Abstract
Diabetic retinopathy (DR) is a disease with an increasing prevalence and the main cause of blindness among working-age population. The risk of severe vision loss can be significantly reduced by timely diagnosis and treatment. Systematic screening for DR has been identified as a cost-effective way to save health services resources. Automatic retinal image analysis is emerging as an important screening tool for early DR detection, which can reduce the workload associated to manual grading as well as save diagnosis costs and time. Many research efforts in the last years have been devoted to developing automatic tools to help in the detection and evaluation of DR lesions. However, there is a large variability in the databases and evaluation criteria used in the literature, which hampers a direct comparison of the different studies. This work is aimed at summarizing the results of the available algorithms for the detection and classification of DR pathology. A detailed literature search was conducted using PubMed. Selected relevant studies in the last 10 years were scrutinized and included in the review. Furthermore, we will try to give an overview of the available commercial software for automatic retinal image analysis.
Keywords: Automated analysis system, diabetic retinopathy, retinal image
Diabetic retinopathy (DR) is the leading cause of blindness in the working-age population.[1] Screening for DR and monitoring disease progression, especially in the early asymptomatic stages, is effective for preventing visual loss and reducing costs for health systems.[2] Most screening programs use nonmydriatic digital color fundus cameras to acquire color photographs of the retina.[3] These photographs are then examined for the presence of lesions indicative of DR, including microaneurysms (MAs), hemorrhages (HEMs), exudates (EXs), and cotton wool spots (CWSs).[4] In any DR screening program, about two-third of patients have no retinopathy.[2] The application of automated image analysis to digital fundus images may reduce the workload and costs by minimizing the number of photographs that need to be manually graded.[5]
Many studies can be found in the literature regarding digital image processing for DR. Most algorithms comprise several steps. First, a preprocessing step is carried out to attenuate image variation by normalizing the original retinal image.[6] Second, anatomical components such as the optic disk (OD) and vessels are removed.[7] Finally, only those remaining pathological features of DR are retained for subsequent classification. This review gives an overview of the available algorithms for DR feature extraction and the automatic retinal image analysis systems based on the aforementioned algorithms.
Materials
The methodological quality of published articles was evaluated, and the following inclusion criteria were defined. Only studies published in English and indexed in PubMed in the last 10 years (2005–2015) were considered. In addition, the results of these studies must be presented using mean sensitivity (SE), mean specificity (SP), or area under the Retinopathy Online Challenge (ROC) curve. Research works using image modalities different from color retinal images or aimed at pathologies different from DR were dismissed.
Methods and results of literature search for EXs segmentation algorithms, red lesions (RLs) segmentation algorithms, and DR screening systems are presented in the following section.
Segmentation of exudates
EXs are lipoprotein intraretinal deposits due to vascular leakage.[8] They appear in retinal images as yellowish lesions with well-defined edges. Their shape, size, brightness, and location vary among different patients.[9] When clusters of EXs are located in the macular region, they are indicative of macular edema (ME), which is the main cause of visual loss in DR patients. For this reason, many researchers introduced the idea of a coordinate system based on the location of the fovea to determine DR grading.[10]
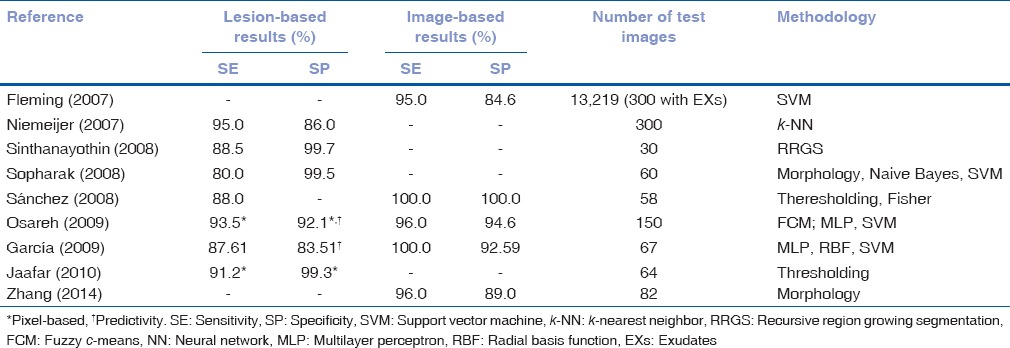
Different techniques have been proposed for EXs detection. In Table 1, information regarding the results of these methods and the databases used in each study is summarized. They can be divided into four categories.[11]
Table 1.
