus revealed a gene repertoire for host-switching permissive lifestyle. Genome Biol. Evol. 7, 3022–3032. 10.1093/gbe/evv199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisio A., Fedele E., Pittaluga A., Olivero G., Grilli M., Chen J., et al. (2014). Isolation of hydroxyoctaprenyl-1′, 4′-hydroquinone, a new octaprenylhydroquinone from the marine sponge Sarcotragus spinosulus and evaluation of its pharmacological activity on acetylcholine and glutamate release in the rat central nervous system. Nat. Prod. Commun. 9, 1581–1584. [PubMed] [Google Scholar]
- Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2015). Marine natural products. Nat. Prod. Rep. 32, 116–211. 10.1039/c4np00144c [DOI] [PubMed] [Google Scholar]
- Britstein M., Devescovi G., Handley K., Malik A., Haber M., Saurav K., et al. (2016). A new N-Acyl homoserine lactone synthase in an uncultured symbiont of the Red Sea sponge Theonella swinhoei. Appl. Environ. Microbiol. 82, 1274–1285. 10.1128/AEM.03111-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl. 1), 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L., Marshall G. R., Eldridge G. R., Hultgren S. J. (2008). The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6, 17–27. 10.1038/nrmicro1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Zhou S., Jiang Y., Zhu W., Zhuang X., Fu J. (2013). Effect of Traditional Chinese Herbal Medicine with Antiquorum Sensing Activity on Pseudomonas aeruginosa. Evid. Based Complement. Alternat. Med. 2013, 7. 10.1155/2013/648257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (2007). CLSI Document M100-S17. Performance Standards for Antimicrobial Susceptibility Testing, 17th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino V., Fattorusso E., Imperatore C., Mangoni A., Freigang S., Teyton L. (2008). Corrugoside, a new immunostimulatory alpha-galactoglycosphingolipid from the marine sponge Axinella corrugata. Bioorg. Med. Chem. 16, 2077–2085. 10.1016/j.bmc.2007.10.098 [DOI] [PubMed] [Google Scholar]
- Costantino V., Fattorusso E., Mangoni A., Perinu C., Cirino G., De Gruttola L., et al. (2009). Tedanol: a potent anti-inflammatory ent-pimarane diterpene from the Caribbean Sponge Tedania ignis. Bioorg. Med. Chem. 17, 7542–7547. 10.1016/j.bmc.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Crowley S. P., O'Gara F., O'sullivan O., Cotter P. D., Dobson A. D. (2014). Marine Pseudovibrio sp. as a novel source of antimicrobials. Mar. Drugs 12, 5916–5929. 10.3390/md12125916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria M. V., Giannini C., Zampella A., Minale L., Debitus C., Roussakis C. (1998). Crellastatin A: a cytotoxic bis-steroid sulfate from the vanuatu marine sponge Crella sp. J. Organ. Chem. 63, 7382–7388. 10.1021/jo981022v [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin B., Qian L., Zeng Z., Yang Z., Li H., et al. (2011). Screening for novel quorum-sensing inhibitors to interfere with the formation of Pseudomonas aeruginosa biofilm. J. Med. Microbiol. 60, 1827–1834. 10.1099/jmm.0.024166-0 [DOI] [PubMed] [Google Scholar]
- Dobretsov S., Teplitski M., Bayer M., Gunasekera S., Proksch P., Paul V. J. (2011). Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 27, 893–905. 10.1080/08927014.2011.609616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. H., Wang L. H., Xu J. L., Zhang H. B., Zhang X. F., Zhang L. H. (2001). Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817. 10.1038/35081101 [DOI] [PubMed] [Google Scholar]
- Dong Y. H., Xu J. L., Li X. Z., Zhang L. H. (2000). AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U.S.A. 97, 3526–3531. 10.1073/pnas.060023897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Fouly M. Z., Sharaf A. M., Shahin A. A. M., El-Bialy H. A., Omara A. M. A. (2015). Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J. Radiat. Res. Appl. Sci. 8, 36–48. 10.1016/j.jrras.2014.10.007 [DOI] [Google Scholar]