Performance of exudates segmentation methods
Region growing methods
With these techniques, images are segmented using the spatial contiguity of gray levels. In the method described by Sinthanayothin et al.[12] adjacent pixels were considered as belonging to the same region if they had a similar gray level or color.[12]
Thresholding methods
With these methods, EXs identification was based on a global or adaptive gray level analysis. As EXs are mainly characterized by their color, Sánchez et al.[13] employed color features to define a feature space. They proposed a modification of the RGB model and used the intensity of various pixels in the new color space to create their training set.[13,14] Other authors divided the image into homogeneous regions and applied an adaptive thresholding method to each region.[15,16]
Mathematical morphology methods
The algorithms based on these methods employed morphological operators to detect structures with defined shapes. Different morphological operators were used for EXs detection in the work of Sopharak et al.[17] Zhang et al.[18] proposed a two-scale segmentation method. To detect large EXs, authors performed a morphological reconstruction followed by a filtering and thresholding operation. Then, authors applied a top-hat operator to the green channel of the original image to recover small EXs.[18]
Classification methods
These studies employed machine learning approaches to separate EX from non-EX regions, including additional types of bright lesions (BLs), such as drusen and CWSs. Although a classification stage was also part of many of the previous studies, we have included in this category only those studies for which classification was the main step.
In Osareh et al.,[19] images were segmented using a combination of color representation in the Luv color space and an efficient coarse to fine segmentation stage based on fuzzy c-means clustering.[19] A similar approach was proposed by García et al.,[20] who combined global and adaptive histogram thresholding methods to coarsely segment bright image regions. Finally, a set of features was extracted from each region and used to assess the performance of three neural network (NN) classifiers: Multilayer perceptron (MLP), radial basis function (RBF), and support vector machine (SVM).[20]
A multi-scale morphological process for candidate EX detection was proposed by Fleming et al.[9] A SVM was subsequently used to classify candidate regions as EX, drusen or background based on their local properties.[9] In the study by Niemeijer et al.,[21] each pixel was assigned a probability of being an EX pixel, resulting in a lesion probability map for each image. Pixels with a high probability were grouped into probable lesion pixel clusters. Then, each pixel was classified as true EX or as non-EX depending on the cluster characteristics. Finally, a BL was classified into EX, CWSs or drusen by means of a k-NN classifier and a linear discriminant classifier.[21]
Segmentation of red lesions (microaneurysms and hemorrhages)
MAs are small saccular bulges in the walls of retinal capillary vessels.[22] In color fundus images, MAs appear like round red dots with a diameter ranging from 10 to 100 µm. MAs are difficult to distinguish from dot-HEMs, which are a little bigger.[23] MAs are normally the first retinal lesions that appear in DR and their number has a direct relationship to DR severity.[24]
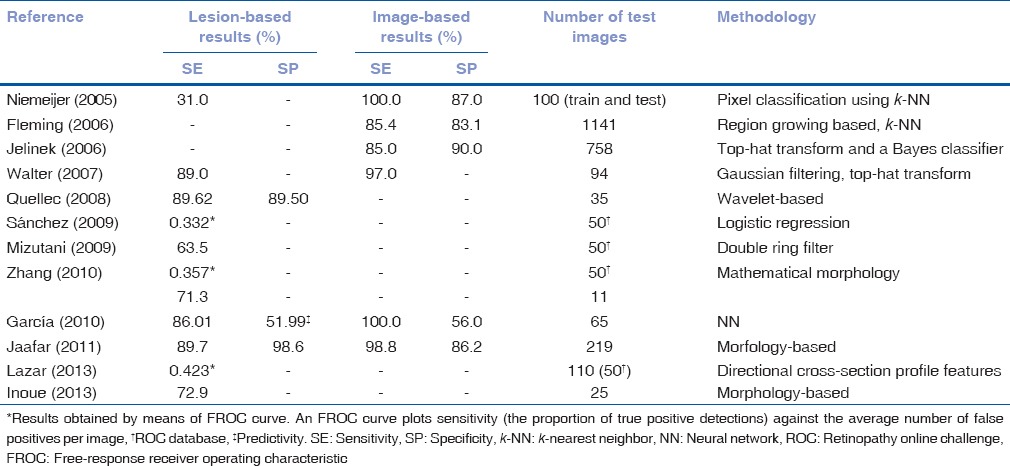
Several approaches have been proposed for MAs segmentation through color image analysis. In Table 2, information regarding results of these methods and the databases used in each study is summarized. The methods for RL detection can be also divided into four categories.[25]
Table 2.
Performance of red lesions segmentation methods
Region growing methods
Fleming et al.[22] evaluated an algorithm where region growing was performed on a watershed gradient image, to identify candidate RL regions.[22] An interesting MA detection algorithm was developed at the University of Waikato and validated by Jelinek et al.[26] It was an automated MA detector inspired by the detectors developed by Cree et al.[27] and Spencer.[24] A top-hat transformation was first used to discriminate between circular, nonconnected RL and the elongated vasculature. Candidate lesions were then segmented by means of a region growing algorithm.
Mathematical morphology methods
A polynomial contrast enhancement operation, based on morphological reconstruction methods, was used by Walter et al.[28] to detect MAs and to discriminate between MAs and vessels.[29]
Wavelet-based methods
The method proposed by Quellec et al.[30] was based on template matching using the wavelet transform. Images were descomposed in subbands, each subband having complementary information to describe MAs.