- El Sayed K. A., Hamann M. T., Hashish N. E., Shier W. T., Kelly M., Khan A. A. (2001). Antimalarial, antiviral, and antitoxoplasmosis norsesterterpene peroxide acids from the Red Sea Sponge Diacarnus erythraeanus. J. Nat. Prod. 64, 522–524. 10.1021/np000529+ [DOI] [PubMed] [Google Scholar]
- Farrand S. K., Qin Y., Oger P. (2002). Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Meth. Enzymol. 358, 452–484. 10.1016/S0076-6879(02)58108-8 [DOI] [PubMed] [Google Scholar]
- Flemer B., Kennedy J., Margassery L. M., Morrissey J. P., O'Gara F., Dobson A. D. (2012). Diversity and antimicrobial activities of microbes from two Irish marine sponges, Suberites carnosus and Leucosolenia sp. J. Appl. Microbiol. 112, 289–301. 10.1111/j.1365-2672.2011.05211.x [DOI] [PubMed] [Google Scholar]
- Fu Y., Hsieh T. C., Guo J., Kunicki J., Lee M. Y., Darzynkiewicz Z., et al. (2004). Licochalcone-A, a novel flavonoid isolated from licorice root (Glycyrrhiza glabra), causes G2 and late-G1 arrests in androgen-independent PC-3 prostate cancer cells. Biochem. Biophys. Res. Commun. 322, 263–270. 10.1016/j.bbrc.2004.07.094 [DOI] [PubMed] [Google Scholar]
- Gardères J., Taupin L., Saïdin J., Dufour A., Le Pennec G. (2012). N-acyl homoserine lactone production by bacteria within the sponge Suberites domuncula (Olivi, 1792) (Porifera, Demospongiae). Mar. Biol. 159, 1685–1692. 10.1007/s00227-012-1956-z [DOI] [Google Scholar]
- Gardiner M., Fernandes N. D., Nowakowski D., Raftery M., Kjelleberg S., Zhong L., et al. (2015). VarR controls colonization and virulence in the marine macroalgal pathogen Nautella italica R11. Front. Microbiol. 6:1130. 10.3389/fmicb.2015.01130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Jansson J. K., Knight R. (2014). The Earth Microbiome project: successes and aspirations. BMC Biol. 12:69. 10.1186/s12915-014-0069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graca A. P., Bondoso J., Gaspar H., Xavier J. R., Monteiro M. C., de la Cruz M., et al. (2013). Antimicrobial activity of heterotrophic bacterial communities from the marine sponge Erylus discophorus (Astrophorida, Geodiidae). PLoS ONE 8:e78992. 10.1371/journal.pone.0078992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington C., Reen F. J., Mooij M. J., Stewart F. A., Chabot J. B., Guerra A. F., et al. (2014). Characterisation of non-autoinducing tropodithietic Acid (TDA) production from marine sponge Pseudovibrio species. Mar. Drugs 12, 5960–5978. 10.3390/md12125960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Schweizer H. P. (1999). Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181, 5489–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J. N. A., Soest R. W. M. V. (2002). Systema Porifera, A Guide to the Classification of the Sponges, Vol. 2 (New York, NY: Springer; ), 1103–1706. [Google Scholar]
- Hwang B. S., Lee K., Yang C., Jeong E. J., Rho J.-R. (2013). Characterization and anti-inflammatory effects of iodinated Acetylenic Acids isolated from the Marine Sponges Suberites mammilaris and Suberites japonicus. J. Nat. Prod. 76, 2355–2359. 10.1021/np400793r [DOI] [PubMed] [Google Scholar]
- Ibrahim S. R. M., Ebel R., Wray V., Müller W. E. G., Edrada-Ebel R., Proksch P. (2008). Diacarperoxides, Norterpene Cyclic Peroxides from the Sponge Diacarnus megaspinorhabdosa. J. Nat. Prod. 71, 1358–1364. 10.1021/np800102u [DOI] [PubMed] [Google Scholar]
- Jiang H., Dong H., Zhang G., Yu B., Chapman L. R., Fields M. W. (2006). Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 72, 3832–3845. 10.1128/AEM.02869-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P. A., Awada A., Twelves C., Yelle L., Perez E. A., Velikova G., et al. (2015). Phase III open-label randomized study of eribulin mesylate versus capecitabine in patients with locally advanced or metastatic breast cancer previously treated with an anthracycline and a taxane. J. Clin. Oncol. 33, 594–601. 10.1200/JCO.2013.52.4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J., Codling C. E., Jones B. V., Dobson A. D., Marchesi J. R. (2008). Diversity of microbes associated with the marine sponge, Haliclona simulans, isolated from Irish waters and identification of polyketide synthase genes from the sponge metagenome. Environ. Microbiol. 