Hybrid methods
A hybrid RL segmentation algorithm was developed by Niemeijer et al.[31] The system combined the candidates detected using a mathematical morphology based algorithm with the candidates of a pixel classification based system.[31] An approach based on multi-scale correlation filtering was evaluated by Zhang et al.[32] For candidate detection, authors calculated the correlation between pixel intensity distributions throughout the image and a Gaussian model of MAs using a sliding window technique.[32] A different approach was based on calculating cross-section profiles along multiple orientations to construct a multi-directional height map.[33]
Another method based on feature classification was proposed by García et al.[34] A set of features was extracted from image regions, and a feature selection algorithm was applied in order to choose the most adequate feature subset for RL detection. Four NN-based classifiers were used to obtain the final segmentation: MLP, RBF, SVM, and a combination of these three NNs using a majority voting schema.[34,35] Sanchez et al.[36] used a three-class Gaussian mixture-model-based on the assumption that each pixel belonged to one of the three classes: Background, foreground (vessels, lesions, and OD), and outlier.[36] The method developed by Mizutani et al. used a modified double ring filter, which extracted MAs along with blood vessels. This method was designed to detect areas of the image in which the average pixel value was lower (inner circle) than the average pixel value in the area surrounding it (outer circle).[37,38]
Due to the numerous MA detection methods published, an international MA detection competition, the retinopathy online challenge (ROC), was created to compare the results of different methods.[4] The dataset used for the competition consisted of 50 training images with available reference standard and 50 test images for which expert annotations were not provided. This permitted a fair comparison between algorithms proposed by different groups. The results of five different methods such as Valladolid,[36] Waikato,[26] Latim,[30] OkMedical,[32] and Fujita Lab[37] were presented by five different teams of researchers. The results of these five MA detection methods were evaluated using the ROC database.[4]
Diabetic retinopathy screening systems
The previously mentioned studies and the related algorithms have enabled different research groups to develop computer-aided diagnosis (CAD) systems for DR screening. Information regarding results of these methods is shown in Table 3. DR severity levels are characterized by the number and type of retinal lesions that appear in the image, as well as by the retinal area in which these lesions appear. Different authors have proposed several DR severity scales to automatically determine the stage of DR in a patient.
Table 3.
Comparison of automatic diabetic retinopathy screening systems
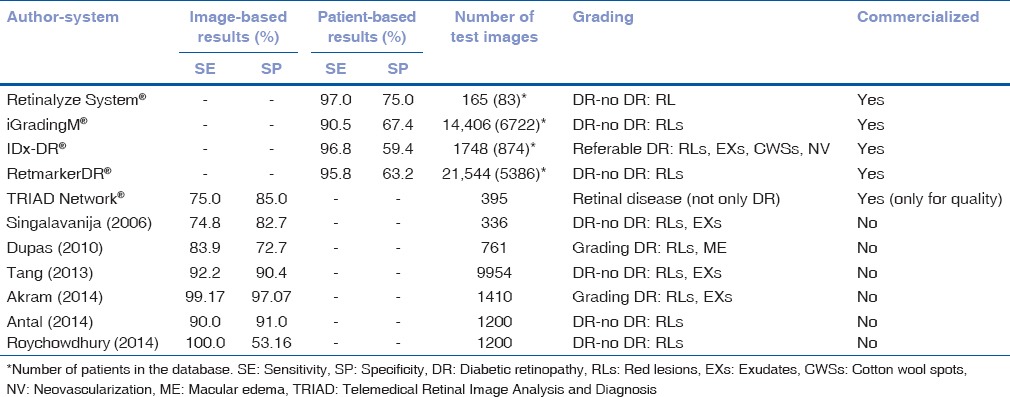
The work developed by Singalavanija et al.[39] tried to differentiate between normal and DR fundus. Although authors detected DR lesions with a good SE and SP, their DR screening system was not sensitive enough to detect early stages of nonproliferative DR (NPDR).[39] In the same way, Dupas et al.[40] determined the severity of DR. Grade 0 was established when no RLs were detected. Besides, Grades 1, 2, and 3 were defined according to the number and type of RLs detected in a retinal image. Authors also evaluated the risk of ME in a patient according to the distance between EXs and the fovea.[40] The automated system proposed by Tang et al.[41] separated normal retinal images from unhealthy images. Images with different quality and resolution were used. No DR severity grading was reported in this study.[41]
A different DR severity grading method was proposed by Usman Akram et al.,[42] based on the type (RLs and EXs) and number of lesions detected. Only images without lesions were considered normal images.[42] In other studies,[43] authors proposed a four different severity grades. Level R0(no DR) corresponded to the case where no RLs were found in an image. Besides, levels R1, R2, and R3(DR images) corresponded, respectively, to the cases where a small, medium and large number of RLs appeared in an image. Authors evaluated the accuracy of their algorithm in distinguishing normal (R0) from pathological (R1, R2, R3) images.[43] Roychowdhury et al.[44] designed a system based on machine learning techniques. Images were classified as with or without DR according to the number of RLs detected.
Several automatic retinal image analysis systems have already become commercially available.[45] These include the Retinalyze System®, which combines the ability of RL detection and image quality control to identify patients with DR and separate them from patients with no signs of DR.[46,47] In the same way, iGradingM® performs “disease/no disease” grading for DR. This software combines image quality assessment algorithms with MA detection methods.[48] SE above 90% for referable retinopathy was achieved[49,50] and showed a manual grading workload reduction of 36.3%.[51] This system was later tested[52] using two-field photographs. In this study, authors found that the inclusion of a second image (disk centered field) did not improve the results. Besides, they also established that including the detection of other types of lesions in the screening system resulted to similar SE but also in a higher number of false positives when compared to the case in which only MAs were considered.[52]
Another available system is IDx-DR®.[53,54,55] It uses several algorithms developed at the University of Iowa for DR lesions detection, such as MAs and HEMs,[31,30] EXs and CWSs.[21] IDx-DR® also includes algorithms for the detection of other types of DR signs, such as neovascularization.[23] The system was validated on a database of 1748 fovea-centered images.[53] The aim of the study was to validate the system in referable DR detection. Referable DR was defined as more than mild NPDR and/or ME. Using IDx-DR®, authors found that the prevalence of referable DR was 21.7%.[53]
The software RetmarkerDR®, developed at the University of Coimbra, should also be mentioned. It is based on combining image quality control with RL detection.[56] The system was able to separate images with no signs of DR or with no evolution of DR compared to previous screening visits, from those with signs of DR pathology or evolution.[56] The system showed a potential reduction of 48.42% in the workload of human graders.[57] Finally, Telemedical Retinal Image Analysis and Diagnosis Network® is a web-based service in which the quality of retinal images is automatically evaluated.[58,59]
These systems have been successfully applied in DR screening scenarios to identify the presence of DR or referable DR. However, to the best of our knowledge, they are not able to identify the high-risk DR or the presence of DME yet.[45]
Discussion
Early DR detection is important to slow down disease progression and avoid severe vision loss in diabetic patients. Regular DR screening is paramount to ensure timely diagnosis and treatment. However, the interpretation and grading of fundus images for this task is actually a manual process. This is a time-consuming approach, which is also subject to inter-observer variability. For this reason, automatic methods to detect DR-related lesions, as well as the development of CAD systems for DR can be a reliable option to cut down DR screening costs, to reduce the workload of ophthalmologists and to improve attention to diabetic patients. In this regard, the British Diabetic Association (BDA) estimates that the rates of any screening program for DR should reach SE >80% and SP >95%.[60] This issue should be considered when comparing the different alternatives for DR lesions detection and screening.[61]
The algorithms for DR lesions detection included in this review were very heterogeneous. The validation methods and test databases of the studies were not uniform or standardized. Therefore, it was not possible to make a direct comparison of their performance. However, some results should be underlined. In the case of EXs, nine studies complied with the inclusion criteria. Most of these studies achieved the BDA figures. As shown in Table 1, the highest lesion-based SE and SP were obtained with the method proposed by Jaafar et al.[15] and the highest image-based SE and SP were obtained by Sánchez et al.[14] However, it should be mentioned that the results of the studies in Table 1 are not directly comparable due to the lack of common measurement criteria and evaluation databases. For example, the studies by Fleming et al.[9] and Niemeijer et al.[21] used a larger database. Besides, these two studies were remarkable because they included the detection and differentiation of several types of BLs.
In the case of RLs, 12 studies met the inclusion criteria for this review. The SE and SP values, in this case, are generally below the figures for EXs detection. This indicates that RLs detection is more challenging. However, the results obtained in some studies [Table 2] were only slightly below the BDA values. It should be noted that not only SE and SP figures are important; the number of images employed in each study must be taken into account. In the case of Fleming et al.[22] and Jelinek et al.[26] a higher number of images were used than in others studies with better results. It should also be mentioned that results of the ROC competition were not measured in terms of SE and SP.[4] Thus, we could not evaluate whether the participants met the requirements of the BDA. However, it is noteworthy that the methods included in the competition could be directly compared using a common database and evaluation criteria.
The lesion detection algorithms have also allowed the development of CAD systems for DR screening. Several of the studies included in Table 3 focus on separating DR and healthy cases.[39,41,44,46,48,56] Other authors also attempt to make a first DR severity grading by distinguishing referable and nonreferable DR cases.[40,42,43,53] This means that there are computationally efficient algorithms that could allow the detection of derivable DR. Moreover, most of commercially available DR screening systems reached 80% SE in detecting DR cases, although none of them achieved the SP values recommended by the BDA. The inclusion of a DR severity grading stage poses additional complexity to these systems since DR severity grades were not standardized and nonuniform severity scales were used.