10, 1888–1902. 10.1111/j.1462-2920.2008.01614 [DOI] [PubMed] [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Kuhl C., Tautenhahn R., Bottcher C., Larson T. R., Neumann S. (2012). CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 84, 283–289. 10.1021/ac202450g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Kannabiran K. (2010). Antifungal activity of Streptomyces VITSVK5 spp. against drug resistant Aspergillus clinical isolates from pulmonary tuberculosis patients. J. Mycol. Méd. J. Med. Mycol. 20, 101–107. 10.1016/j.mycmed.2010.04.005 [DOI] [Google Scholar]
- LaSarre B., Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdunski A. M., Ventre I., Sturgis J. N. (2004). Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Micro. 2, 581–592. 10.1038/nrmicro924 [DOI] [PubMed] [Google Scholar]
- Li J. W.-H., Vederas J. C. (2009). Drug discovery and natural products: end of an Era or an Endless Frontier? Science 325, 161–165. 10.1126/science.1168243 [DOI] [PubMed] [Google Scholar]
- Liu H., Edrada-Ebel R., Ebel R., Wang Y., Schulz B., Draeger S., et al. (2009). Drimane Sesquiterpenoids from the Fungus Aspergillus ustus Isolated from the Marine Sponge Suberites domuncula. J. Nat. Prod. 72, 1585–1588. 10.1021/np900220r [DOI] [PubMed] [Google Scholar]
- Liu Y., Bae B. H., Alam N., Hong J., Sim C. J., Lee C.-O., et al. (2001). New Cytotoxic Sesterterpenes from the Sponge Sarcotragus species. J. Nat. Prod. 64, 1301–1304. 10.1021/np0101494 [DOI] [PubMed] [Google Scholar]
- Liu Y., Hong J., Lee C.-O., Im K. S., Kim N. D., Choi J. S., et al. (2002). Cytotoxic Pyrrolo- and Furanoterpenoids from the Sponge Sarcotragus species. J. Nat. Prod. 65, 1307–1314. 10.1021/np020145e [DOI] [PubMed] [Google Scholar]
- Liu Y., Mansoor T. A., Hong J., Lee C.-O., Sim C. J., Im K. S., et al. (2003). New cytotoxic sesterterpenoids and norsesterterpenoids from two sponges of the genus Sarcotragus. J. Nat. Prod. 66, 1451–1456. 10.1021/np030268e [DOI] [PubMed] [Google Scholar]
- Ma W. S., Mutka T., Vesley B., Amsler M. O., McClintock J. B., Amsler C. D., et al. (2009). Norselic Acids A−E, highly oxidized anti-infective steroids that deter mesograzer predation, from the Antarctic Sponge Crella sp. J. Nat. Prod. 72, 1842–1846. 10.1021/np900382x [DOI] [PubMed] [Google Scholar]
- Macintyre L., Zhang T., Viegelmann C., Martinez I. J., Cheng C., Dowdells C., et al. (2014). Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 12, 3416–3448. 10.3390/md12063416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai T., Tintillier F., Lucasson A., Moriou C., Bonno E., Petek S., et al. (2015). Quorum sensing inhibitors from Leucetta chagosensis Dendy, 1863. Lett. Appl. Microbiol. 61, 311–317. 10.1111/lam.12461 [DOI] [PubMed] [Google Scholar]
- Manefield M., Welch M., Givskov M., Salmond G. P., Kjelleberg S. (2001). Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora. FEMS Microbiol. Lett. 205, 131–138. 10.1111/j.1574-6968.2001.tb10936.x [DOI] [PubMed] [Google Scholar]
- Margassery L. M., Kennedy J., O'Gara F., Dobson A. D., Morrissey J. P. (2012). Diversity and antibacterial activity of bacteria isolated from the coastal marine sponges Amphilectus fucorum and Eurypon major. Lett. Appl. Microbiol. 55, 2–8. 10.1111/j.1472-765X.2012.03256.x [DOI] [PubMed] [Google Scholar]
- Martinelli D., Grossmann G., Sequin U., Brandl H., Bachofen R. (2004). Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum. BMC Microbiol. 4:25. 10.1186/1471-2180-4-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S., Miyata Y., van Soest R. W. M., Fusetani N. (2004). Tetradehydrohalicyclamine A and 22-Hydroxyhalicyclamine a, new cytotoxic bis-piperidine Alkaloids from a Marine Sponge Amphimedon sp.1. J. Nat. Prod. 67, 1758–1760. 10.1021/np049824a [DOI] [PubMed] [Google Scholar]