Although the results of the developed algorithms are promising, challenges still remain. Further work is necessary to improve the proposed CAD systems so they can efficiently reduce the workload of ophthalmologists. First, the proposed methods should be tested on larger databases to ensure that they are capable of preventing visual loss in DR patients in a cost-effective way. Although there are some publicly available databases designed for automatic retinal image analysis, algorithms should be tested in a larger datasets representative from screening scenarios. In addition, inter- and intra-observer variability should be addressed in studies related to DR lesions detection or DR screening software development. Finally, DR severity grading systems should be consistent with clinically approved DR severity scales and thus, consider the different signs of DR. Despite these difficulties; several research groups are working toward the improvement and validation of CAD systems to efficiently diagnose DR and determine the DR severity grade in a patient. The final goal would be to develop an automatic system for DR screening with enough accuracy to be incorporated in the daily clinical practice.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgement
“Our research group was partly supported by the research projects RTC-2015-3467-1 from “Ministerio de Economía y Competitividad” and FEDER and BIO/VA09/15 from “Consejería de Sanidad de la Junta de Castilla y León”.
References
- 1.Trento M, Bajardi M, Borgo E, Passera P, Maurino M, Gibbins R, et al. Perceptions of diabetic retinopathy and screening procedures among diabetic people. Diabet Med. 2002;19:810–3. doi: 10.1046/j.1464-5491.2002.00784.x. [DOI] [PubMed] [Google Scholar]
- 2.Singer DE, Nathan DM, Fogel HA, Schachat AP. Screening for diabetic retinopathy. Ann Intern Med. 1992;116:660–71. doi: 10.7326/0003-4819-116-8-660. [DOI] [PubMed] [Google Scholar]
- 3.Bursell SE, Cavallerano JD, Cavallerano AA, Clermont AC, Birkmire-Peters D, Aiello LP, et al. Stereo nonmydriatic digital-video color retinal imaging compared with early treatment diabetic retinopathy study seven standard field 35-mm stereo color photos for determining level of diabetic retinopathy. Ophthalmology. 2001;108:572–85. doi: 10.1016/s0161-6420(00)00604-7. [DOI] [PubMed] [Google Scholar]
- 4.Niemeijer M, van Ginneken B, Cree MJ, Mizutani A, Quellec G, Sanchez CI, et al. Retinopathy online challenge: Automatic detection of microaneurysms in digital color fundus photographs. IEEE Trans Med Imaging. 2010;29:185–95. doi: 10.1109/TMI.2009.2033909. [DOI] [PubMed] [Google Scholar]
- 5.Scotland GS, McNamee P, Fleming AD, Goatman KA, Philip S, Prescott GJ, et al. Costs and consequences of automated algorithms versus manual grading for the detection of referable diabetic retinopathy. Br J Ophthalmol. 2010;94:712–9. doi: 10.1136/bjo.2008.151126. [DOI] [PubMed] [Google Scholar]
- 6.Winder RJ, Morrow PJ, McRitchie IN, Bailie JR, Hart PM. Algorithms for digital image processing in diabetic retinopathy. Comput Med Imaging Graph. 2009;33:608–22. doi: 10.1016/j.compmedimag.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Li B, Li HK. Automated analysis of diabetic retinopathy images: Principles, recent developments, and emerging trends. Curr Diab Rep. 2013;13:453–9. doi: 10.1007/s11892-013-0393-9. [DOI] [PubMed] [Google Scholar]
- 8.Ali S, Sidibé D, Adal KM, Giancardo L, Chaum E, Karnowski TP, et al. Statistical atlas based exudate segmentation. Comput Med Imaging Graph. 2013;37:358–68. doi: 10.1016/j.compmedimag.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Fleming AD, Philip S, Goatman KA, Williams GJ, Olson JA, Sharp PF. Automated detection of exudates for diabetic retinopathy screening. Phys Med Biol. 2007;52:7385–96. doi: 10.1088/0031-9155/52/24/012. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Chutatape O. Automated feature extraction in color retinal images by a model based approach. IEEE Trans Biomed Eng. 2004;51:246–54. doi: 10.1109/TBME.2003.820400. [DOI] [PubMed] [Google Scholar]
- 11.Giancardo L, Meriaudeau F, Karnowski TP, Li Y, Garg S, Tobin KW, Jr, et al. Exudate-based diabetic macular edema detection in fundus images using publicly available datasets. Med Image Anal. 2012;16:216–26. doi: 10.1016/j.media.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinthanayothin C, Boyce JF, Williamson TH, Cook HL, Mensah E, Lal S, et al. Automated detection of diabetic retinopathy on digital fundus images. Diabet Med. 2002;19:105–12. doi: 10.1046/j.1464-5491.2002.00613.x. [DOI] [PubMed] [Google Scholar]
- 13.Sánchez CI, Mayo A, García M, López MI, Hornero R. Automatic image processing algorithm to detect hard exudates based on mixture models. Conf Proc IEEE Eng Med Biol Soc. 2006;1:4453–6. doi: 10.1109/IEMBS.2006.260434. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez CI, Hornero R, López MI, Aboy M, Poza J, Abásolo D. A novel automatic image processing algorithm for detection of hard exudates based on retinal image analysis. Med Eng Phys. 2008;30:350–7. doi: 10.1016/j.medengphy.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Jaafar HF, Nandi AK, Al-Nuaimy W. Detection of exudates in retinal images using a pure splitting technique. Conf Proc IEEE Eng Med Biol Soc 2010. 2010:6745–8. doi: 10.1109/IEMBS.2010.5626014. [DOI] [PubMed] [Google Scholar]
- 16.Jaafar HF, Nandi AK, Al-Nuaimy W. Decision support system for the detection and grading of hard exudates from color fundus photographs. J Biomed Opt. 2011;16:116001. doi: 10.1117/1.3643719. [DOI] [PubMed] [Google Scholar]
- 17.Sopharak A, Uyyanonvara B, Barman S, Williamson TH. Automatic detection of diabetic retinopathy exudates from non-dilated retinal images using mathematical morphology methods. Comput Med Imaging Graph. 2008;32:720–7. doi: 10.1016/j.compmedimag.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Thibault G, Decencière E, Marcotegui B, Laÿ B, Danno R, et al. Exudate detection in color retinal images for mass screening of diabetic retinopathy. Med Image Anal. 2014;18:1026–43. doi: 10.1016/j.media.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Osareh A, Shadgar B, Markham R. A computational-intelligence-based approach for detection of exudates in diabetic retinopathy images. IEEE Trans Inf Technol Biomed. 2009;13:535–45. doi: 10.1109/TITB.2008.2007493. [DOI] [PubMed] [Google Scholar]
- 20.García M, Sánchez CI, López MI, Abásolo D, Hornero R. Neural network based detection of hard exudates in retinal images. Comput Methods Programs Biomed. 2009;93:9–19. doi: 10.1016/j.cmpb.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Niemeijer M, van Ginneken B, Russell SR, Suttorp-Schulten MS, Abràmoff MD. Automated detection and differentiation of drusen, exudates, and cotton-wool spots in digital color fundus photographs for diabetic retinopathy diagnosis. Invest Ophthalmol Vis Sci. 2007;48:2260–7. doi: 10.1167/iovs.06-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming AD, Philip S, Goatman KA, Olson JA, Sharp PF. Automated microaneurysm detection using local contrast normalization and local vessel detection. IEEE Trans Med Imaging. 2006;25:1223–32. doi: 10.1109/tmi.2006.879953. [DOI] [PubMed] [Google Scholar]
- 23.Tang L, Niemeijer M, Reinhardt JM, Garvin MK, Abràmoff MD. Splat feature classification with application to retinal hemorrhage detection in fundus images. IEEE Trans Med Imaging. 2013;32:364–75. doi: 10.1109/TMI.2012.2227119. [DOI] [PubMed] [Google Scholar]
- 24.Spencer T, Olson JA, McHardy KC, Sharp PF, Forrester JV. An image-processing strategy for the segmentation and quantification of microaneurysms in fluorescein angiograms of the ocular fundus. Comput Biomed Res. 1996;29:284–302. doi: 10.1006/cbmr.1996.0021. [DOI] [PubMed] [Google Scholar]
- 25.Mookiah MR, Acharya UR, Chua CK, Lim CM, Ng EY, Laude A. Computer-aided diagnosis of diabetic retinopathy: A review. Comput Biol Med. 2013;43:2136–55. doi: 10.1016/j.compbiomed.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Jelinek HJ, Cree MJ, Worsley D, Luckie A, Nixon P. An automated microaneurysm detector as a tool for identification of diabetic retinopathy in rural optometric practice. Clin Exp Optom. 2006;89:299–305. doi: 10.1111/j.1444-0938.2006.00071.x. [DOI] [PubMed] [Google Scholar]
- 27.Cree MJ, Olson JA, McHardy KC, Sharp PF, Forrester JV. A fully automated comparative microaneurysm digital detection system. Eye (Lond) 1997;11(Pt 5):622–8. doi: 10.1038/eye.1997.166. [DOI] [PubMed] [Google Scholar]