- Mayer A. M., Glaser K. B., Cuevas C., Jacobs R. S., Kem W., Little R. D., et al. (2010). The odyssey of marine pharmaceuticals: a current pipeline perspective. Trends Pharmacol. Sci. 31, 255–265. 10.1016/j.tips.2010.02.005 [DOI] [PubMed] [Google Scholar]
- McClean K. H., Winson M. K., Fish L., Taylor A., Chhabra S. R., Camara M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143 (Pt 12), 3703–3711. 10.1099/00221287-143-12-3703 [DOI] [PubMed] [Google Scholar]
- Mitova M., Popov S., De Rosa S. (2004). Cyclic Peptides from a Ruegeria Strain of Bacteria Associated with the Sponge Suberites domuncula. J. Nat. Prod. 67, 1178–1181. 10.1021/np049900+ [DOI] [PubMed] [Google Scholar]
- Mohamed N. M., Cicirelli E. M., Kan J., Chen F., Fuqua C., Hill R. T. (2008). Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 10, 75–86. 10.1111/j.1462-2920.2007.01431 [DOI] [PubMed] [Google Scholar]
- Ovenden S. P. B., Capon R. J., Lacey E., Gill J. H., Friedel T., Wadsworth D. (1999). Amphilactams A–D: Novel Nematocides from Southern Australian Marine Sponges of the Genus Amphimedon. J. Org. Chem. 64, 1140–1144. 10.1021/jo981377e [DOI] [Google Scholar]
- Pejin B., Talevska A., Ciric A., Glamoclija J., Nikolic M., Talevski T., et al. (2014). Anti-quorum sensing activity of selected sponge extracts: a case study of Pseudomonas aeruginosa. Nat. Prod. Res. 28, 2330–2333. 10.1080/14786419.2014.934239 [DOI] [PubMed] [Google Scholar]
- Prince J. T., Marcotte E. M. (2006). Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Anal. Chem. 78, 6140–6152. 10.1021/ac0605344 [DOI] [PubMed] [Google Scholar]
- Pruesse E., Peplies J., Glockner F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey F. A., Ward-Rainey N., Kroppenstedt R. M., Stackebrandt E. (1996). The genus Norcardiopsis represents a hylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocaridiopsaceae family. Int. J. Syst. Bacteriol. 46, 1088–1092. [DOI] [PubMed] [Google Scholar]
- Rasko D. A., Sperandio V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. 10.1038/nrd3013 [DOI] [PubMed] [Google Scholar]
- Rasmussen T. B., Bjarnsholt T., Skindersoe M. E., Hentzer M., Kristoffersen P., Kote M., et al. (2005a). Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187, 1799–1814. 10.1128/JB.187.5.1799-1814.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. B., Skindersoe M. E., Bjarnsholt T., Phipps R. K., Christensen K. B., Jensen P. O., et al. (2005b). Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151, 1325–1340. 10.1099/mic.0.27715-0 [DOI] [PubMed] [Google Scholar]
- Rohde S., Nietzer S., Schupp P. J. (2015). Prevalence and mechanisms of dynamic chemical defenses in tropical sponges. PLoS ONE 10:e0132236. 10.1371/journal.pone.0132236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero M., Martin-Cuadrado A.-B., Roca-Rivada A., Cabello A. M., Otero A. (2011). Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 75, 205–217. 10.1111/j.1574-6941.2010.01011.x [DOI] [PubMed] [Google Scholar]
- Rutherford S. T., Bassler B. L. (2012). Bacterial Quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar Ganesh P., Rai Vittal R. (2015). In vitro antibiofilm activity of Murraya koenigii essential oil extracted using supercritical fluid CO method against Pseudomonas aeruginosa PAO1. Nat. Prod. Res. 29, 2295–2298. 10.1080/14786419.2015.1004673 [DOI] [PubMed] [Google Scholar]
- Saurav K., Kannabiran K. (2012). In vitro activity of 5-(2,4-dimethylbenzyl) pyrrolidin-2-one extracted from marine Streptomyces VITSVK5 spp. against fungal and bacterial human pathogens. Rev. Iberoam. Micol. 29, 29–33. 10.1016/j.riam.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skindersoe M., Ettinger-Epstein P., Rasmussen T., Bjarnsholt T., de Nys R., Givskov M. (2008). Quorum sensing antagonism from Marine Organisms. Mar. Biotechnol. 10, 56–63. 10.1007/s10126-007-9036-y [DOI] [PubMed] [Google Scholar]