- 28.Walter T, Massin P, Erginay A, Ordonez R, Jeulin C, Klein JC. Automatic detection of microaneurysms in color fundus images. Med Image Anal. 2007;11:555–66. doi: 10.1016/j.media.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Jaafar HF, Nandi AK, Al-Nuaimy W. Automated detection of red lesions from digital colour fundus photographs. Conf Proc IEEE Eng Med Biol Soc 2011. 2011:6232–5. doi: 10.1109/IEMBS.2011.6091539. [DOI] [PubMed] [Google Scholar]
- 30.Quellec G, Lamard M, Josselin PM, Cazuguel G, Cochener B, Roux C. Optimal wavelet transform for the detection of microaneurysms in retina photographs. IEEE Trans Med Imaging. 2008;27:1230–41. doi: 10.1109/TMI.2008.920619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemeijer M, van Ginneken B, Staal J, Suttorp-Schulten MS, Abràmoff MD. Automatic detection of red lesions in digital color fundus photographs. IEEE Trans Med Imaging. 2005;24:584–92. doi: 10.1109/TMI.2005.843738. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Wu X, You J, Li Q, Karray F. Detection of microaneurysms using multi-scale correlation coefficients. Pattern Recognit. 2010;43:2237–48. [Google Scholar]
- 33.Lazar I, Hajdu A. Retinal microaneurysm detection through local rotating cross-section profile analysis. IEEE Trans Med Imaging. 2013;32:400–7. doi: 10.1109/TMI.2012.2228665. [DOI] [PubMed] [Google Scholar]
- 34.García M, Sánchez CI, López MI, Díez A, Hornero R. Automatic detection of red lesions in retinal images using a multilayer perceptron neural network. Conf Proc IEEE Eng Med Biol Soc 2008. 2008:5425–8. doi: 10.1109/IEMBS.2008.4650441. [DOI] [PubMed] [Google Scholar]
- 35.García M, López MI, Alvarez D, Hornero R. Assessment of four neural network based classifiers to automatically detect red lesions in retinal images. Med Eng Phys. 2010;32:1085–93. doi: 10.1016/j.medengphy.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Sánchez CI, Hornero R, Mayo A, García M. Mixture model based clustering and logistic regression for automatic detection of microaneurysms in retinal images. Proc SPIE. 2009;7260(72601M) DOI: 10.1117/12.812088. [Google Scholar]
- 37.Mizutani A, Muramatsu C, Hatanaka Y, Suemori S, Hara T, Fujita H. Automated microaneurysm detection method based on double ring filter in retinal fundus images. Proc SPIE. 2009 7260:72601N DOI: 10.1117/12.813468. [Google Scholar]
- 38.Inoue T, Hatanaka Y, Okumura S, Muramatsu C, Fujita H. Automated microaneurysm detection method based on eigenvalue analysis using Hessian matrix in retinal fundus images. Conf Proc IEEE Eng Med Biol Soc 2013. 2013:5873–6. doi: 10.1109/EMBC.2013.6610888. [DOI] [PubMed] [Google Scholar]
- 39.Singalavanija A, Supokavej J, Bamroongsuk P, Sinthanayothin C, Phoojaruenchanachai S, Kongbunkiat V. Feasibility study on computer-aided screening for diabetic retinopathy. Jpn J Ophthalmol. 2006;50:361–6. doi: 10.1007/s10384-005-0328-3. [DOI] [PubMed] [Google Scholar]
- 40.Dupas B, Walter T, Erginay A, Ordonez R, Deb-Joardar N, Gain P, et al. Evaluation of automated fundus photograph analysis algorithms for detecting microaneurysms, haemorrhages and exudates, and of a computer-assisted diagnostic system for grading diabetic retinopathy. Diabetes Metab. 2010;36:213–20. doi: 10.1016/j.diabet.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Tang HL, Goh J, Peto T, Ling BW, Al Turk LI, Hu Y, et al. The reading of components of diabetic retinopathy: An evolutionary approach for filtering normal digital fundus imaging in screening and population based studies. PLoS One. 2013;8:e66730. doi: 10.1371/journal.pone.0066730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Usman Akram M, Khalid S, Tariq A, Khan SA, Azam F. Detection and classification of retinal lesions for grading of diabetic retinopathy. Comput Biol Med. 2014;45:161–71. doi: 10.1016/j.compbiomed.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Antal B, Hajdu A. An ensemble-based system for automatic screening of diabetic retinopathy. Knowl Based Syst. 2014;60:20–7. [Google Scholar]
- 44.Roychowdhury S, Koozekanani DD, Parhi KK. DREAM: Diabetic retinopathy analysis using machine learning. IEEE J Biomed Health Inform. 2014;18:1717–28. doi: 10.1109/JBHI.2013.2294635. [DOI] [PubMed] [Google Scholar]