- Smith C. A., Want E. J., O'Maille G., Abagyan R., Siuzdak G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787. 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- Steindler L., Venturi V. (2007). Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266, 1–9. 10.1111/j.1574-6968.2006.00501.x [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Kubota T., Fromont J., Kobayashi J. I. (2009). Zamamidines A and B, New Manzamine Alkaloids from the Sponge Amphimedon Species. Org. Lett. 11, 21–24. 10.1021/ol802251q [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Kubota T., Shibazaki A., Gonoi T., Fromont J., Kobayashi J. i. (2011). Nakijinamines C–E, New Heteroaromatic Alkaloids from the Sponge Suberites Species. Org. Lett. 13, 3016–3019. 10.1021/ol2008473 [DOI] [PubMed] [Google Scholar]
- Takekawa Y., Matsunaga S., van Soest R. W. M., Fusetani N. (2006). Amphimedosides, 3-Alkylpyridine Glycosides from a Marine Sponge Amphimedon sp. J. Nat. Prod. 69, 1503–1505. 10.1021/np060122q [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R., Bottcher C., Neumann S. (2008). Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9:504. 10.1186/1471-2105-9-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R., Patti G. J., Rinehart D., Siuzdak G. (2012). XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039. 10.1021/ac300698c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfike A. F., Viegelmann C., Edrada-Ebel R. (2013). Metabolomics and dereplication strategies in natural products. Methods Mol. Biol. 1055, 227–244. 10.1007/978-1-62703-577-4_17 [DOI] [PubMed] [Google Scholar]
- Taylor M. W., Schupp P. J., Baillie H. J., Charlton T. S., de Nys R., Kjelleberg S., et al. (2004). Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 70, 4387–4389. 10.1128/AEM.70.7.4387-4389.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta R., Irollo E., Della Sala G., Pirozzi G., Mangoni A., Costantino V. (2013). Smenamides A and B, chlorinated peptide/polyketide hybrids containing a dolapyrrolidinone unit from the Caribbean sponge Smenospongia aurea. Evaluation of their role as leads in antitumor drug research. Mar. Drugs 11, 4451–4463. 10.3390/md11114451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T. R. A., Kavlekar D. P., LokaBharathi P. A. (2010). Marine Drugs from Sponge-Microbe Association—a review. Mar. Drugs 8, 1417–1468. 10.3390/md8041417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Song J., Jang K. H., Lee H.-S., Li X., Oh K.-B., et al. (2008). Sesterterpenoids from the Sponge Sarcotragus sp. J. Nat. Prod. 71, 551–557. 10.1021/np0780147 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D., Tsuda M., Kobayashi J. I. (1998). Three new manzamine congeners from Amphimedon Sponge. J. Natl. Products 61, 689–692. 10.1021/np970564p [DOI] [PubMed] [Google Scholar]
- Webster N. S., Wilson K. J., Blackall L. L., Hill R. T. (2001). Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67, 434–444. 10.1128/AEM.67.1.434-444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Milligan K. E., Gerwick W. H. (1997). Three new malyngamides from the marine cyanobacterium Lyngbya majuscula. Tetrahedron 53, 15983–15990. 10.1016/S0040-4020(97)10067-9 [DOI] [Google Scholar]
- Yang L., Rybtke M. T., Jakobsen T. H., Hentzer M., Bjarnsholt T., Givskov M., et al. (2009). Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 53, 2432–2443. 10.1128/AAC.01283-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef D. T. A. (2004). Tasnemoxides A-C, new cytotoxic cyclic norsesterterpene peroxides from the Red Sea Sponge Diacarnus erythraenus. J. Nat. Prod. 67, 112–114. 10.1021/np0340192 [DOI] [PubMed] [Google Scholar]
- Youssef D. T. A., Yoshida W. Y., Kelly M., Scheuer P. J. (2001). Cytotoxic cyclic norterpene peroxides from a Red Sea Sponge Diacarnus erythraenus. J. Nat. Prod. 64, 1332–1335. 10.1021/np010184a [DOI] [PubMed] [Google Scholar]
- Zan J., Cicirelli E. M., Mohamed N. M., Sibhatu H., Kroll S., Choi O., et al. (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85, 916–933. 10.1111/j.1365-2958.2012.08149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.