- 45.Sim DA, Keane PA, Tufail A, Egan CA, Aiello LP, Silva PS. Automated retinal image analysis for diabetic retinopathy in telemedicine. Curr Diab Rep. 2015;15:14. doi: 10.1007/s11892-015-0577-6. [DOI] [PubMed] [Google Scholar]
- 46.Larsen N, Godt J, Grunkin M, Lund-Andersen H, Larsen M. Automated detection of diabetic retinopathy in a fundus photographic screening population. Invest Ophthalmol Vis Sci. 2003;44:767–71. doi: 10.1167/iovs.02-0417. [DOI] [PubMed] [Google Scholar]
- 47.Hansen AB, Hartvig NV, Jensen MS, Borch-Johnsen K, Lund-Andersen H, Larsen M. Diabetic retinopathy screening using digital non-mydriatic fundus photography and automated image analysis. Acta Ophthalmol Scand. 2004;82:666–72. doi: 10.1111/j.1600-0420.2004.00350.x. [DOI] [PubMed] [Google Scholar]
- 48.Philip S, Fleming AD, Goatman KA, Fonseca S, McNamee P, Scotland GS, et al. The efficacy of automated “disease/no disease” grading for diabetic retinopathy in a systematic screening programme. Br J Ophthalmol. 2007;91:1512–7. doi: 10.1136/bjo.2007.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleming AD, Goatman KA, Philip S, Williams GJ, Prescott GJ, Scotland GS, et al. The role of haemorrhage and exudate detection in automated grading of diabetic retinopathy. Br J Ophthalmol. 2010;94:706–11. doi: 10.1136/bjo.2008.149807. [DOI] [PubMed] [Google Scholar]
- 50.Soto-Pedre E, Navea A, Millan S, Hernaez-Ortega MC, Morales J, Desco MC, et al. Evaluation of automated image analysis software for the detection of diabetic retinopathy to reduce the ophthalmologists’ workload. Acta Ophthalmol. 2015;93:e52–6. doi: 10.1111/aos.12481. [DOI] [PubMed] [Google Scholar]
- 51.Fleming AD, Goatman KA, Philip S, Prescott GJ, Sharp PF, Olson JA. Automated grading for diabetic retinopathy: A large-scale audit using arbitration by clinical experts. Br J Ophthalmol. 2010;94:1606–10. doi: 10.1136/bjo.2009.176784. [DOI] [PubMed] [Google Scholar]
- 52.Goatman K, Charnley A, Webster L, Nussey S. Assessment of automated disease detection in diabetic retinopathy screening using two-field photography. PLoS One. 2011;6:e27524. doi: 10.1371/journal.pone.0027524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abràmoff MD, Folk JC, Han DP, Walker JD, Williams DF, Russell SR, et al. Automated analysis of retinal images for detection of referable diabetic retinopathy. JAMA Ophthalmol. 2013;131:351–7. doi: 10.1001/jamaophthalmol.2013.1743. [DOI] [PubMed] [Google Scholar]
- 54.Sánchez CI, Niemeijer M, Dumitrescu AV, Suttorp-Schulten MS, Abràmoff MD, van Ginneken B. Evaluation of a computer-aided diagnosis system for diabetic retinopathy screening on public data. Invest Ophthalmol Vis Sci. 2011;52:4866–71. doi: 10.1167/iovs.10-6633. [DOI] [PubMed] [Google Scholar]
- 55.Abràmoff MD, Reinhardt JM, Russell SR, Folk JC, Mahajan VB, Niemeijer M, et al. Automated early detection of diabetic retinopathy. Ophthalmology. 2010;117:1147–54. doi: 10.1016/j.ophtha.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliveira CM, Cristóvão LM, Ribeiro ML, Abreu JR. Improved automated screening of diabetic retinopathy. Ophthalmologica. 2011;226:191–7. doi: 10.1159/000330285. [DOI] [PubMed] [Google Scholar]
- 57.Ribeiro L, Oliveira CM, Neves C, Ramos JD, Ferreira H, Cunha-Vaz J. Screening for diabetic retinopathy in the central region of Portugal. Added value of automated ‘Disease/No Disease’ grading. Ophthalmologica. 2015;233:96–103. doi: 10.1159/000368426. DOI: 10.1159/000368426. [DOI] [PubMed] [Google Scholar]
- 58.Karnowski TP, Aykac D, Giancardo L, Li Y, Nichols T, Tobin KW, Jr, et al. Automatic detection of retina disease: Robustness to image quality and localization of anatomy structure. Conf Proc IEEE Eng Med Biol Soc 2011. 2011:5959–64. doi: 10.1109/IEMBS.2011.6091473. [DOI] [PubMed] [Google Scholar]
- 59.Chaum E, Karnowski TP, Govindasamy VP, Abdelrahman M, Tobin KW. Automated diagnosis of retinopathy by content-based image retrieval. Retina. 2008;28:1463–77. doi: 10.1097/IAE.0b013e31818356dd. [DOI] [PubMed] [Google Scholar]
- 60.Mead A, Burnett S, Davey C. Diabetic retinal screening in the UK. J R Soc Med. 2001;94:127–9. doi: 10.1177/014107680109400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Squirrell DM, Talbot JF. Screening for diabetic retinopathy. J R Soc Med. 2003;96:273–6. doi: 10.1258/jrsm.96.6.273. [DOI] [PMC free article] [PubMed] [Google Scholar]


